1. General notes about information in this file
: All background rate plots that are contained in this file in which only the non tracking data was used, used the wrong time to normalize by. Instead of using the active shutter open time of the non tracking part, it used the active time of all the data. As such the background rates are about 1/20 too low. Those that are generated with all data (e.g. sec. 29.1.11.5 for the latest at the moment) use the correct numbers. As does any produced after the above date.
2. Reminder about data taking & detector properties
Recap the data InGrid data taking campaign.
Run-2: October 2017 - March 2018
Run-3: October 2018 - December 2018
| Solar tracking [h] | Background [h] | Active tracking [h] | Active tracking (eventDuration) [h] | Active background [h] | Total time [h] | Active time [h] | Active [%] | |
|---|---|---|---|---|---|---|---|---|
| Run-2 | 106.006 | 2401.43 | 93.3689 | 93.3689 | 2144.67 | 2507.43 | 2238.78 | 0.89285842 |
| Run-3 | 74.2981 | 1124.93 | 67.0066 | 67.0066 | 1012.68 | 1199.23 | 1079.6 | 0.90024432 |
| Total | 180.3041 | 3526.36 | 160.3755 | 160.3755 | 3157.35 | 3706.66 | 3318.38 | 0.89524801 |
Ratio of tracking to background: 3156.8 / 159.8083 = 19.7536673627
Calibration data:
| Calibration [h] | Active calibration [h] | Total time [h] | Active time [h] | |
|---|---|---|---|---|
| Run-2 | 107.422 | 2.60139 | 107.422 | 2.60139 |
| Run-3 | 87.0632 | 3.52556 | 87.0632 | 3.52556 |
| solar tracking | background | calibration | |
|---|---|---|---|
| Run-2 | 106 h | 2401 h | 107 h |
| Run-3 | 74 h | 1125 h | 87 h |
| Total | 180 h | 3526 h | 194 h |
These numbers can be obtained for example with ./../../CastData/ExternCode/TimepixAnalysis/Tools/writeRunList/writeRunList.nim by running it on Run-2 and Run-3 files. They correspond to the total time and not the active detector time!
The following detector features were used:
- \(\SI{300}{\nano\meter} \ce{SiN}\) entrance window available in Run-2 and Run-3
central InGrid surrounded by 6 additional InGrids for background suppression of events
available in Run-2 and Run-3
recording analog grid signals from central chip with an FADC for background suppression based on signal shapes and more importantly as trigger for events above \(\mathcal{O}(\SI{1.2}{\kilo\electronvolt})\) (include FADC spectrum somewhere?)
available in Run-2 and Run-3
- two veto scintillators:
- SCL (large "horizontal" scintillator pad) to veto events from cosmics or induced X-ray fluorescence photons (available in Run-3)
- SCS (small scintillator behind anode plane) to veto possible cosmics orthogonal to readout plane (available in Run-3)
As a table: Overview of working (\green{o}), mostly working (\orange{m}), not working (\red{x}) features
| Feature | Run 2 | Run 3 |
|---|---|---|
| Septemboard | \green{o} | \green{o} |
| FADC | \orange{m} | \green{o} |
| Veto scinti | \red{x} | \green{o} |
| SiPM | \red{x} | \green{o} |
2.1. Calculate total tracking and background times used above
UPDATE: These numbers in this section are also now outdated. The most up to date are in ./../../phd/thesis.html. Those numbers appear in the table in the section above now!
The table above is generated by using the ./../../CastData/ExternCode/TimepixAnalysis/Tools/writeRunList/writeRunList.nim tool:
writeRunList -b ~/CastData/data/DataRuns2017_Reco.h5 -c ~/CastData/data/CalibrationRuns2017_Reco.h5
writeRunList -b ~/CastData/data/DataRuns2018_Reco.h5 -c ~/CastData/data/CalibrationRuns2018_Reco.h5
This produces the following table:
| Solar tracking [h] | Background [h] | Active tracking [h] | Active background [h] | Total time [h] | Active time [h] | Active % | |
|---|---|---|---|---|---|---|---|
| Run-2 | 106.006 | 2391.16 | 92.8017 | 2144.12 | 2497.16 | 2238.78 | 0.89653046 |
| Run-3 | 74.2981 | 1124.93 | 67.0066 | 1012.68 | 1199.23 | 1079.6 | 0.90024432 |
| Total | 180.3041 | 3516.09 | 159.8083 | 3156.8 | 3696.39 | 3318.38 | 0.89773536 |
(use org-table-sum C-c + on each column to compute the total).
2.1.1. Outdated numbers
The numbers below were the ones obtained from a faulty calculation. See ./../journal.org#sec:journal:2023_07_08:missing_time
These numbers yielded the following table:
| Solar tracking [h] | Background [h] | Active tracking [h] | Active background [h] | Total time [h] | Active time [h] | Active % | |
|---|---|---|---|---|---|---|---|
| Run-2 | 106.006 | 2401.43 | 94.1228 | 2144.67 | 2507.43 | 2238.78 | 0.89285842 |
| Run-3 | 74.2981 | 1124.93 | 66.9231 | 1012.68 | 1199.23 | 1079.60 | 0.90024432 |
| Total | 180.3041 | 3526.36 | 161.0460 | 3157.35 | 3706.66 | 3318.38 | 0.89524801 |
Run-2:
./writeRunList -b ~/CastData/data/DataRuns2017_Reco.h5 -c ~/CastData/data/CalibrationRuns2017_Reco.h5
Type: rtBackground total duration: 14 weeks, 6 days, 11 hours, 25 minutes, 59 seconds, 97 milliseconds, 615 microseconds, and 921 nanoseconds In hours: 2507.433082670833 active duration: 2238.783333333333 trackingDuration: 4 days, 10 hours, and 20 seconds In hours: 106.0055555555556 active tracking duration: 94.12276972527778 nonTrackingDuration: 14 weeks, 2 days, 1 hour, 25 minutes, 39 seconds, 97 milliseconds, 615 microseconds, and 921 nanoseconds In hours: 2401.427527115278 active background duration: 2144.666241943055
| Solar tracking [h] | Background [h] | Active tracking [h] | Active background [h] | Total time [h] | Active time [h] |
|---|---|---|---|---|---|
| 106.006 | 2401.43 | 94.1228 | 2144.67 | 2507.43 | 2238.78 |
Type: rtCalibration total duration: 4 days, 11 hours, 25 minutes, 20 seconds, 453 milliseconds, 596 microseconds, and 104 nanoseconds In hours: 107.4223482211111 active duration: 2.601388888888889 trackingDuration: 0 nanoseconds In hours: 0.0 active tracking duration: 0.0 nonTrackingDuration: 4 days, 11 hours, 25 minutes, 20 seconds, 453 milliseconds, 596 microseconds, and 104 nanoseconds In hours: 107.4223482211111 active background duration: 2.601391883888889
| Solar tracking [h] | Background [h] | Active tracking [h] | Active background [h] | Total time [h] | Active time [h] |
|---|---|---|---|---|---|
| 0 | 107.422 | 0 | 2.60139 | 107.422 | 2.60139 |
Run-3:
./writeRunList -b ~/CastData/data/DataRuns2018_Reco.h5 -c ~/CastData/data/CalibrationRuns2018_Reco.h5
Type: rtBackground total duration: 7 weeks, 23 hours, 13 minutes, 35 seconds, 698 milliseconds, 399 microseconds, and 775 nanoseconds In hours: 1199.226582888611 active duration: 1079.598333333333 trackingDuration: 3 days, 2 hours, 17 minutes, and 53 seconds In hours: 74.29805555555555 active tracking duration: 66.92306679361111 nonTrackingDuration: 6 weeks, 4 days, 20 hours, 55 minutes, 42 seconds, 698 milliseconds, 399 microseconds, and 775 nanoseconds In hours: 1124.928527333056 active background duration: 1012.677445774444
| Solar tracking [h] | Background [h] | Active tracking [h] | Active background [h] | Total time [h] | Active time [h] |
|---|---|---|---|---|---|
| 74.2981 | 1124.93 | 66.9231 | 1012.68 | 1199.23 | 1079.6 |
Type: rtCalibration total duration: 3 days, 15 hours, 3 minutes, 47 seconds, 557 milliseconds, 131 microseconds, and 279 nanoseconds In hours: 87.06321031416667 active duration: 3.525555555555556 trackingDuration: 0 nanoseconds In hours: 0.0 active tracking duration: 0.0 nonTrackingDuration: 3 days, 15 hours, 3 minutes, 47 seconds, 557 milliseconds, 131 microseconds, and 279 nanoseconds In hours: 87.06321031416667 active background duration: 3.525561761944445
| Solar tracking [h] | Background [h] | Active tracking [h] | Active background [h] | Total time [h] | Active time [h] |
|---|---|---|---|---|---|
| 0 | 87.0632 | 0 | 3.52556 | 87.0632 | 3.52556 |
2.2. Shutter settings
The data taken in 2017 uses Timepix shutter settings of 2 / 32 (very long / 32), which results in frames of length ~2.4 s.
From 2018 on this was reduced to 2 / 30 (very long / 30), which is closer to 2.2 s. The exact reason for the change is not clear to me in hindsight.
[ ]*NOTE: Add event mean duration by run (e.g. from ./../../CastData/ExternCode/TimepixAnalysis/Tools/outerChipActivity/outerChipActivity.nim) here to showcase!
2.3. Data backups
Data is found in the following places:
/datadirectory on tpc19- on tpc00
- tpc06 is the lab computer that was used for testing etc., contains
data for the development, sparking etc.
Under
/data/tpc/datait contains a huge amount of backed up runs, including the whole sparking history etc. It's about 400 GB of data and should be fully backed up soon. Otherwise we might lose it forever. - my laptop & desktop at home contain most data
2.4. Detector documentation
The relevant IMPACT form, which contains the detector documentation is
https://impact.cern.ch/impact/secure/?place=editActivity:101629 A PDF
version of this document can be found at
The version uploaded indeed matches the latest status of the document in ./Detector/CastDetectorDocumentation.html, including the funny notes, comments and TODOs. :)
2.5. Timeline of CAST data taking
[-]add dates of each calibration[X]add Geometer measurements here[X]add time of scintillator calibration- ref:
https://espace.cern.ch/cast-share/elog/Lists/Posts/Post.aspx?ID=3420
and
- June/July detector brought to CERN
- before alignment of LLNL telescope by Jaime
- laser alignment (see
 )
) - vacuum leak tests & installation of detector
(see:
 )
) - after installation of lead shielding
- Geometer measurement of InGrid alignment for X-ray finger run
- - : first X-ray finger run (not useful to determine position of detector, due to dismount after)
- after: dismounted to make space for KWISP
- ref:
https://espace.cern.ch/cast-share/elog/Lists/Posts/Post.aspx?ID=3420
and
- Remount in September 2017 -
- installation from to
- Alignment with geometers for data taking, magnet warm and under vacuum.
- weekend: (ref: ./../Talks/CCM_2017_Sep/CCM_2017_Sep.html)
- calibration (but all wrong)
- water cooling stopped working
- next week: try fix water cooling
- quick couplings: rubber disintegrating causing cooling flow to go to zero
- attempt to clean via compressed air
- final cleaning : wrong tube, compressed detector…
- detector window exploded…
- show image of window and inside detector
- detector investigation in CAST CDL
see
 images & timestamps of images
images & timestamps of images - study of contamination & end of Sep CCM
- detector back to Bonn, fixed
- weekend: (ref: ./../Talks/CCM_2017_Sep/CCM_2017_Sep.html)
- detector installation before first data taking
- reinstall in October for start of data taking in 30th Oct 2017
- remount start
- Alignment with Geometers (after removal & remounting due to window accident) for data taking. Magnet cold and under vacuum.
- calibration of scintillator veto paddle in RD51 lab
- remount installation finished incl. lead shielding (mail "InGrid status update" to Satan Forum on )
- <data taking period from to in
2017>
- between runs 85 & 86: fix of
src/waitconditions.cppTOS bug, which caused scinti triggers to be written in all files up to next FADC trigger - run 101 was the first with FADC noise
significant enough to make me change settings:
- Diff: 50 ns -> 20 ns (one to left)
- Coarse gain: 6x -> 10x (one to right)
- run 109: crazy amounts of noise on FADC
- run 111: stopped early. tried to debug noise and blew a fuse in gas interlock box by connecting NIM crate to wrong power cable
- run 112: change FADC settings again due to noise:
- integration: 50 ns -> 100 ns This was done at around
- integration: 100 ns -> 50 ns again at around .
- run 121: Jochen set the FADC main amplifier integration time from 50 -> 100 ns again, around
- between runs 85 & 86: fix of
- <data taking period from to
beginning 2018>
- start of 2018 period: temperature sensor broken!
- to issues with moving THL values & weird detector behavior. Changed THL values temporarily as an attempted fix, but in the end didn't help, problem got worse. (ref: gmail "Update 17/02" and ./../Mails/cast_power_supply_problem_thlshift/power_supply_problem.html) issue with power supply causing severe drop in gain / increase in THL (unclear, #hits in 55Fe dropped massively ; background eventually only saw random active pixels). Fixed by replugging all power cables and improving the grounding situation. iirc: this was later identified to be an issue with the grounding between the water cooling system and the detector.
- by everything was fixed and detector was running correctly again.
2 runs:
were missed because of this.
- removal of veto scintillator and lead shielding
- X-ray finger run 2 on . This run is actually useful to determine the position of the detector.
- Geometer measurement after warming up magnet and not under vacuum. Serves as reference for difference between vacuum & cold on !
- detector fully removed and taken back to Bonn
- installation started . Mounting due to lead shielding support was more complicated than intended (see mails "ingrid installation" including Damien Bedat)
- shielding fixed by and detector installed the next couple of days
- Alignment with Geometers for data taking. Magnet warm and not under vacuum.
- data taking was supposed to start end of September, but delayed.
- detector had issue w/ power supply, finally fixed on . Issue was a bad soldering joint on the Phoenix connector on the intermediate board. Note: See chain of mails titled "Unser Detektor…" starting on for more information. Detector behavior was weird from beginning Oct. Weird behavior seen on the voltages of the detector. Initial worry: power supply dead or supercaps on it. Replaced power supply (Phips brought it a few days after), but no change.
- data taking starts
- run 297, 298 showed lots of noise again, disabled FADC on (went to CERN next day)
- data taking ends
runs that were missed:
The last one was not a full run.
[ ]CHECK THE ELOG FOR WHAT THE LAST RUN WAS ABOUT
- detector mounted in CAST Detector Lab
- data taking from to .
- detector dismounted and taken back to Bonn
- ref: ./../outerRingNotes.html
- calibration measurements of outer chips with a 55Fe source using a custom anode & window
- between and calibrations of each outer chip using Run 2 and Run 3 detector calibrations
- start of a new detector calibration
- another set of measurements between to with a new set of calibrations
2.6. Detector alignment at CAST [/]
There were 3 different kinds of alignments:
- laser alignment. Done in July 2017 and 27/04/2018 (see mail of
Theodoros for latter "alignment of LLNL telescope")
- images:
 the spot is the one on the vertical line from the center down!
The others are just refractions. Was easier visible by eye.
the spot is the one on the vertical line from the center down!
The others are just refractions. Was easier visible by eye. The right one is the alignment as it was after data taking in
Apr 2018. The left is after a slight realignment by loosening
the screws and moving a bit. Theodoros explanation about it
from the mail listed above:
The right one is the alignment as it was after data taking in
Apr 2018. The left is after a slight realignment by loosening
the screws and moving a bit. Theodoros explanation about it
from the mail listed above:
Hello,
After some issues the geometres installed the aligned laser today. Originally Jaime and I saw the spot as seen at the right image. It was +1mm too high. We rechecked Sebastian’s images from the Xray fingers and confirmed that his data indicated a parallel movement of ~1.4 mm (detector towards airport). We then started wondering whether there are effects coming from the target itself or the tolerances in the holes of the screws. By unscrewing it a bit it was clear that one can easily reposition it with an uncertainty of almost +-1mm. For example in the left picture you can see the new position we put it in, in which the spot is almost perfectly aligned.
We believe that the source of these shifts is primarily the positioning of the detector/target on the plexiglass drum. As everything else seems to be aligned, we do not need to realign. On Monday we will lock the manipulator arms and recheck the spot. Jaime will change his tickets to leave earlier.
Thursday-Friday we can dismount the shielding support to send it for machining and the detector can go to Bonn.
With this +-1mm play in the screw holes in mind (and the possible delays from the cavities) we should seriously consider doing an X-ray finger run right after the installation of InGRID which may need to be shifted accordingly. I will try to adjust the schedule next week.
Please let me know if you have any further comments.
Cheers,
Theodoros
- images:
geometer measurements. 4 measurements performed, with EDMS links (the links are fully public!):
- 11.07.2017 https://edms.cern.ch/document/1827959/1
- 14.09.2017 https://edms.cern.ch/document/2005606/1
- 26.10.2017 https://edms.cern.ch/document/2005690/1
- 23.07.2018 https://edms.cern.ch/document/2005895/1
For geometer measurements in particular search gmail archive for Antje Behrens (Antje.Behrens@cern.ch) or "InGrid alignment" The reports can also be found here: ./CAST_Alignment/
- X-ray finger measurements, 2 runs:
[ ]13.07.2017, run number 21 LINK DATA[ ]20.04.2018, run number 189, after first part data taking in 2018. LINK DATA
2.7. X-ray finger
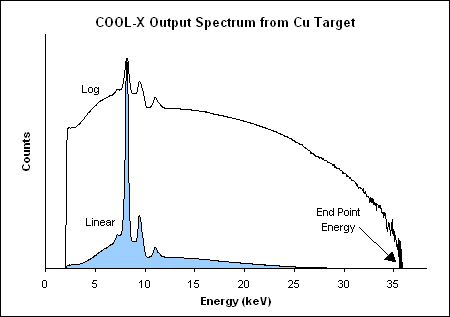
The X-ray finger used at CAST is an Amptek COOL-X:
https://www.amptek.com/internal-products/obsolete-products/cool-x-pyroelectric-x-ray-generator
The relevant plots for our purposes are shown in:
In addition the simple Monte Carlo simulation of the expected signal (written in Clojure) is found in: ./../Code/CAST/XrayFinderCalc/
2 X-ray finger runs:
[ ]13.07.2017, run number 21 LINK DATA[ ]20.04.2018, run number 189, after first part data taking in 2018. LINK DATA
Important note: The detector was removed directly after the first of these X-ray measurements! As such, the measurement has no bearing on the real position the detector was in during the first data taking campaign.
The X-ray finger run is used both to determine a center position of the detector, as well as determine the rotation of the graphite spacer of the LLNL telescope, i.e. the rotation of the telescope.
[X]Determine the rotation angle of the graphite spacer from the X-ray finger data -> do now. X-ray finger run:->
 -> It comes out to 14.17°!
But for run 21 (between which detector was dismounted of course):
-> It comes out to 14.17°!
But for run 21 (between which detector was dismounted of course):
 -> Only 11.36°!
That's a huge uncertainty given the detector was only dismounted!
3°.
-> Only 11.36°!
That's a huge uncertainty given the detector was only dismounted!
3°.
NOTE: For more information including simulations, for now see here: ./../journal.html from the day of , sec. [BROKEN LINK: sec:journal:2023_09_05_xray_finger].
2.7.1. Run 189
The below is copied from thesis.org.
I copied the X-ray finger runs from tpc19 over to ./../../CastData/data/XrayFingerRuns/. The run of interest is mainly the run 189, as it's the run done with the detector installed as in 2017/18 data taking.
cd /dev/shm # store here for fast access & temporary cp ~/CastData/data/XrayFingerRuns/XrayFingerRun2018.tar.gz . tar xzf XrayFingerRun2018.tar.gz raw_data_manipulation -p Run_189_180420-09-53 --runType xray --out xray_raw_run189.h5 reconstruction -i xray_raw_run189.h5 --out xray_reco_run189.h5 # make sure `config.toml` for reconstruction uses `default` clustering! reconstruction -i xray_reco_run189.h5 --only_charge reconstruction -i xray_reco_run189.h5 --only_gas_gain reconstruction -i xray_reco_run189.h5 --only_energy_from_e plotData --h5file xray_reco_run189.h5 --runType=rtCalibration -b bGgPlot --ingrid --occupancy --config plotData.toml
which gives us the following plot:
With many more plots here: ./../Figs/statusAndProgress/xrayFingerRun/run189/
One very important plot:
-> So the peak is at around 3 keV instead of about 8 keV, as the plot
from Amptek in the section above pretends.
[ ]Maybe at CAST they changed the target?
2.8. Detector window
The window layout is shown in fig. 2.
The sizes are thus:
- Diameter: \(\SI{14}{\mm}\)
- 4 strongbacks of:
- width: \(\SI{0.5}{\mm}\)
- thickness: \(\SI{200}{\micro\meter}\)
- \(\SI{20}{\nm}\) Al coating
- they get wider towards the very outside
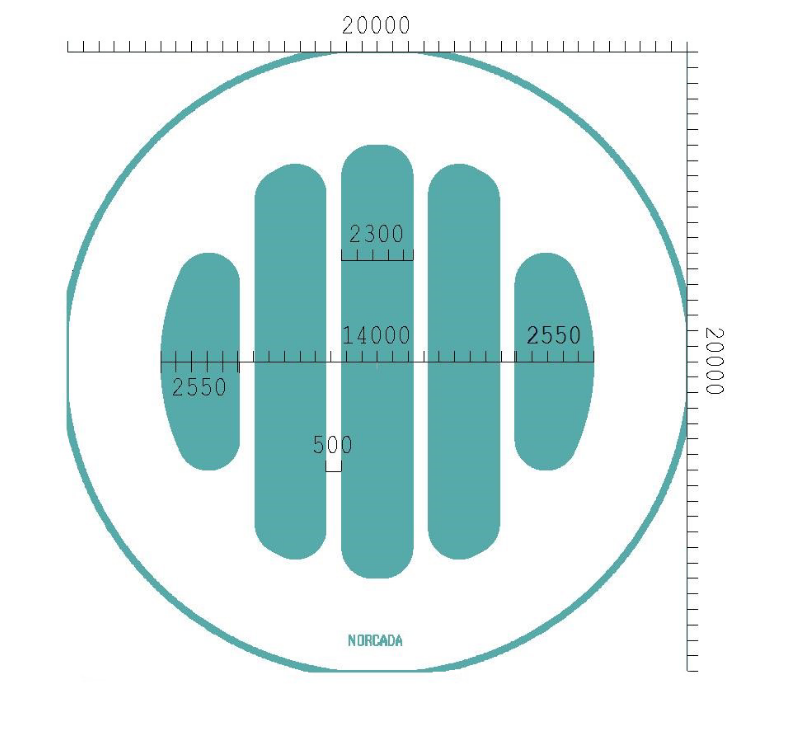
Let's compute the amount of occlusion by the strongbacks. Using code based on Johanna's raytracer:
## Super dumb MC sampling over the entrance window using the Johanna's code from `raytracer2018.nim` ## to check the coverage of the strongback of the 2018 window import ggplotnim, random, chroma proc colorMe(y: float): bool = const stripDistWindow = 2.3 #mm stripWidthWindow = 0.5 #mm if abs(y) > stripDistWindow / 2.0 and abs(y) < stripDistWindow / 2.0 + stripWidthWindow or abs(y) > 1.5 * stripDistWindow + stripWidthWindow and abs(y) < 1.5 * stripDistWindow + 2.0 * stripWidthWindow: result = true else: result = false proc sample() = randomize(423) const nmc = 100_000 let black = color(0.0, 0.0, 0.0) var dataX = newSeqOfCap[float](nmc) var dataY = newSeqOfCap[float](nmc) var inside = newSeqOfCap[bool](nmc) for idx in 0 ..< nmc: let x = rand(-7.0 .. 7.0) let y = rand(-7.0 .. 7.0) if x*x + y*y < 7.0 * 7.0: dataX.add x dataY.add y inside.add colorMe(y) let df = toDf(dataX, dataY, inside) echo "A fraction of ", df.filter(f{`inside` == true}).len / df.len, " is occluded by the strongback" let dfGold = df.filter(f{abs(idx(`dataX`, float)) <= 2.25 and abs(idx(`dataY`, float)) <= 2.25}) echo "Gold region: A fraction of ", dfGold.filter(f{`inside` == true}).len / dfGold.len, " is occluded by the strongback" ggplot(df, aes("dataX", "dataY", fill = "inside")) + geom_point() + # draw the gold region as a black rectangle geom_linerange(aes = aes(y = 0, x = 2.25, yMin = -2.25, yMax = 2.25), color = some(black)) + geom_linerange(aes = aes(y = 0, x = -2.25, yMin = -2.25, yMax = 2.25), color = some(black)) + geom_linerange(aes = aes(x = 0, y = 2.25, xMin = -2.25, xMax = 2.25), color = some(black)) + geom_linerange(aes = aes(x = 0, y = -2.25, xMin = -2.25, xMax = 2.25), color = some(black)) + xlab("x [mm]") + ylab("y [mm]") + ggsave("/home/basti/org/Figs/statusAndProgress/detector/SiN_window_occlusion.png", width = 1150, height = 1000) sample()
A fraction of 0.16170429252782195 is occluded by the strongback Gold region: A fraction of 0.2215316951907448 is occluded by the strongback (exact should be 22.2 % based on two \SI{0.5}{\mm} strongbacks within a square of \SI{4.5}{\mm} long sides).
So to summarize it in a table, tab 1 and as a figure in fig. 3.
| Region | Occlusion / % |
|---|---|
| Full | 16.2 |
| Gold | 22.2 |

The X-ray absorption properties were obtained using the online calculator from here: https://henke.lbl.gov/optical_constants/
The relevant resource files are found in:
- 200μm Si strongback: ./../resources/Si_density_2.33_thickness_200microns.txt
- 300nm SiN: ./../resources/Si3N4_density_3.44_thickness_0.3microns.txt
- 20nm Al: ./../resources/Al_20nm_transmission_10keV.txt
- 3cm Ar: ./../resources/transmission-argon-30mm-1050mbar-295K.dat
Let's create a plot of:
- window transmission
- gas absorption
- convolution of both
import ggplotnim let al = readCsv("/home/basti/org/resources/Al_20nm_transmission_10keV.txt", sep = ' ', header = "#") let siN = readCsv("/home/basti/org/resources/Si3N4_density_3.44_thickness_0.3microns.txt", sep = ' ') let si = readCsv("/home/basti/org/resources/Si_density_2.33_thickness_200microns.txt", sep = ' ') let argon = readCsv("/home/basti/org/resources/transmission-argon-30mm-1050mbar-295K.dat", sep = ' ') var df = newDataFrame() df["300nm SiN"] = siN["Transmission", float] df["200μm Si"] = si["Transmission", float] df["30mm Ar"] = argon["Transmission", float][0 .. argon.high - 1] df["20nm Al"] = al["Transmission", float] df["Energy [eV]"] = siN["PhotonEnergy(eV)", float] df = df.mutate(f{"Energy [keV]" ~ idx("Energy [eV]") / 1000.0}, f{"30mm Ar Abs." ~ 1.0 - idx("30mm Ar")}, f{"Efficiency" ~ idx("30mm Ar Abs.") * idx("300nm SiN") * idx("20nm Al")}, f{"Eff • SB • ε" ~ `Efficiency` * 0.78 * 0.8}) # strongback occlusion of 22% and ε = 80% .drop(["Energy [eV]", "Ar"]) .gather(["300nm SiN", "Efficiency", "Eff • SB • ε", "30mm Ar Abs.", "200μm Si", "20nm Al"], key = "Type", value = "Efficiency") echo df ggplot(df, aes("Energy [keV]", "Efficiency", color = "Type")) + geom_line() + ggtitle("Detector efficiency of combination of 300nm SiN window and 30mm of Argon absorption, including ε = 80% and strongback occlusion of 22%") + margin(top = 1.5) + ggsave("/home/basti/org/Figs/statusAndProgress/detector/window_plus_argon_efficiency.pdf", width = 800, height = 600)
Fig. 4 shows the combined efficiency of the SiN window, the \SI{20}{\nm} of Al coating and the gas \SI{30}{\mm} of Argon absorption and in addition the software efficiency (at ε = 80%) and strongback occlusion (22% in gold region).
The following code exists to plot the window transmissions for the window material in combination with the axion flux in:
It produces the combined plot as shown in fig. 5.
2.8.1. Window layout with correct window rotation
## Super dumb MC sampling over the entrance window using the Johanna's code from `raytracer2018.nim` ## to check the coverage of the strongback of the 2018 window import ggplotnim, chroma, unchained proc hitsStrongback(y: float): bool = const stripDistWindow = 2.3 #mm stripWidthWindow = 0.5 #mm if abs(y) > stripDistWindow / 2.0 and abs(y) < stripDistWindow / 2.0 + stripWidthWindow or abs(y) > 1.5 * stripDistWindow + stripWidthWindow and abs(y) < 1.5 * stripDistWindow + 2.0 * stripWidthWindow: result = true else: result = false proc sample() = let black = color(0.0, 0.0, 0.0) let nPoints = 256 var xs = linspace(-7.0, 7.0, nPoints) var dataX = newSeqOfCap[float](nPoints^2) var dataY = newSeqOfCap[float](nPoints^2) var inside = newSeqOfCap[bool](nPoints^2) for x in xs: for y in xs: if x*x + y*y < 7.0 * 7.0: when false: dataX.add x * cos(30.°.to(Radian)) + y * sin(30.°.to(Radian)) dataY.add y * cos(30.°.to(Radian)) - x * sin(30.°.to(Radian)) inside.add hitsStrongback(y) else: dataX.add x dataY.add y # rotate current y back, such that we can analyze in a "non rotated" coord. syst let yRot = y * cos(-30.°.to(Radian)) - x * sin(-30.°.to(Radian)) inside.add hitsStrongback(yRot) let df = toDf(dataX, dataY, inside) ggplot(df, aes("dataX", "dataY", fill = "inside")) + geom_point() + # draw the gold region as a black rectangle geom_linerange(aes = aes(y = 0, x = 2.25, yMin = -2.25, yMax = 2.25), color = some(black)) + geom_linerange(aes = aes(y = 0, x = -2.25, yMin = -2.25, yMax = 2.25), color = some(black)) + geom_linerange(aes = aes(x = 0, y = 2.25, xMin = -2.25, xMax = 2.25), color = some(black)) + geom_linerange(aes = aes(x = 0, y = -2.25, xMin = -2.25, xMax = 2.25), color = some(black)) + xlab("x [mm]") + ylab("y [mm]") + xlim(-7, 7) + ylim(-7, 7) + ggsave("/home/basti/org/Figs/statusAndProgress/detector/SiN_window_occlusion_rotated.png", width = 1150, height = 1000) sample()
Which gives us:

2.9. General event & outer chip information
Running ./../../CastData/ExternCode/TimepixAnalysis/Tools/outerChipActivity/outerChipActivity.nim we can extract information about the total number of events and the activity on the center chip vs. the outer chips.
For both the 2017/18 data (run 2) and the end of 2018 data (run 3) we will now look at:
- number of total events
- number of events with any activity (> 3 hits)
- number of events with activity only on center chip
- number of events with activity on center and outer chips (but not only center)
- number of events with activity only on outer chips
UPDATE: The reason for the two peaks in the Run 2 data of the event duration histogram is that we accidentally used run settings 2/32 in 2017 and 2/30 in 2018! (This does not explain the 0 time events of course)
2.9.1. 2017/18 (Run 2)
Number of total events: 3758960 Number of events without center: 1557934 | 41.44587864728542% Number of events only center: 23820 | 0.633685913124907% Number of events with center activity and outer: 984319 | 26.185939728009878% Number of events any hit events: 2542253 | 67.6318183752953% Mean of event durations: 2.144074329358038
Interestingly, the histogram of event durations looks as follows, fig. 6.
We can cut to the range between 0 and 2.2 s, fig. 7.
The peak at 0 is plain and simply a peak at exact 0 values (the previous figure only removed exact 0 values).
What does the energy distribution look like for these events? Fig. 8.
And the same split up per run (to make sure it's not one bad run), fig. 9.
Hmm. I suppose it's a bug in the firmware that the event duration is not correctly returned? Could happen if FADC triggers and for some reason 0 clock cycles are returned. This could be connected to the weird "hiccups" the readout sometimes does (when the FADC doesn't actually trigger for a full event). Maybe these are the events right after?
- Noisy pixels
In this run there are a few noisy pixels that need to be removed before background rates are calculated. These are listed in tab. 2.
Table 2: Number of counts noisy pixels in 2017/18 dataset contribute to the number of background clusters remaining. The total number of noise clusters amounts to 1265 in this case (depends on the clustering algorithm potentially). These must be removed for a sane background level (and the area must be removed from from the size of active area in this dataset. NOTE: When using these numbers, make sure the x and y coordinates are not accidentally inverted. x y Count after logL 64 109 7 64 110 9 65 108 30 66 108 50 67 108 33 65 109 74 66 109 262 67 109 136 68 109 29 65 110 90 66 110 280 67 110 139 65 111 24 66 111 60 67 111 34 67 112 8 \clearpage
2.9.2. End of 2018 (Run 3)
NOTE: In Run 3 we only used 2/30 as run settings! Hence a single peak in event duration.
And the same plots and numbers for 2018.
Number of total events: 1837330 Number of events without center: 741199 | 40.34109278137297% Number of events only center: 9462 | 0.514986420512374% Number of events with center activity and outer: 470188 | 25.590830172043127% Number of events any hit events: 1211387 | 65.9319229534161% Mean of event durations: 2.1157526632342307
2.10. CAST maximum angle from the sun
A question that came up today. What is the maximum difference in grazing angle that we could see on the LLNL telescope behind CAST for an axion coming from the Sun?
The Sun has an apparent size of ~32 arcminutes https://en.wikipedia.org/wiki/Sun.
If the dominant axion emission comes from the inner 10% of the radius, that's still 3 arcminutes, which is \(\SI{0.05}{°}\).
The first question is whether the magnet bore appears larger or smaller than this size from one end to the other:
import unchained, math const L = 9.26.m # Magnet length const d = 4.3.cm # Magnet bore echo "Maximum angle visible through bore = ", arctan(d / L).Radian.to(°)
so \SI{0.266}{°}, which is larger than the apparent size of the solar core.
That means the maximum angle we can see at a specific point on the telescope is up to the apparent size of the core, namely \(\SI{0.05}{°}\).
2.11. LLNL telescope
IMPORTANT: The multilayer coatings of the LLNL telescope are carbon
at the top and platinum at the bottom, despite "Pt/C" being used to
refer to them. See fig. 4.11 in the PhD thesis
 .
.
UPDATE: I randomly stumbled on a PhD thesis about the NuSTAR telescope! It validates some things I have been wondering about. See sec. 2.11.2.
UPDATE: Jaime sent me two text files today:
- ./../resources/LLNL_telescope/cast20l4_f1500mm_asDesigned.txt
- ./../resources/LLNL_telescope/cast20l4_f1500mm_asBuilt.txt
both of which are quite different from the numbers in Anders Jakobsen's thesis! These do reproduce a focal length of \(\SI{1500}{mm}\) instead of \(\SI{1530}{mm}\) when calculating it using the Wolter equation (when not using \(R_3\), but rather the virtual reflection point!).
This section covers details about the telescope design, i.e. the mirror angles, radii and all that stuff as well as information about it from external sources (e.g. the raytracing results from LLNL about it). For more information about our raytracing results, see sec. 11.
Further, for more information about the telescope see ./LLNL_def_REST_format/llnl_def_rest_format.html.
Some of the most important information is repeated here.
The information for the LLNL telescope can best be found in the PhD thesis of Anders Clemen Jakobsen from DTU in Denmark: https://backend.orbit.dtu.dk/ws/portalfiles/portal/122353510/phdthesis_for_DTU_orbit.pdf
in particular page 58 (59 in the PDF) for the following table: UPDATE: The numbers in this table are wrong. See update at the top of this section.
| Layer | Area [mm²] | Relative area [%] | Cumulative area [mm²] | α [°] | α [mrad] | R1 [mm] | R5 [mm] |
|---|---|---|---|---|---|---|---|
| 1 | 13.863 | 0.9546 | 13.863 | 0.579 | 10.113 | 63.006 | 53.821 |
| 2 | 48.175 | 3.3173 | 62.038 | 0.603 | 10.530 | 65.606 | 56.043 |
| 3 | 69.270 | 4.7700 | 131.308 | 0.628 | 10.962 | 68.305 | 58.348 |
| 4 | 86.760 | 5.9743 | 218.068 | 0.654 | 11.411 | 71.105 | 60.741 |
| 5 | 102.266 | 7.0421 | 320.334 | 0.680 | 11.877 | 74.011 | 63.223 |
| 6 | 116.172 | 7.9997 | 436.506 | 0.708 | 12.360 | 77.027 | 65.800 |
| 7 | 128.419 | 8.8430 | 564.925 | 0.737 | 12.861 | 80.157 | 68.474 |
| 8 | 138.664 | 9.5485 | 703.589 | 0.767 | 13.382 | 83.405 | 71.249 |
| 9 | 146.281 | 10.073 | 849.87 | 0.798 | 13.921 | 86.775 | 74.129 |
| 10 | 150.267 | 10.347 | 1000.137 | 0.830 | 14.481 | 90.272 | 77.117 |
| 11 | 149.002 | 10.260 | 1149.139 | 0.863 | 15.062 | 93.902 | 80.218 |
| 12 | 139.621 | 9.6144 | 1288.76 | 0.898 | 15.665 | 97.668 | 83.436 |
| 13 | 115.793 | 7.973 | 1404.553 | 0.933 | 16.290 | 101.576 | 86.776 |
| 14 | 47.648 | 3.2810 | 1452.201 | 0.970 | 16.938 | 105.632 | 90.241 |
Further information can be found in the JCAP paper about the LLNL telescope for CAST: https://iopscience.iop.org/article/10.1088/1475-7516/2015/12/008/meta
in particular table 1 (extracted with caption):
| Property | Value |
|---|---|
| Mirror substrates | glass, Schott D263 |
| Substrate thickness | 0.21 mm |
| L, length of upper and lower mirrors | 225 mm |
| Overall telescope length | 454 mm |
| f , focal length | 1500 mm |
| Layers | 13 |
| Total number of individual mirrors in optic | 26 |
| ρmax , range of maximum radii | 63.24–102.4 mm |
| ρmid , range of mid-point radii | 62.07–100.5 mm |
| ρmin , range of minimum radii | 53.85–87.18 mm |
| α, range of graze angles | 0.592–0.968 degrees |
| Azimuthal extent | Approximately 30 degrees |
2.11.1. Information (raytracing, effective area etc) from CAST Nature paper
Jaime finally sent the information about the raytracing results from the LLNL telescope to Cristina . She shared it with me: https://unizares-my.sharepoint.com/personal/cmargalejo_unizar_es/_layouts/15/onedrive.aspx?ga=1&id=%2Fpersonal%2Fcmargalejo%5Funizar%5Fes%2FDocuments%2FDoctorado%20UNIZAR%2FCAST%20official%2FLimit%20calculation%2FJaime%27s%20data
I downloaded and extracted the files to here: ./../resources/llnl_cast_nature_jaime_data/
Things to note:
- the CAST2016Dec* directories contain
.fitsfiles for the axion image for different energies - the same directories also contain text files for the effective area!
- the
./../resources/llnl_cast_nature_jaime_data/2016_DEC_Final_CAST_XRT/
directory contains the axion images actually used for the limit - I
presume - in form of
.txtfiles. that directory also contains a "final" ? effective area file! excerpt from that file: UPDATE: In the meeting with Jaime and Julia on Jaime mentioned this is the final effective area that they calculated and we should use this!
E(keV) Area(cm^2) Area_lower_limit(cm^2) Area_higher_limit(cm^2) 0.000000 9.40788 8.93055 9.87147 0.100000 2.51070 1.76999 3.56970 0.200000 5.96852 5.06843 6.93198 0.300000 4.05163 3.55871 4.60069 0.400000 5.28723 4.70362 5.92018 0.500000 6.05037 5.50801 6.63493 0.600000 5.98980 5.44433 6.56380 0.700000 6.33760 5.81250 6.86565 0.800000 6.45533 5.97988 6.94818 0.900000 6.68399 6.22210 7.15994 1.00000 6.87400 6.42313 7.32568 1.10000 7.01362 6.57078 7.44991 1.20000 7.11297 6.68403 7.53477 1.30000 7.18784 6.76026 7.60188 1.40000 7.23464 6.82698 7.65152 1.50000 7.26598 6.85565 7.66851 1.60000 7.28027 6.86977 7.67453 1.70000 7.26311 6.86645 7.66171 1.80000 7.22509 6.83192 7.61740 1.90000 7.14513 6.76611 7.52503 2.00000 6.96418 6.58820 7.32984 2.10000 5.28441 5.00942 5.55890 2.20000 3.64293 3.45370 3.82893 2.30000 5.17823 4.90664 5.44582 2.40000 5.29972 5.02560 5.57611 2.50000 5.29166 5.02555 5.57095 2.60000 5.17942 4.91425 5.43329 2.70000 4.92675 4.67978 5.18098 2.80000 4.92422 4.66858 5.17432 2.90000 4.83265 4.58795 5.08459 3.00000 4.64834 4.41387 4.89098i.e. it peaks at ~7.3.
Plot the "final" effective area against the extracted data from the JCAP paper:
Note that we do not know with certainty that this is indeed the effective area used for the CAST Nature limit. That's just my assumption!
import ggplotnim const path = "/home/basti/org/resources/llnl_cast_nature_jaime_data/2016_DEC_Final_CAST_XRT/EffectiveArea.txt" const pathJCAP = "/home/basti/org/resources/llnl_xray_telescope_cast_effective_area.csv" let dfJcap = readCsv(pathJCAP) let df = readCsv(path, sep = ' ') .rename(f{"Energy[keV]" <- "E(keV)"}, f{"EffectiveArea[cm²]" <- "Area(cm^2)"}) .select("Energy[keV]", "EffectiveArea[cm²]") let dfC = bind_rows([("JCAP", dfJcap), ("Nature", df)], "Type") ggplot(dfC, aes("Energy[keV]", "EffectiveArea[cm²]", color = "Type")) + geom_line() + ggsave("/tmp/effective_area_jcap_vs_nature_llnl.pdf")
So it seems like the effective area here is even lower than the
effective area in the JCAP LLNL paper! That's ridiculous.
HOWEVER the shape seems to match much better with the shape we get
from computing the effective area ourselves!
-> UPDATE: No, not really. I ran the code in journal.org with
makePlot and makeRescaledPlot using dfJaimeNature as a rescaling
reference using the 3 arcmin code.
So the shape is very different after all.
[ ]Is there a chance the difference is due toxrayAttenuation? Note the weird energy dependent linear offset comparingxrayAttenuationreflectivity compared to the DarpanX numbers! Could that shift be the reason?
- LLNL raytracing for axion image and CoolX X-ray finger
The DTU thesis contains raytracing images (from page 78) for the X-ray finger run and for the axion image.
- X-ray finger
The image (as a screenshot) from the X-ray finger:

where we can see a few things:
- the caption mentions the source was 14.2 m away from the optic. This is nonsensical. The magnet is 9.26m long and even with the cryo housing etc. we won't get to much more than 10 m from the telescope. The X-ray finger was installed in the bore of the magnet!
- it mentions the source being 6 mm diameter (text mentions diameter
explicitly). All we know about it is from the manufacturer that the
size is given as 15 mm. But there is nothing about the actual size
of the emission surface.
- the resulting raytraced image has a size of only slightly less than 3 mm in the short axis and maybe about 3 mm in the long axis.
About 3: Our own X-ray finger is the following: file:///home/basti/phd/Figs/CAST_Alignment/xray_finger_centers_run_189.pdf (Note: it needs to be rotated of course) We can see that our real image is much larger! Along "x" it goes from about 5.5 to 10 mm or so! Quite a bit larger. And along y from less than 4 to maybe 10!
Given that we have the raytracing data from Jaime, let's plot their data to see if it actually looks like that:
import ggplotnim, sequtils, seqmath let df = readCsv("/home/basti/org/resources/llnl_cast_nature_jaime_data/2016_DEC_Final_CAST_XRT/3.00keV_2Dmap_CoolX.txt", sep = ' ', skipLines = 2, colNames = @["x", "y", "z"]) .mutate(f{"x" ~ `x` - mean(`x`)}, f{"y" ~ `y` - mean(`y`)}) var customInferno = inferno() customInferno.colors[0] = 0 # transparent ggplot(df, aes("x", "y", fill = "z")) + geom_raster() + scale_fill_gradient(customInferno) + xlab("x [mm]") + ylab("y [mm]") + ggtitle("LLNL raytracing of X-ray finger (Jaime)") + ggsave("~/org/Figs/statusAndProgress/rayTracing/raytracing_xray_finger_llnl_jaime.pdf") ggplot(df.filter(f{`x` >= -7.0 and `x` <= 7.0 and `y` >= -7.0 and `y` <= 7.0}), aes("x", "y", fill = "z")) + geom_raster() + scale_fill_gradient(customInferno) + xlab("x [mm]") + ylab("y [mm]") + xlim(-7.0, 7.0) + ylim(-7.0, 7.0) + ggtitle("LLNL raytracing of X-ray finger zoomed (Jaime)") + ggsave("~/org/Figs/statusAndProgress/rayTracing/raytracing_xray_finger_llnl_jaime_gridpix_size.pdf")
This yields the following figure:
and cropped to the range of the GridPix:
This is MUCH bigger than the plot from the paper indicates. And the shape is also much more elongated! More in line with what we really see.
Let's use our raytracer to produce the X-ray finger according to the specification of 14.2 m first and then a more reasonable estimate.
Make sure to put the following into the
config.tomlfile:[TestXraySource] useConfig = true # sets whether to read these values here. Can be overriden here or useng flag `--testXray` active = true # whether the source is active (i.e. Sun or source?) sourceKind = "classical" # whether a "classical" source or the "sun" (Sun only for position *not* for energy) parallel = false energy = 3.0 # keV The energy of the X-ray source distance = 14200 # 9260.0 #106820.0 #926000 #14200 #9260.0 #2000.0 # mm Distance of the X-ray source from the readout radius = 3.0 #21.5 #44.661 #8.29729 #46.609 #4.04043 #3.0 #4.04043 #21.5 # #21.5 # mm Radius of the X-ray source offAxisUp = 0.0 # mm offAxisLeft = 0.0 # mm activity = 0.125 # GBq The activity in `GBq` of the source lengthCol = 0.0 #0.021 # mm Length of a collimator in front of the source
./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_xrayFinger_14.2m_3mm"which more or less matches the size of our real data.
Now the same with a source that is 10 m away:
[TestXraySource] useConfig = true # sets whether to read these values here. Can be overriden here or useng flag `--testXray` active = true # whether the source is active (i.e. Sun or source?) sourceKind = "classical" # whether a "classical" source or the "sun" (Sun only for position *not* for energy) parallel = false energy = 3.0 # keV The energy of the X-ray source distance = 10000 # 9260.0 #106820.0 #926000 #14200 #9260.0 #2000.0 # mm Distance of the X-ray source from the readout radius = 3.0 #21.5 #44.661 #8.29729 #46.609 #4.04043 #3.0 #4.04043 #21.5 # #21.5 # mm Radius of the X-ray source offAxisUp = 0.0 # mm offAxisLeft = 0.0 # mm activity = 0.125 # GBq The activity in `GBq` of the source lengthCol = 0.0 #0.021 # mm Length of a collimator in front of the source
./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_xrayFinger_10m_3mm"which is quite a bit bigger than our real data. Maybe we allow some angles that we shouldn't, i.e. the X-ray finger has a collimator? Or our reflectivities are too good for too large angles?
Without good knowledge of the real size of the X-ray finger emission this is hard to get right.
- Axion image
The axion image as mentioned in the PhD thesis is the following:

First of all let's note that the caption talks about emission of a 3 arcminute source. Let's check the apparent size of the sun and the typical emission, which is from the inner 30%:
import unchained, math let Rsun = 696_342.km # SOHO mission 2003 & 2006 # use the tangent to compute based on radius of sun: # tan α = Rsun / 1.AU echo "Apparent size of the sun = ", arctan(Rsun / 1.AU).Radian.to(ArcMinute) echo "Typical emission sun from inner 30% = ", arctan(Rsun * 0.3 / 1.AU).Radian.to(ArcMinute) let R3arc = (tan(3.ArcMinute.to(Radian)) * 1.AU).to(km) echo "Used radius for 3' = ", R3arc echo "As fraction of solar radius = ", R3arc / RSun
So 3' correspond to about 18.7% of the radius. All in all that seems reasonable at least.
Let's plot the axion image as we have it from Jaime's data:
import ggplotnim, seqmath import std / [os, sequtils, strutils] proc readRT(p: string): DataFrame = result = readCsv(p, sep = ' ', skipLines = 4, colNames = @["x", "y", "z"]) result["File"] = p proc meanData(df: DataFrame): DataFrame = result = df.mutate(f{"x" ~ `x` - mean(col("x"))}, f{"y" ~ `y` - mean(col("y"))}) proc plots(df: DataFrame, title, outfile: string) = var customInferno = inferno() customInferno.colors[0] = 0 # transparent ggplot(df, aes("x", "y", fill = "z")) + geom_raster() + scale_fill_gradient(customInferno) + xlab("x [mm]") + ylab("y [mm]") + ggtitle(title) + ggsave(outfile) ggplot(df.filter(f{`x` >= -7.0 and `x` <= 7.0 and `y` >= -7.0 and `y` <= 7.0}), aes("x", "y", fill = "z")) + geom_raster() + scale_fill_gradient(customInferno) + xlab("x [mm]") + ylab("y [mm]") + xlim(-7.0, 7.0) + ylim(-7.0, 7.0) + ggtitle(title & " (zoomed)") + ggsave(outfile.replace(".pdf", "_gridpix_size.pdf")) block Single: let df = readRT("/home/basti/org/resources/llnl_cast_nature_jaime_data/2016_DEC_Final_CAST_XRT/3.00keV_2Dmap.txt") .meanData() df.plots("LLNL raytracing of axion image @ 3 keV (Jaime)", "~/org/Figs/statusAndProgress/rayTracing/raytracing_axion_image_llnl_jaime_3keV.pdf") block All: var dfs = newSeq[DataFrame]() for f in walkFiles("/home/basti/org/resources/llnl_cast_nature_jaime_data/2016_DEC_Final_CAST_XRT/*2Dmap.txt"): echo "Reading: ", f dfs.add readRT(f) echo "Summarize" var df = dfs.assignStack() df = df.group_by(@["x", "y"]) .summarize(f{float: "z" << sum(`z`)}, f{float: "zMean" << mean(`z`)}) df.writeCsv("/tmp/llnl_raytracing_jaime_all_energies_raw_sum.csv") df = df.meanData() df.writeCsv("/tmp/llnl_raytracing_jaime_all_energies.csv") plots(df, "LLNL raytracing of axion image (sum all energies) (Jaime)", "~/org/Figs/statusAndProgress/rayTracing/raytracing_axion_image_llnl_jaime_all_energies.pdf")
The 3 keV data for the axion image:
and cropped again:
And the sum of all energies:
and cropped again:
Both clearly show the symmetric shape that is so weird but also - again - does NOT reproduce the raytracing seen in the screenshot above! That one clearly has a very stark tiny center with the majority of the flux, which is gone and replaced by a much wider region of significant flux!
Both are in strong contrast to our own axion image. Let's compute that using the Primakoff only (make sure to disable the X-ray test source in the config file!):
./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_axionImagePrimakoff_focal_point"and for a more realistic image at the expected conversion point:
[DetectorInstallation] useConfig = true # sets whether to read these values here. Can be overriden here or using flag `--detectorInstall` # Note: 1500mm is LLNL focal length. That corresponds to center of the chamber! distanceDetectorXRT = 1487.93 # mm distanceWindowFocalPlane = 0.0 # mm lateralShift = 0.0 # mm lateral ofset of the detector in repect to the beamline transversalShift = 0.0 # mm transversal ofset of the detector in repect to the beamline #0.0.mm #
./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_axionImagePrimakoff_conversion_point"which yields:
which is not that far off in size of the LLNL raytraced image. The shape is just quite different!
- X-ray finger
- Reply to Igor about LLNL telescope raytracing
Igor wrote me the following mail:
Hi Sebastian, Now that we are checking with Cristina the shape of the signal after the LLNL telescope for the SRMM analysis, I got two questions on your analysis:
- The signal spot shape that you present is different from the one we have for the Nature physics paper. Do you understand why? There was a change in the Ingrid setup wrt the SRMM setup that explains it, maybe?
- Do you have a spot calibration data that allows to crosscheck the position (and rotation) of the signal spot in the Ingrid chip coordinates?
Best, Igor
as a reply to my "Limit method for 7-GridPix @ CAST" mail on .
I ended up writing a lengthy reply.
The reply is also found here: ./../Mails/igorReplyLLNL/igor_reply_llnl_axion_image.html
- My reply
Hey,
sorry for the late reply. I didn't want to reply with one sentence for each question. While looking into the questions in more details more things came up.
One thing - embarrassingly - is that I completely forgot to apply the rotation of my detector in the limit calculation (in our case the detector is rotated by 90° compared to the "data" x-y plane). Added to that is the slight rotation of the LLNL axis, which I also need to include (here I simply forgot that we never added it to the raytracer. Given that the spacer is not visible in the axion image, it didn't occur to me).
Let's start with your second question
Do you have a spot calibration data that allows to crosscheck the position (and rotation) of the signal spot in the Ingrid chip coordinates?
Yes, we have two X-ray finger runs. Unfortunately, one of them is not useful, as it was taken in July 2017 after our detector had to be removed again to make space for a short KWISP data taking. We have a second one from April 2018, which is partially useful. However, the detector was again dismounted between April and October 2018 and we don't have an X-ray finger run for the last data taking between Oct 2018 to Dec 2018.
Fig. 14 shows the X-ray finger run shows the latter X-ray finger run. The two parallel lines with few clusters are two of the window strongbacks. The other line is the graphite spacer of the telescope. The center positions of the clusters are at
- (x, y) = (7.43, 6.59)
(the chip center is at (7, 7). This is what makes up the basis of our position systematic uncertainty of 5%. The 5% correspond to 0.05*7 mm = 0.35 mm.
Figure 14: X-ray finger run from April 2018, which can be used as a rough guide for the spot center. Center position is at \((x, y) = (\SI{7.43}{mm}, \SI{6.59}{mm})\). I decided not to move the actual center of the solar axion image because the X-ray finger data is hard to interpret for three different reasons:
- The entire CAST setup is "modified" in between normal data takings and installation of the X-ray finger. Who knows the effect warming up the magnet etc. is on the spot position?
- determining the actual center position of the axion spot based on the X-ray finger cluster centers is problematic due to the fact that the LLNL telescope is only a portion of a full telescope. With the resulting shape of the X-ray finger signal, combined with the missing data due to the window strongback and graphite spacer and relatively low statistics in the first place, makes trusting the numbers problematic.
- as I said before, we don't even have an X-ray finger run for the last part of the data taking. While we have the geometer measurements from the targets, I don't have the patience to learn about the coordinate system they use and attempt to reconstruct the possible movement based on those measured coordinates.
Given that we take into account the possible movement in the systematics, I believe this is acceptable.
The signal spot shape that you present is different from the one we have for the Nature physics paper. Do you understand why? There was a change in the Ingrid setup wrt the SRMM setup that explains it, maybe?
Here we now come to the actual part that is frustrating for me, too. Unfortunately, due to the "black box" nature of the LLNL telescope, Johanna and me never fully understood this. We don't understand how the raytracing calculations done by Michael Pivovaroff can ever produce a symmetric image given that the LLNL telescope is a) not a perfect Wolter design, but has cone shaped mirrors, b) is only a small portion of a full telescope and c) the incoming X-rays are not perfectly parallel. Intuitively I don't expect to have a symmetric image there. And our raytracing result does not produce anything like that.
A couple of years ago Johanna tried to find out more information about the LLNL raytracing results, but back then when Julia and Jaime were still at LLNL, the answer was effectively a "it's a secret, we can't provide more information".
As such all I can do is try to reproduce the results as well as possible. If they don't agree all I can do is provide explanations about what we compute and give other people access to my data, code and results. Then at least we can all hopefully figure out if there's something wrong with our approach.
Fig. 15 is the raytracing result as it is presented on page 78 of the PhD thesis of A. Jakobsen. It mentions that the Sun is considered as a 3' source, implying the inner ~18% of the Sun are contributing to axion emission.

Figure 15: Raytracing result as shown on page 78 (79 in PDF) of the PhD thesis of A. Jakobsen. Mentions a 3' source and has a very pronounced tiny focal spot. If I compute this with our own raytracer for the focal spot, I get the plot shown in fig. \ref{fig:axion_image_primakoff_focal_spot}. Fig. \ref{fig:axion_image_primakoff_median_conv} then corresponds to the point that sees the median of all conversions in the gas based on X-ray absorption in the gas. This is now for the case of a pure Primakoff emission and not for dominant axion-electron coupling, as I showed in my presentation (this changes the dominant contributions by radius slightly, see fig. \ref{fig:radial_production_primakoff} { Primakoff } and fig. \ref{fig:radial_production_axion_electron} { axion-electron }). They look very similar, but there are slight changes between the two axion images.
This is one of the big reasons I want to have my own raytracing simulation. Different emission models result in different axion images!
\begin{figure}[htbp] \centering \begin{subfigure}{0.5\linewidth} \centering \includegraphics[width=0.95\textwidth]{/home/basti/org/Figs/statusAndProgress/rayTracing/raytracing_axion_image_primakoff_focal_point.pdf} \caption{Focal spot} \label{fig:axion_image_primakoff_focal_spot} \end{subfigure}% \begin{subfigure}{0.5\linewidth} \centering \includegraphics[width=0.95\textwidth]{/home/basti/org/Figs/statusAndProgress/rayTracing/raytracing_axion_image_primakoff_conversion_point.pdf} \caption{Median conversion point} \label{fig:axion_image_primakoff_median_conv} \end{subfigure} \label{fig:axion_image} \caption{\subref{fig:axion_image_primakoff_focal_spot} Axion image for Primakoff emission from the Sun, computed for the exact LLNL focal spot. (Ignore the title) \subref{fig:axion_image_primakoff_median_conv} Axion image for the median conversion point of the X-rays actually entering the detector. } \end{figure} \begin{figure}[htbp] \centering \begin{subfigure}{0.5\linewidth} \centering \includegraphics[width=0.95\textwidth]{~/org/Figs/statusAndProgress/axionProduction/sampled_radii_primakoff.pdf} \caption{Primakoff radii} \label{fig:radial_production_primakoff} \end{subfigure}% \begin{subfigure}{0.5\linewidth} \centering \includegraphics[width=0.95\textwidth]{~/org/Figs/statusAndProgress/axionProduction/sampled_radii_axion_electron.pdf} \caption{Axion-electron radii} \label{fig:radial_production_axion_electron} \end{subfigure} \label{fig:radial_production} \caption{\subref{fig:radial_production_primakoff} Radial production in the Sun for Primakoff emission. \subref{fig:radial_production_axion_electron} Radial production for axion-electron emission. } \end{figure}Note that this currently does not yet take into account the slight rotation of the telescope. I first need to extract the rotation angle from the X-ray finger run.
Fig. 16 is the sum of all energies of the raytracing results that Jaime finally sent to Cristina a couple of weeks ago. In this case cropped to the size of our detector, placed at the center. These should be - as far as I understand - the ones that the contours used in the Nature paper are based on. However, these clearly do not match the results shown in the PhD thesis of Jakobsen. The extremely small focus area in black is gone and replaced by a much more diffuse area. But again, it is very symmetric, which I don't understand.
Figure 16: Raytracing image (sum of all energies) presumabely from LLNL. Likely what the Nature contours are based on. And while I was looking into this I also thought I should try to (attempt to) reproduce the X-ray finger raytracing result. Here came another confusion, because the raytracing results for that shown in the PhD thesis, fig. 17, mention that the X-ray finger was placed \SI{14.2}{m} away from the optic with a diameter of \SI{6}{mm}. That seems very wrong, given that the magnet bore is only \SI{9.26}{m} long. In total the entire magnet is - what - maybe \SI{10}{m}? At most it's maybe \SI{11}{m} to the telescope when the X-ray finger is installed in the bore? Unfortunately, the website about the X-ray finger from Amptek is not very helpful either:
https://www.amptek.com/internal-products/obsolete-products/cool-x-pyroelectric-x-ray-generator
as the only thing it says about the size is:
Miniature size: 0.6 in dia x 0.4 in (15 mm dia x 10 mm)

Figure 17: X-ray finger raytracing simulation from PhD thesis of A. Jakobsen. Mentions a distance of \(\SI{14.2}{m}\) and a source diameter of \(\SI{6}{mm}\), but size is only a bit more than \(2·\SI{2}{mm²}\). Nothing about the actual size of the area that emits X-rays. Neither do I know anything about a possible collimator used.
Furthermore, the spot size seen here is only about \(\sim 2.5·\SI{3}{mm²}\) or so. Comparing it to the spot size seen with our detector it's closer to \(\sim 5·\SI{5}{mm²}\) or even a bit larger!
So I decided to run a raytracing following these numbers, i.e. \(\SI{14.2}{m}\) and a \(\SI{3}{mm}\) radius disk shaped source without a collimator. That yields fig. 18. As we can see the size is more in line with our actually measured data.
Figure 18: Raytracing result of an "X-ray finger" at a distance of \(\SI{14.2}{m}\) and diameter of \(\SI{6}{mm}\). Results in a size closer to our real X-ray finger result. (Ignore the title) Again, I looked at the raytracing results that Jaime sent to Cristina, which includes a file with suffix "CoolX". That plot is shown in fig. 19. As we can see, it is also much larger suddenly than shown in the PhD thesis (more than \(4 · \SI{4}{mm²}\)), slightly smaller than ours.
Note that the Nature paper mentions the source is about \(\SI{12}{m}\) away. I was never around when the X-ray finger was installed, nor do I have any good data about the real magnet size or lengths of the pipes between magnet and telescope.
Figure 19: LLNL raytracing image from Jaime. Shows a much larger size now than presented in the PhD thesis. So, uhh, yeah. This is all very confusing. No matter where one looks regarding this telescope, one is bound to find contradictions or just confusing statements… :)
2.11.2. Information from NuSTAR PhD thesis
I found the following PhD thesis:
 which is about the NuSTAR optic and also from DTU. It explains a lot
of things:
which is about the NuSTAR optic and also from DTU. It explains a lot
of things:
- in the introductory part about multilayers it expands on why the low density material is at the top!
- Fig. 1.11 shows that indeed the spacers are 15° apart from one another.
- Fig. 1.11 mentions the graphite spacers are only 1.2 mm wide instead of 2 mm! But the DTU LLNL thesis explicitly mentions \(x_{gr} = \SI{2}{mm}\) on page 64.
- it has a plot of energy vs angle of the reflectivity similar to what we produce! It looks very similar.
- for the NuSTAR telescope they apparently have measurements of the surface roughness to μm levels, which are included in their simulations!
2.11.3. X-ray raytracers
Other X-ray raytracers:
- McXtrace from DTU and Synchrotron SOLEIL: https://www.mcxtrace.org/about/ https://github.com/McStasMcXtrace/McCode
- MTRAYOR (mentioned in DTU NuSTAR PhD thesis):
written in Yorick https://github.com/LLNL/yorick
https://en.wikipedia.org/wiki/Yorick_(programming_language)
a language developed at LLNL!
->
https://web.archive.org/web/20170102091157/http://www.jeh-tech.com/yorick.html
for an 'introduction'
https://ftp.spacecenter.dk/pub/njw/MT_RAYOR/mt_rayor_man4.pdf
We have the MTRAYOR code here: ./../../src/mt_rayor/ it needs Yorick, which can be found here:
2.11.4. DTU FTP server [/]
The DTU has a publicly accessible FTP server with a lot of useful information. I found it by googling for MTRAYOR, because the manual is found there.
https://ftp.spacecenter.dk/pub/njw/
I have a mirror of the entire FTP here: ./../../Documents/ftpDTU/
[ ]Remove all files larger than X MB if they appear uninteresting to us.
2.11.5. Michael Pivovaroff talk about Axions, CAST, IAXO
Michael Pivovaroff giving a talk about axions, CAST, IAXO at LLNL: https://youtu.be/H_spkvp8Qkk
First he mentions: https://youtu.be/H_spkvp8Qkk?t=2372 "Then we took the telescope to PANTER" -> implying yes the CAST optic really was at PANTER. Then he says wrongly there was a 55Fe source at the other end of the magnet, showing the X-ray finger data + simulation below that title. And finally in https://youtu.be/H_spkvp8Qkk?t=2468 he says ABOUT HIS OWN RAYTRACING SIMULATION that it was a simulation for a source at infinity…
https://youtu.be/H_spkvp8Qkk?t=3134 He mentions Jaime and Julia wanted to write a paper about using NuSTAR data to set an ALP limit for reconversion of axions etc in the solar corona by looking at the center…
3. Theory
3.1. Solar axion flux
From ./../Papers/first_cast_results_physrevlett.94.121301.pdf
There are different analytical expressions for the solar axion flux for Primakoff production. These stem from the fact that a solar model is used to model the internal density, temperature, etc. in the Sun to compute the photon distribution (essentially the blackbody radiation) near the core. From it (after converting via the Primakoff effect) we get the axion flux.
Different solar models result in different expressions for the flux. The first one uses an older model, while the latter ones use newer models.
Analytical flux from first CAST result paper: g₁₀ = gaγ • 10¹⁰ GeV dΦa/dEa = g²₁₀ 3.821•10¹⁰ cm⁻²•s⁻¹•keV⁻¹ (Ea / keV)³ / (exp(Ea / (1.103 keV)) - 1) results in an integrated flux: Φa = g²₁₀ 3.67•10¹¹ cm⁻²•s⁻¹
In comparison I used in my master thesis:
def axion_flux_primakoff(w, g_ay): # axion flux produced by the Primakoff effect # in units of m^(-2) year^(-1) keV^(-1) val = 2.0 * 10**18 * (g_ay / 10**(-12) )**2 * w**(2.450) * np.exp(-0.829 * w) return val
(./../../Documents/Masterarbeit/PyAxionFlux/PyAxionFlux.py / ./../Code/CAST/PyAxionFlux/PyAxionFlux.py) The version I use is from the CAST paper about the axion electron coupling: ./../Papers/cast_axion_electron_jcap_2013_pnCCD.pdf eq. 3.1 on page 7.
Another comparison from here:
- Weighing the solar axion
Contains, among others, a plot and (newer) description for the solar axion flux (useful as a comparison)
ΦP₁₀ = 6.02e10.cm⁻²•s⁻¹•keV⁻¹
dΦa/dEa = ΦP₁₀ (gaγ / 1e-10.GeV⁻¹) * pow(Ea / 1.keV, 2.481) / (exp(Ea / (1.205.keV)))
3.1.1. Solar axion-electron flux
We compute the differential axion flux using ./../../CastData/ExternCode/AxionElectronLimit/src/readOpacityFile.nim
We have a version of the plot that is generated by it here:
but let's generate one from the setup we use as a "base" at CAST, namely the file: ./../resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv which uses a distance Sun ⇔ Earth of 0.989 AU, corresponding to the mean of all solar trackings we took at CAST.
import ggplotnim const path = "~/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv" let df = readCsv(path) .filter(f{`type` notin ["LP Flux", "TP Flux", "57Fe Flux"]}) echo df ggplot(df, aes("Energy", "diffFlux", color = "type")) + geom_line() + xlab(r"Energy [$\si{keV}$]", margin = 1.5) + ylab(r"Flux [$\si{keV^{-1}.cm^{-2}.s^{-1}}$]", margin = 2.75) + ggtitle(r"Differential solar axion flux for $g_{ae} = \num{1e-13}, g_{aγ} = \SI{1e-12}{GeV^{-1}}$") + xlim(0, 10) + margin(top = 1.5, left = 3.25) + theme_transparent() + ggsave("~/org/Figs/statusAndProgress/differential_flux_sun_earth_distance/differential_solar_axion_fluxg_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.pdf", width = 800, height = 480, useTeX = true, standalone = true)
3.1.2. Radial production
Part of the raytracer are now also plots about the radial emission for the production.
With our default file (axion-electron)
solarModelFile = "solar_model_dataframe.csv"
running via:
./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_axion_electron" --sanity
yields
And for the Primakoff flux, using the new file:
solarModelFile = "solar_model_dataframe_fluxKind_fkAxionPhoton_0.989AU.csv" #solar_model_dataframe.csv"
running:
./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_primakoff" --sanity
we get
3.2. Axion conversion probability
Ref:
Biljana's and Kreso's notes on the axion-photon interaction here:
Further see the notes on the IAXO gas phase:
which contains the explicit form of \(P\) in the next equation!
I think it should be straight forward to derive this one from what's
given in the former PDF in eq. (3.41) (or its derivation).
[ ]Investigate this There is a chance it is non trivial due to Γ. The first PDF includes \(m_γ\), but does not mention gas in any way. So I'm not sure how one ends up at the latter. Potentially by 'folding' with the losses after the conversion?
The axion-photon conversion probability \(P_{a\rightarrow\gamma}\) in general is given by:
\begin{equation} \label{eq_conversion_prob} P_{a\rightarrow\gamma} = \left(\frac{g_{a\gamma} B}{2}\right)^2 \frac{1}{q^2 + \Gamma^2 / 4} \left[ 1 + e^{-\Gamma L} - 2e^{-\frac{\Gamma L}{2}} \cos(qL)\right], \end{equation}where \(\Gamma\) is the inverse absorption length for photons (or attenuation length).
The coherence condition for axions is
with \(L\) the length of the magnetic field (20m for IAXO, 10m for BabyIAXO), \(m_a\) the axion mass and \(E_a\) the axion energy (taken from solar axion spectrum).
In the presence of a low pressure gas, the photon receives an effective mass \(m_{\gamma}\), resulting in a new \(q\):
Thus, we first need some values for the effective photon mass in a low pressure gas, preferably helium.
From this we can see that coherence in the gas is restored if \(m_{\gamma} = m_a\), \(q \rightarrow 0\) for \(m_a \rightarrow m_{\gamma}\). This means that in those cases the energy of the incoming axion is irrelevant for the sensitivity!
Analytically the vacuum conversion probability can be derived from the expression eq. \eqref{eq_conversion_prob} by simplifying \(q\) for \(m_{\gamma} \rightarrow 0\) and \(\Gamma = 0\):
\begin{align} \label{eq_conversion_prob_vacuum} P_{a\rightarrow\gamma, \text{vacuum}} &= \left(\frac{g_{a\gamma} B}{2}\right)^2 \frac{1}{q^2} \left[ 1 + 1 - 2 \cos(qL) \right] \\ P_{a\rightarrow\gamma, \text{vacuum}} &= \left(\frac{g_{a\gamma} B}{2}\right)^2 \frac{2}{q^2} \left[ 1 - \cos(qL) \right] \\ P_{a\rightarrow\gamma, \text{vacuum}} &= \left(\frac{g_{a\gamma} B}{2}\right)^2 \frac{2}{q^2} \left[ 2 \sin^2\left(\frac{qL}{2}\right) \right] \\ P_{a\rightarrow\gamma, \text{vacuum}} &= \left(g_{a\gamma} B\right)^2 \frac{1}{q^2} \sin^2\left(\frac{qL}{2}\right) \\ P_{a\rightarrow\gamma, \text{vacuum}} &= \left(\frac{g_{a\gamma} B L}{2} \right)^2 \left(\frac{\sin\left(\frac{qL}{2}\right)}{ \left( \frac{qL}{2} \right)}\right)^2 \\ P_{a\rightarrow\gamma, \text{vacuum}} &= \left(\frac{g_{a\gamma} B L}{2} \right)^2 \left(\frac{\sin\left(\delta\right)}{\delta}\right)^2 \\ \end{align}The conversion probability in the simplified case amounts to:
\[ P(g_{aγ}, B, L) = \left(\frac{g_{aγ} \cdot B \cdot L}{2}\right)^2 \] in natural units, where the relevant numbers for the CAST magnet are:
- \(B = \SI{8.8}{T}\)
- \(L = \SI{9.26}{m}\)
and in the basic axion-electron analysis a fixed axion-photon coupling of \(g_{aγ} = \SI{1e-12}{\per\giga\electronvolt}\).
This requires either conversion of the equation into SI units by adding the "missing" constants or converting the SI units into natural units. As the result is a unit less number, the latter approach is simpler.
The conversion factors from Tesla and meter to natural units are as follows:
import unchained echo "Conversion factor Tesla: ", 1.T.toNaturalUnit() echo "Conversion factor Meter: ", 1.m.toNaturalUnit()
Conversion factor Tesla: 195.353 ElectronVolt² Conversion factor Meter: 5.06773e+06 ElectronVolt⁻¹
As such, the resulting conversion probability ends up as:
import unchained, math echo "9 T = ", 9.T.toNaturalUnit() echo "9.26 m = ", 9.26.m.toNaturalUnit() echo "P = ", pow( 1e-12.GeV⁻¹ * 9.T.toNaturalUnit() * 9.26.m.toNaturalUnit() / 2.0, 2.0)
9 T = 1758.18 ElectronVolt² 9.26 m = 4.69272e+07 ElectronVolt⁻¹ P = 1.701818225891982e-21
\begin{align} P(g_{aγ}, B, L) &= \left(\frac{g_{aγ} \cdot B \cdot L}{2}\right)^2 \\ &= \left(\frac{\SI{1e-12}{GeV^{-1}} \cdot \SI{1758.18}{eV^2} \cdot \SI{4.693e7}{eV^{-1}}}{2}\right)^2 \\ &= \num{1.702e-21} \end{align}Note that this is of the same (inverse) order of magnitude as the flux of solar axions (\(\sim10^{21}\) in some sensible unit of time), meaning the experiment expects \(\mathcal{O}(1)\) counts, which is sensible.
import unchained, math echo "9 T = ", 9.T.toNaturalUnit() echo "9.26 m = ", 9.26.m.toNaturalUnit() echo "P(natural) = ", pow( 1e-12.GeV⁻¹ * 9.T.toNaturalUnit() * 9.26.m.toNaturalUnit() / 2.0, 2.0) echo "P(SI) = ", ε0 * (hp / (2*π)) * (c^3) * (1e-12.GeV⁻¹ * 9.T * 9.26.m / 2.0)^2
3.2.1. Deriving the missing constants in the conversion probability
The conversion probability is given in natural units. In order to plug in SI units directly without the need for a conversion to natural units for the magnetic field and length, we need to reconstruct the missing constants.
The relevant constants in natural units are:
\begin{align*} ε_0 &= \SI{8.8541878128e-12}{A.s.V^{-1}.m^{-1}} \\ c &= \SI{299792458}{m.s^{-1}} \\ \hbar &= \frac{\SI{6.62607015e-34}{J.s}}{2π} \end{align*}which are each set to 1.
If we plug in the definition of a volt we get for \(ε_0\) units of:
\[ \left[ ε_0 \right] = \frac{\si{A^2.s^4}}{\si{kg.m^3}} \]
The conversion probability naively in natural units has units of:
\[ \left[ P_{aγ, \text{natural}} \right] = \frac{\si{T^2.m^2}}{J^2} = \frac{1}{\si{A^2.m^2}} \]
where we use the fact that \(g_{aγ}\) has units of \(\si{GeV^{-1}}\) which is equivalent to units of \(\si{J^{-1}}\) (care has to be taken with the rest of the conversion factors of course!) and Tesla in SI units:
\[ \left[ B \right] = \si{T} = \frac{\si{kg}}{\si{s^2.A}} \]
From the appearance of \(\si{A^2}\) in the units of \(P_{aγ, \text{natural}}\) we know a factor of \(ε_0\) is missing. This leaves the question of the correct powers of \(\hbar\) and \(c\), which come out to:
\begin{align*} \left[ ε_0 \hbar c^3 \right] &= \frac{\si{A^2.s^4}}{\si{kg.m^3}} \frac{\si{kg.m^2}}{\si{s}} \frac{\si{m^3}}{\si{s^3}} \\ &= \si{A^2.m^2}. \end{align*}So the correct expression in SI units is:
\[ P_{aγ} = ε_0 \hbar c^3 \left( \frac{g_{aγ} B L}{2} \right)^2 \]
where now only \(g_{aγ}\) needs to be expressed in units of \(\si{J^{-1}}\) for a correct result using tesla and meter.
3.3. Gaseous detector physics
I have a big confusion.
In the Bethe equation there is the factor I, the mean excitation
energy. It is roughly \(I(Z) = 10 Z\), where \(Z\) is the charge of the
element.
To determine the number of primary electrons however we have the distinction between:
- the actual excitation energy of the element / the molecules, e.g. ~15 eV for Argon gas
- the "average ionization energy per ion" \(w\), which is the well known 26 eV for Argon gas
- where does the difference between \(I\) and \(w\) come from? What does one mean vs. the other? They are different by a factor of 10 after all!
- why the large distinction between excitation energy and average energy per ion? Is it only because of rotational / vibrational modes of the molecules?
Relevant references:
- PDG chapter 33 (Bethe, losses) and 34 (Gaseous detector)
- Mean excitation energies for the stopping power of atoms and molecules evaluated from oscillator-strength spectra https://aip.scitation.org/doi/10.1063/1.2345478 about ionization energy I
- A method to improve tracking and particle identification in TPCs and silicon detectors https://doi.org/10.1016/j.nima.2006.03.009 About more correct losses in gases
This is all very confusing.
3.3.1. Average distance X-rays travel in Argon at CAST conditions [/]
In order to be able to compute the correct distance to use in the raytracer for the position of the axion image, we need a good understanding of where the average X-ray will convert in the gas.
By combining the expected axion flux (or rather that folded with the telescope and window transmission to get the correct energy distribution) with the absorption length of X-rays at different energies we can compute a weighted mean of all X-rays and come up with a single number.
For that reason we wrote xrayAttenuation.
Let's give it a try.
- Analytical approach
import xrayAttenuation, ggplotnim, unchained # 1. read the file containing efficiencies var effDf = readCsv("/home/basti/org/resources/combined_detector_efficiencies.csv") .mutate(f{"NoGasEff" ~ idx("300nm SiN") * idx("20nm Al") * `LLNL`}) # 2. compute the absorption length for Argon let ar = Argon.init() let ρ_Ar = density(1050.mbar.to(Pascal), 293.K, ar.molarMass) effDf = effDf .filter(f{idx("Energy [keV]") > 0.05}) .mutate(f{float: "l_abs" ~ absorptionLength(ar, ρ_Ar, idx("Energy [keV]").keV).float}) # compute the weighted mean of the effective flux behind the window with the # absorption length, i.e. # `<x> = Σ_i (ω_i x_i) / Σ_i ω_i` let weightedMean = (effDf["NoGasEff", float] *. effDf["l_abs", float]).sum() / effDf["NoGasEff", float].sum() echo "Weighted mean of distance: ", weightedMean.Meter.to(cm) # for reference the effective flux: ggplot(effDf, aes("Energy [keV]", "NoGasEff")) + geom_line() + ggsave("/tmp/combined_efficiency_no_gas.pdf") ggplot(effDf, aes("Energy [keV]", "l_abs")) + geom_line() + ggsave("/tmp/absorption_length_argon_cast.pdf")
This means the "effective" position of the axion image should be 0.0122 m or 1.22 cm in the detector. This is (fortunately) relatively close to the 1.5 cm (center of the detector) that we used so far.
[X]Is the above even correct? The absorption length describes the distance at which only 1/e particles are left. That means at that distance (1 - 1/e) have disappeared. To get a number don't we need to do a monte carlo (or some kind of integral) of the average? -> Well, the mean of an exponential distribution is 1/λ (if defined as \(\exp(-λx)\)!), from that point of view I think the above is perfectly adequate! Note however that the median of the distribution is \(\frac{\ln 2}{λ}\)! When looking at the distribution of our transverse RMS values for example the peak corresponds to something that is closer to the median (but is not exactly the median either; the peak is the 'mode' of the distribution). Arguably more interesting is the cutoff we see in the data as that corresponds to the largest possible diffusion (but again that is being folded with the statistics of getting a larger RMS! :/ )
UPDATE: See the section below for the numerical approach. As it turns out the above unfortunately is not correct for 3 important reasons (2 of which we were aware of):
- It does not include the axion spectrum, it changes the location of the mean slightly.
- It implicitly assumes all X-rays of all energies will be detected. This implies an infinitely long detector and not our detector limited by a length of 3 cm! This skews the actual mean to lower values, because the mean of those that are detected are at smaller values.
- Point 2 implies not only that some X-rays won't be detected, but effectively it gives a higher weight to energies that are absorbed with certainty compared to those that sometimes are not absorbed! This further reduces the mean. This can be interpreted as reducing the input flux by the percentage of the absorption probability for each energy. In this sense the above needs to be multiplied by the absorption probability to be more correct! Yet this still does not make it completely right, as that just assumes the fraction of photons of a given energy are reduced, but not that all detected ones have lengths consistent with a 3cm long volume!
- (minor) does not include isobutane.
A (shortened and) improved version of the above (but still not quite correct!):
import xrayAttenuation, ggplotnim, unchained # 1. read the file containing efficiencies var effDf = readCsv("/home/basti/org/resources/combined_detector_efficiencies.csv") .mutate(f{"NoGasEff" ~ idx("300nm SiN") * idx("20nm Al") * `LLNL` * idx("30mm Ar Abs.")}) # 2. compute the absorption length for Argon let ar = Argon.init() let ρ_Ar = density(1050.mbar.to(Pascal), 293.K, ar.molarMass) effDf = effDf.filter(f{idx("Energy [keV]") > 0.05}) .mutate(f{float: "l_abs" ~ absorptionLength(ar, ρ_Ar, idx("Energy [keV]").keV).float}) let weightedMean = (effDf["NoGasEff", float] *. effDf["l_abs", float]).sum() / effDf["NoGasEff", float].sum() echo "Weighted mean of distance: ", weightedMean.Meter.to(cm)
We could further multiply in the axion flux of course, but as this cannot be fully correct in this way, we'll do it numerically. We would have to calculate the real mean of the exponential distribution for each energy based on the truncated exponential distribution. Effectively we have a bonded exponential between 0 and 3 cm, whose mean is of course going to differ from the parameter \(λ\).
- Numerical approach
Let's write a version of the above code that computes the result by sampling from the exponential distribution for the conversion point.
What we need:
- our sampling logic
- sampling from exponential distribution depending on energy
- the axion flux
Let's start by importing the modules we need:
import helpers / sampling_helper # sampling distributions import unchained # sane units import ggplotnim # see something! import xrayAttenuation # window efficiencies import math, sequtils
where the
sampling_helpersis a small module to sample from a procedure or a sequence.In addition let's define some helpers:
from os import `/` const ResourcePath = "/home/basti/org/resources" const OutputPath = "/home/basti/org/Figs/statusAndProgress/axion_conversion_point_sampling/"
Now let's read the LLNL telescope efficiency as well as the axion flux model. Note that we may wish to calculate the absorption points not only for a specific axion flux model, but potentially any other kind of signal. We'll build in functionality to disable different contributions.
let dfAx = readCsv(ResourcePath / "solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15.csv") .filter(f{`type` == "Total flux"}) let dfLLNL = readCsv(ResourcePath / "llnl_xray_telescope_cast_effective_area_parallel_light_DTU_thesis.csv") .mutate(f{"Efficiency" ~ idx("EffectiveArea[cm²]") / (PI * 2.15 * 2.15)})
Note: to get the differential axion flux use
readOpacityFilefrom https://github.com/jovoy/AxionElectronLimit. It generates the CSV file.Next up we need to define the material properties of the detector window in order to compute its transmission.
let Si₃N₄ = compound((Si, 3), (N, 4)) # actual window const ρSiN = 3.44.g•cm⁻³ const lSiN = 300.nm # window thickness let Al = Aluminium.init() # aluminium coating const ρAl = 2.7.g•cm⁻³ const lAl = 20.nm # coating thickness
With these numbers we can compute the transmission at an arbitrary energy. In order to compute the correct inputs for the calculation we now have everything. We wish to compute the following, the intensity \(I(E)\) is the flux that enters the detector
\[ I(E) = f(E) · ε_{\text{LLNL}} · ε_{\ce{Si3.N4}} · ε_{\ce{Al}} \]
where \(f(E)\) is the solar axion flux and the \(ε_i\) are the efficiencies associated with the telescope and transmission of the window. The idea is to sample from this intensity distribution to get a realistic set of X-rays as they would be experienced in the experiment. One technical aspect still to be done is an interpolation of the axion flux and LLNL telescope efficiency to evaluate the data at an arbitrary energy as to define a function that yields \(I(E)\).
Important note: We fully neglect here the conversion probability and area of the magnet bore. These (as well as a potential time component) are purely constants and do not affect the shape of the distribution \(I(E)\). We want to sample from it to get the correct weighting of the different energies, but do not care about absolute numbers. So differential fluxes are fine.
The idea is to define the interpolators and then create a procedure that captures the previously defined properties and interpolators.
from numericalnim import newLinear1D, eval let axInterp = newLinear1D(dfAx["Energy", float].toSeq1D, dfAx["diffFlux", float].toSeq1D) let llnlInterp = newLinear1D(dfLLNL["Energy[keV]", float].toSeq1D, dfLLNL["Efficiency", float].toSeq1D)
With the interpolators defined let's write the implementation for \(I(E)\):
proc I(E: keV): float = ## Compute the intensity of the axion flux after telescope & window eff. ## ## Axion flux and LLNL efficiency can be disabled by compiling with ## `-d:noAxionFlux` and `-d:noLLNL`, respectively. result = transmission(Si₃N₄, ρSiN, lSiN, E) * transmission(Al, ρAl, lAl, E) when not defined(noAxionFlux): result *= axInterp.eval(E.float) when not defined(noLLNL): result *= llnlInterp.eval(E.float)
Let's test it and see what we get for e.g. \(\SI{1}{keV}\):
echo I(1.keV)
yields \(1.249e20\). Not the most insightful, but it seems to work. Let's plot it:
let energies = linspace(0.01, 10.0, 1000).mapIt(it.keV) let Is = energies.mapIt(I(it)) block PlotI: let df = toDf({ "E [keV]" : energies.mapIt(it.float), "I" : Is }) ggplot(df, aes("E [keV]", "I")) + geom_line() + ggtitle("Intensity entering the detector gas") + ggsave(OutputPath / "intensity_axion_conversion_point_simulation.pdf")
shown in fig. 20. It looks exactly as we would expect.
Figure 20: Intensity that enters the detector taking into account LLNL telescope and window efficiencies as well as the solar axion flux Now we define the sampler for the intensity distribution \(I(E)\), which returns an energy weighted by \(I(E)\):
let Isampler = sampler( (proc(x: float): float = I(x.keV)), # wrap `I(E)` to take `float` 0.01, 10.0, num = 1000 # use 1000 points for EDF & sample in 0.01 to 10 keV )
and define a random number generator:
import random var rnd = initRand(0x42)
First we will sample 100,000 energies from the distribution to see if we recover the intensity plot from before.
block ISampled: const nmc = 100_000 let df = toDf( {"E [keV]" : toSeq(0 ..< nmc).mapIt(rnd.sample(Isampler)) }) ggplot(df, aes("E [keV]")) + geom_histogram(bins = 200, hdKind = hdOutline) + ggtitle("Energies sampled from I(E)") + ggsave(OutputPath / "energies_intensity_sampled.pdf")
This yields fig. 21, which clearly shows the sampling works as intended.
Figure 21: Energies sampled from the distribution \(I(E)\) using 100k samples. The shape is nicely reproduced, here plotted using a histogram of 200 bins. The final piece now is to use the same sampling logic to generate energies according to \(I(E)\), which correspond to X-rays of said energy entering the detector. For each of these energies then sample from the Beer-Lambert law
\[ I(z) = I_0 \exp\left[ - \frac{z}{l_{\text{abs}} } \right] \] where \(I_0\) is some initial intensity and \(l_\text{abs}\) the absorption length. The absorption length is computed from the gas mixture properties of the gas used at CAST, namely Argon/Isobutane 97.7/2.3 at \(\SI{1050}{mbar}\). It is the inverse of the attenuation coefficient \(μ_M\)
\[ l_{\text{abs}} = \frac{1}{μ_M} \]
where the attenuation coefficient is computed via
\[ μ_m = \frac{N_A}{M * σ_A} \]
with \(N_A\) Avogadro's constant, \(M\) the molar mass of the compound and \(σ_A\) the atomic absorption cross section. The latter again is defined by
\[ σ_A = 2 r_e λ f₂ \]
with \(r_e\) the classical electron radius, \(λ\) the wavelength of the X-ray and \(f₂\) the second scattering factor. Scattering factors are tabulated for different elements, for example by NIST and Henke. For a further discussion of this see the README and implementation of
xrayAttenuation.We will now go ahead and define the CAST gas mixture:
proc initCASTGasMixture(): GasMixture = ## Returns the absorption length for the given energy in keV for CAST ## gas conditions: ## - Argon / Isobutane 97.7 / 2.3 % ## - 20°C ( for this difference in temperature barely matters) let arC = compound((Ar, 1)) # need Argon gas as a Compound let isobutane = compound((C, 4), (H, 10)) # define the gas mixture result = initGasMixture(293.K, 1050.mbar, [(arC, 0.977), (isobutane, 0.023)]) let gm = initCASTGasMixture()
To sample from the Beer-Lambert law with a given absorption length we also define a helper that returns a sampler for the target energy using the definition of a normalized exponential distribution
\[ f_e(x, λ) = \frac{1}{λ} \exp \left[ -\frac{x}{λ} \right] \]
The sampling of the conversion point is the crucial aspect of this. Naively we might want to sample between the detector volume from 0 to \(\SI{3}{cm}\). However, this skews our result. Our calculation depends on the energy distribution of the incoming X-rays. If the absorption length is long enough the probability of reaching the readout plane and thus not being detected is significant. Restricting the sampler to \(\SI{3}{cm}\) would pretend that independent of absorption length we would always convert within the volume, giving too large a weight to the energies that should sometimes not be detected!
Let's define the sampler now. It takes the gas mixture and the target energy. A constant
SampleTois defined to adjust the position to which we sample at compile time (to play around with different numbers).proc generateSampler(gm: GasMixture, targetEnergy: keV): Sampler = ## Generate the exponential distribution to sample from based on the ## given absorption length # `xrayAttenuation` `absorptionLength` returns number in meter! let λ = absorptionLength(gm, targetEnergy).to(cm) let fnSample = (proc(x: float): float = result = expFn(x, λ.float) # expFn = 1/λ · exp(-x/λ) ) const SampleTo {.intdefine.} = 20 ## `SampleTo` can be set via `-d:SampleTo=<int>` let num = (SampleTo.float / 3.0 * 1000).round.int # number of points to sample at result = sampler(fnSample, 0.0, SampleTo, num = num)
Note that this is inefficient, because we generate a new sampler from which we only sample a single point, namely the conversion point of that X-ray. If one intended to perform a more complex calculation or wanted to sample orders of magnitude more X-rays, one should either restructure the code (i.e. sample from known energies and then reorder based on the weight defined by \(I(E)\) or cache the samplers and pre-bin the energies.
For reference let's compute the absorption length as a function of energy for the CAST gas mixture:
block GasAbs: let df = toDf({ "E [keV]" : linspace(0.03, 10.0, 1000), "l_abs [cm]" : linspace(0.03, 10.0, 1000).mapIt(absorptionLength(gm, it.keV).m.to(cm).float) }) ggplot(df, aes("E [keV]", "l_abs [cm]")) + geom_line() + ggtitle("Absorption length of X-rays in CAST gas mixture: " & $gm) + margin(top = 1.5) + ggsave(OutputPath / "cast_gas_absorption_length.pdf")
which yields fig. 22
Figure 22: Absorption length in the CAST gas mixture as a function of X-ray energy. So, finally: let's write the MC sampling!
const nmc = 500_000 # start with 100k samples var Es = newSeqOfCap[keV](nmc) var zs = newSeqOfCap[cm](nmc) while zs.len < nmc: # 1. sample an energy according to `I(E)` let E = rnd.sample(Isampler).keV # 2. get the sampler for this energy let distSampler = generateSampler(gm, E) # 3. sample from it var z = Inf.cm when defined(Equiv3cmSampling): ## To get the same result as directly sampling ## only up to 3 cm use the following code while z > 3.0.cm: z = rnd.sample(distSampler).cm elif defined(UnboundedVolume): ## This branch pretends the detection volume ## is unbounded if we sample within 20cm z = rnd.sample(distSampler).cm else: ## This branch is the physically correct one. If an X-ray reaches the ## readout plane it is _not_ recorded, but it was still part of the ## incoming flux! z = rnd.sample(distSampler).cm if z > 3.0.cm: continue # just drop this X-ray zs.add z Es.add E
Great, now we have sampled the conversion points according to the correct intensity. We can now ask for statistics or create different plots (e.g. conversion point by energies etc.).
import stats, seqmath # mean, variance and percentile let zsF = zs.mapIt(it.float) # for math echo "Mean conversion position = ", zsF.mean().cm echo "Median conversion position = ", zsF.percentile(50).cm echo "Variance of conversion position = ", zsF.variance().cm
This prints the following:
Mean conversion position = 0.556813 cm Median conversion position = 0.292802 cm Variance of conversion position = 0.424726 cm
As we can see (unfortunately) our initial assumption of a mean distance of \(\SI{1.22}{cm}\) are quite of the mark. The more realistic number is only \(\SI{0.56}{cm}\). And if we were to use the median it's only \(\SI{0.29}{cm}\).
Let's plot the conversion points of all sampled (and recorded!) X-rays as well as what their distribution against energy looks like.
let dfZ = toDf({ "E [keV]" : Es.mapIt(it.float), "z [cm]" : zs.mapIt(it.float) }) ggplot(dfZ, aes("z [cm]")) + geom_histogram(bins = 200, hdKind = hdOutline) + ggtitle("Conversion points of all sampled X-rays according to I(E)") + ggsave(OutputPath / "sampled_axion_conversion_points.pdf") ggplot(dfZ, aes("E [keV]", "z [cm]")) + geom_point(size = 1.0, alpha = 0.2) + ggtitle("Conversion points of all sampled X-rays according to I(E) against their energy") + ggsave(OutputPath / "sampled_axion_conversion_points_vs_energy.png", width = 1200, height = 800)
The former is shown in fig. 23. The overlapping exponential distribution is obvious, as one would expect. The same data is shown in fig. 24, but in this case not as a histogram, but by their energy as a scatter plot. We can clearly see the impact of the absorption length on the conversion points for each energy!
Figure 23: Distribution of the conversion points of all sampled X-rays for which conversion in the detector took place as sampled from \(I(E)\). 
Figure 24: Distribution of the conversion points of all sampled X-rays for which conversion in the detector took place as sampled from \(I(E)\) as a scatter plot against the energy for each X-ray. - Compiling and running the code
The code above is written in literate programming style. To compile and run it we use
ntangleto extract it from the Org file:ntangle <file>
which generates ./../../../../tmp/sample_axion_xrays_conversion_points.nim.
Compiling and running it can be done via:
nim r -d:danger /tmp/sample_axion_xrays_conversion_points.nim
which compiles and runs it as an optimized build.
We have the following compilation flags to compute different cases:
-d:noLLNL: do not include the LLNL efficiency into the input intensity-d:noAxionFlux: do not include the axion flux into the input intensity-d:SampleTo=<int>: change to where we sample the position (only to 3cm for example)-d:UnboundedVolume: if used together with the defaultSampleTo(or any large value) will effectively compute the case of an unbounded detection volume (i.e. every X-ray recorded with 100% certainty).-d:Equiv3cmSampling: Running this with the defaultSampleTo(or any large value) will effectively change the sampling to a maximum \SI{3}{cm} sampling. This can be used as a good crossheck to verify the sampling behavior is independent of the sampling range.
Configurations of note:
nim r -d:danger -d:noAxionFlux /tmp/sample_axion_xrays_conversion_points.nim
\(⇒\) realistic case for a flat input spectrum Yields:
Mean conversion position = 0.712102 cm Median conversion position = 0.445233 cm Variance of conversion position = 0.528094 cm
nim r -d:danger -d:noAxionFlux -d:UnboundedVolume /tmp/sample_axion_xrays_conversion_points.nim\(⇒\) the closest analogue to the analytical calculation from section 3.3.1.1 (outside of including isobutane here) Yields:
Mean conversion position = 1.25789 cm Median conversion position = 0.560379 cm Variance of conversion position = 3.63818 cm
nim r -d:danger /tmp/sample_axion_xrays_conversion_points.nim
\(⇒\) the case we most care about and of which the numbers are mentioned in the text above.
- Absorption edge in data
Question:
[X]Can we see the absorption edge of Argon in our data? E.g. in the transverse RMS of the CDL data? In theory we should see a huge jump in the transverse nature (and cluster size) of the clusters above and below that point. MAYBE this could also relate to the strong cutoff we see in our background rate at 3 keV due to some effect of the efficiency of our cuts changing significantly there?
If my "theory" is correct it would mean that the transverse RMS should be significantly different if I cut to the energy for e.g. the photo peak and escape peak?
Update : As explained in multiple places since the above two TODOs were written. It's not as straightforward, because the exponential distribution still implies that a large fraction of events convert close to the cathode. The result is a smoothed out distribution of the RMS data, making the difference between escape and photo peak for example not as extreme as one might imagine. See the simulations below and the related FADC rise time simulations for more insight.
3.3.2. Simulating longitudinal and transverse cluster sizes using MC
Sample from distribution:
import std / [random, sequtils, algorithm] import seqmath, ggplotnim template toEDF(data: seq[float], isCumSum = false): untyped = ## Computes the EDF of binned data var dataCdf = data if not isCumSum: seqmath.cumsum(dataCdf) let integral = dataCdf[^1] let baseline = min(data) # 0.0 dataCdf.mapIt((it - baseline) / (integral - baseline)) proc sample(cdf: seq[float], ys: seq[float]): float = let point = rand(1.0) let idx = cdf.lowerBound(point) if idx < cdf.len: result = ys[idx] else: result = Inf proc expFn(x: float, λ: float): float = result = 1.0 / λ * exp(- x / λ) const Upper = 3.0 const λ = 2.0 let xs = linspace(0.0, Upper, 1000) let ys = xs.mapIt(expFn(it, λ)) # now sample 100,000 points let cdf = ys.toEdf() let ySampled = toSeq(0 ..< 1_000_000).mapIt(sample(cdf, xs)) let dfS = toDf(ySampled) ggplot(toDf(xs, cdf), aes("xs", "cdf")) + geom_line() + ggsave("/t/test_cdf.pdf") echo dfS # rescale according to normalization of the range we use # normalize by y = y / (∫_Lower^Upper f(x) dx) = # Lower = 0, Upper = 3.0 (`Upper`) # y = y / (∫_0^Upper 1/λ exp(-x/λ) dx = y / [ ( -exp(-x/λ) )|^Upper_0 ] # y = y / [ (-exp(-Upper/λ) - (-exp(-Lower/λ) ) ] # y = y / [ (-exp(-3.0/λ)) + 1 ] ^--- 1 = exp(0) let df = toDf(xs, ys) .mutate(f{"ys" ~ `ys` / (-exp(-Upper / λ) + 1.0)}) ggplot(df, aes("xs")) + geom_line(aes = aes(y = "ys")) + geom_histogram(data = dfS, aes = aes(x = "ySampled"), bins = 100, density = true, alpha = 0.5, hdKind = hdOutline, fillColor = "red") + ggsave("/t/test_sample.pdf")
The below is also in: ./../../CastData/ExternCode/TimepixAnalysis/NimUtil/helpers/sampling_helper.nim
import std / [random, sequtils, algorithm] import seqmath, ggplotnim template toEDF*(data: seq[float], isCumSum = false): untyped = ## Computes the EDF of binned data var dataCdf = data if not isCumSum: seqmath.cumsum(dataCdf) let integral = dataCdf[^1] ## XXX: why min? let baseline = min(data) # 0.0 dataCdf.mapIt((it - baseline) / (integral - baseline)) proc sample*(cdf: seq[float], ys: seq[float]): float = let point = rand(1.0) let idx = cdf.lowerBound(point) if idx < cdf.len: result = ys[idx] else: result = Inf proc expFn(x: float, λ: float): float = result = 1.0 / λ * exp(- x / λ) proc sampleFrom*(fn: proc(x: float): float, low, high: float, num = 1000, samples = 1_000_000): seq[float] = ## Note: it may be useful to hand a closure with wrapped arguments! let xs = linspace(low, high, num) let ys = xs.mapIt( fn(it) ) # now sample 100,000 points let cdf = ys.toEdf() result = toSeq(0 ..< samples).mapIt(sample(cdf, xs)) when isMainModule: ## Mini test: Compare with plot output from /tmp/test_sample.nim! let λ = 2.0 let fnSample = (proc(x: float): float = result = expFn(x, λ) ) let ySampled = sampleFrom(fnSample, 0.0, 3.0) let ySampled2 = sampleFrom(fnSample, 0.0, 10.0) proc toHisto(xs: seq[float]): DataFrame = const binSize = 0.1 let binNum = ((xs.max - xs.min) / binSize).round.int let (hist, bins) = histogram(xs, binNum) let maxH = hist.max result = toDf({"x" : bins[0 ..< ^2], "y" : hist.mapIt(it / maxH)}) let dfC = bind_rows([("1", ySampled.toHisto()), ("2", ySampled2.toHisto())], "val") ggplot(dfC, aes("x", "y", fill = "val")) + #geom_histogram(bins = 100, density = true, alpha = 0.5, hdKind = hdOutline, fillColor = "red") + geom_histogram(bins = 100, alpha = 0.5, hdKind = hdOutline, stat = "identity", position = "identity") + ggsave("/t/test_sample_from.pdf")
Now use that to sample from our exponential to determine typical conversion points of X-rays. The exponential decay according to the Lambert-Beer (attenuation) law tells us something about the inverse decay likelihood?
Effectively it's the same as radioactive decay, where for each distance in the medium it is a Poisson process depending on the elements still present.
So the idea is to MC N samples that enter the cathode. At each step Δx we sample the Poisson process to find the likelihood of a decay. If it stays, cool. If not, its position is added to our decay (or in this case photoelectron origin) positions.
The result of that is precisely the percentage of the exponential distribution of course! This means we can use the exponential distribution as the starting point for our sampling of the diffusion for each event. We sample from the exponential, get a position each time where a particle may have converted, then based on that position we compute a target size, which we do by drawing from a normal distribution centered around the longitudinal / transverse diffusion coefficients, as these after all represent the 1σ sizes of the diffusion. So in effect what we're actually just computing is the exponential distribution of our data folded with a normal distribution. In theory we could just compute that somehow.
import std / [random, sequtils, algorithm, strformat] import seqmath, ggplotnim, unchained import /tmp/sampling_helper proc expFn(x: float, λ: float): float = result = 1.0 / λ * exp(- x / λ) proc main(λ: float) = let σT = 640.0 # μm·√cm let fnSample = (proc(x: float): float = result = expFn(x, λ) ) proc rmsTrans(x: float): float = let toDrift = (3.0 - x) result = sqrt(toDrift) * σT # sample from our exponential distribution describing absorption let ySampled = sampleFrom(fnSample, 0.0, 3.0) # now compute the long and trans RMS for each let yRmsTrans = ySampled.mapIt(rmsTrans(it)) ggplot(toDf(yRmsTrans), aes("yRmsTrans")) + geom_histogram(bins = 100, density = true, alpha = 0.5, hdKind = hdOutline, fillColor = "red") + ggsave(&"/t/sample_transverse_rms_{λ}_cm_absorption_length.pdf") #let sampleTransFn = (proc(x: float): float = # result = gaus(x = x, mean = σT, when isMainModule: import cligen dispatch main
The above already produces quite decent results in terms of the transverse RMS for known absorption lengths!
basti at voidRipper in /t λ ./simulate_rms_transverse_simple --λ 3.0 # below Ar absorption edge basti at voidRipper in /t λ ./simulate_rms_transverse_simple --λ 2.2 # 5.9 keV basti at voidRipper in /t λ ./simulate_rms_transverse_simple --λ 0.5 # 3.x keV above Ar absorption edge
yields:
These need to be compared to equivalent plots from CAST / CDL data.
- CAST 5.9 keV (Photo):
- CAST 3.0 keV (Escape):
- CDL C-EPIC-0.6 (~250 eV, extremely low λ):
- CDL Ag-Ag-6kV (3 keV, λ > 3cm):
- CDL Ti-Ti-9kV (4.5 keV, λ ~ 1cm):
- CDL Mn-Cr-12kV (5.9 keV, λ ~ 2.2cm):
For all the plots:
cd /tmp/ mkdir RmsTransversePlots && cd RmsTransversePlots
For the CAST plots:
plotData --h5file ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --runType rtCalibration \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --ingrid \ --cuts '("rmsTransverse", 0.1, 1.5)' \ --cuts '("energyFromCharge", 2.5, 3.2)' \ --applyAllCuts \ --region crSilver
plotData --h5file ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --runType rtCalibration \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --ingrid \ --cuts '("rmsTransverse", 0.1, 1.5)' \ --cuts '("energyFromCharge", 5.5, 6.5)' \ --applyAllCuts \ --region crSilver
For the CDL plots:
cdl_spectrum_creation -i ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 --dumpAccurate --hideNloptFit
yields the plots initially in
/tmp/RmsTransversePlots/out/CDL_2019_Raw_<SomeDate>
The main takeaway from these plots is: Especially for the cases with longer absorption the shape actually matches quite nicely already! Of course the hard cutoff in the simulation is not present in the real data, which makes sense (we use the same transverse value only dependent on the height; max height = max value). However, for the C-EPIC & the 0.5 absorption length the differences are quite big. Likely because the diffusion is not actually fixed, but itself follows some kind of normal distribution around the mean value. That latter at least is what we will implement now, using the width of the somewhat gaussian distribution of the C-EPIC 0.6kV data as a reference.
The next code snippet does exactly that, it adds sampling from a normal distribution with mean of the transverse diffusion and a width described roughly by the width from the C-EPIC 0.6kV data above so that each sample is spread somewhat.
import std / [random, sequtils, algorithm, strformat] import seqmath, ggplotnim, unchained import /tmp/sampling_helper proc expFn(x: float, λ: float): float = result = 1.0 / λ * exp(- x / λ) import random / mersenne import alea / [core, rng, gauss] proc main(E = 5.9, λ = 0.0) = ## Introduce sampling of a gaussian around σT with something like this ## which is ~150 = 1σ for a √3cm drift (seen in C-EPIC 0.6 kV CDL line ## rmsTransverse data) ## Note: another number we have for ΔσT is of course the simulation error ## on σT, but I suspect that's not a good idea (also it's large, but still ## much smaller than this). let ΔσT = 86.0 # / 2.0 ## XXX: Implement calculation of absorption length from `xrayAttenuation` # let dfAbs = ## XXX: Implement extraction of diffusion values from data: let dfGas = readCsv("/home/basti/org/resources/ar_iso_97_7_2_3_septemboard_cast_different_temps.csv") let σT = 640.0 # μm/√cm let fnSample = (proc(x: float): float = result = expFn(x, λ) ) var rnd = wrap(initMersenneTwister(1337)) var gaus = gaussian(0.0, 1.0) # we will modify this gaussian for every draw! proc rmsTrans(x: float): float = let toDrift = (3.0 - x) # adjust the gaussian to Diffusion = σ_T · √(drift distance) # and width of Sigma = ΔσT · √(drift distance) (at 3 cm we want Δ of 150) gaus.mu = sqrt(toDrift) * σT gaus.sigma = ΔσT * sqrt(toDrift) #echo "DRAWING AROUND: ", gaus.mu, " WITH SIGMA: ", gaus.sigma result = rnd.sample(gaus) # sample from our exponential distribution describing absorption let ySampled = sampleFrom(fnSample, 0.0, 3.0) # now compute the long and trans RMS for each let yRmsTrans = ySampled.mapIt(rmsTrans(it)) let GoldenMean = (sqrt(5.0) - 1.0) / 2.0 # Aesthetic ratio FigWidth = 1200.0 # width in pixels FigHeight = FigWidth * GoldenMean # height in pixels ggplot(toDf(yRmsTrans), aes("yRmsTrans")) + geom_histogram(bins = 100, density = true, alpha = 0.5, hdKind = hdOutline, fillColor = "red") + ggsave(&"/t/sample_gauss_transverse_rms_{λ}_cm_absorption_length.pdf", width = FigWidth, height = FigHeight) when isMainModule: import cligen dispatch main
Let's generate the same cases we already generated with the simple version before:
basti at voidRipper in /t λ ./simulate_rms_transverse_gauss --λ 3.0 basti at voidRipper in /t λ ./simulate_rms_transverse_gauss --λ 2.2 basti at voidRipper in /t λ ./simulate_rms_transverse_gauss --λ 2.0 basti at voidRipper in /t λ ./simulate_rms_transverse_gauss --λ 1.0 basti at voidRipper in /t λ ./simulate_rms_transverse_gauss --λ 0.5 basti at voidRipper in /t λ ./simulate_rms_transverse_gauss --λ 0.1
First of all we can see that the 0.1 and 0.5 cm absorption length case are almost fully gaussian. The other cases have the typical asymmetric shape we expect.
Let's generate raw CDL plots (from plotData with minimal cuts, esp no
rmsTransverse cut):
For C-EPIC-0.6kV (~250 eV, extremely low λ)
plotData --h5file ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --runType rtCalibration \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --ingrid \ --cuts '("rmsTransverse", 0.1, 1.5)' \ --applyAllCuts \ --runs 342 --runs 343 \ --region crSilver
For Ag-Ag-6kV (3 keV, λ > 3cm):
plotData --h5file ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --runType rtCalibration \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --ingrid \ --cuts '("rmsTransverse", 0.1, 1.5)' \ --applyAllCuts \ --runs 328 --runs 329 --runs 351 \ --region crSilver
For Ti-Ti-9kV (4.5 keV, λ ~ 1cm):
plotData --h5file ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --runType rtCalibration \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --ingrid \ --cuts '("rmsTransverse", 0.1, 1.5)' \ --applyAllCuts \ --runs 328 --runs 329 --runs 351 \ --region crSilver
which are:
where we see that the very short absorption length case of C-EPIC
0.6kV is indeed also almost gaussian and has a long tail even to the
right side. As a matter of fact it seems like it even has a skewness
towards the right instead of left. However, that is likely due to
double hits etc. in the data, which we did not filter out in this
version (compare with the cdl_spectrum_creation version above).
So to summarize comparing these 'raw' plots against our simulation, especially the higher absorption length plots do actually fit quite nicely, if one considers the simplicity of our simulation and the fact that the width of the gaussian we smear with is pretty much just guessed. The differences that are still present are very likely due to all sorts of other reasons that affect the size of the clusters and how our detector resolves them beyond simply assuming the diffusion coefficient is different! This is more of an "effective theory" for the problem, incorporating the real variances that happen at a fixed transverse diffusion by merging them into a variance onto the diffusion itself, which is clearly lacking as a method.
Anything else to do here?
[ ]could simulate the same for the longitudinal case[ ]could simulate expected rise times base don longitudinal data.
3.4. Polarization of X-rays and relation to axions
See the discussion in ./../void_settings.html.
4. General information
4.1. X-ray fluorescence lines
X-Ray Data Booklet Table 1-2. Photon energies, in electron volts, of principal K-, L-, and M-shell emission lines. (from: https://xdb.lbl.gov/Section1/Table_1-2.pdf)
| Z | Element | Kα1 | Kα2 | Kβ1 | Lα1 | Lα2 | Lβ1 | Lβ2 | Lγ1 | Mα1 |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | Li | 54.3 | ||||||||
| 4 | Be | 108.5 | ||||||||
| 5 | B | 183.3 | ||||||||
| 6 | C | 277 | ||||||||
| 7 | N | 392.4 | ||||||||
| 8 | O | 524.9 | ||||||||
| 9 | F | 676.8 | ||||||||
| 10 | Ne | 848.6 | 848.6 | |||||||
| 11 | Na | 1,040.98 | 1,040.98 | 1,071.1 | ||||||
| 12 | Mg | 1,253.60 | 1,253.60 | 1,302.2 | ||||||
| 13 | Al | 1,486.70 | 1,486.27 | 1,557.45 | ||||||
| 14 | Si | 1,739.98 | 1,739.38 | 1,835.94 | ||||||
| 15 | P | 2,013.7 | 2,012.7 | 2,139.1 | ||||||
| 16 | S | 2,307.84 | 2,306.64 | 2,464.04 | ||||||
| 17 | Cl | 2,622.39 | 2,620.78 | 2,815.6 | ||||||
| 18 | Ar | 2,957.70 | 2,955.63 | 3,190.5 | ||||||
| 19 | K | 3,313.8 | 3,311.1 | 3,589.6 | ||||||
| 20 | Ca | 3,691.68 | 3,688.09 | 4,012.7 | 341.3 | 341.3 | 344.9 | |||
| 21 | Sc | 4,090.6 | 4,086.1 | 4,460.5 | 395.4 | 395.4 | 399.6 | |||
| Z | Element | Kα1 | Kα2 | Kβ1 | Lα1 | Lα2 | Lβ1 | Lβ2 | Lγ1 | Mα1 |
| 22 | Ti | 4,510.84 | 4,504.86 | 4,931.81 | 452.2 | 452.2 | 458.4 | |||
| 23 | V | 4,952.20 | 4,944.64 | 5,427.29 | 511.3 | 511.3 | 519.2 | |||
| 24 | Cr | 5,414.72 | 5,405.509 | 5,946.71 | 572.8 | 572.8 | 582.8 | |||
| 25 | Mn | 5,898.75 | 5,887.65 | 6,490.45 | 637.4 | 637.4 | 648.8 | |||
| 26 | Fe | 6,403.84 | 6,390.84 | 7,057.98 | 705.0 | 705.0 | 718.5 | |||
| 27 | Co | 6,930.32 | 6,915.30 | 7,649.43 | 776.2 | 776.2 | 791.4 | |||
| 28 | Ni | 7,478.15 | 7,460.89 | 8,264.66 | 851.5 | 851.5 | 868.8 | |||
| 29 | Cu | 8,047.78 | 8,027.83 | 8,905.29 | 929.7 | 929.7 | 949.8 | |||
| 30 | Zn | 8,638.86 | 8,615.78 | 9,572.0 | 1,011.7 | 1,011.7 | 1,034.7 | |||
| 31 | Ga | 9,251.74 | 9,224.82 | 10,264.2 | 1,097.92 | 1,097.92 | 1,124.8 | |||
| 32 | Ge | 9,886.42 | 9,855.32 | 10,982.1 | 1,188.00 | 1,188.00 | 1,218.5 | |||
| 33 | As | 10,543.72 | 10,507.99 | 11,726.2 | 1,282.0 | 1,282.0 | 1,317.0 | |||
| 34 | Se | 11,222.4 | 11,181.4 | 12,495.9 | 1,379.10 | 1,379.10 | 1,419.23 | |||
| 35 | Br | 11,924.2 | 11,877.6 | 13,291.4 | 1,480.43 | 1,480.43 | 1,525.90 | |||
| 36 | Kr | 12,649 | 12,598 | 14,112 | 1,586.0 | 1,586.0 | 1,636.6 | |||
| 37 | Rb | 13,395.3 | 13,335.8 | 14,961.3 | 1,694.13 | 1,692.56 | 1,752.17 | |||
| 38 | Sr | 14,165 | 14,097.9 | 15,835.7 | 1,806.56 | 1,804.74 | 1,871.72 | |||
| 39 | Y | 14,958.4 | 14,882.9 | 16,737.8 | 1,922.56 | 1,920.47 | 1,995.84 | |||
| 40 | Zr | 15,775.1 | 15,690.9 | 17,667.8 | 2,042.36 | 2,039.9 | 2,124.4 | 2,219.4 | 2,302.7 | |
| 41 | Nb | 16,615.1 | 16,521.0 | 18,622.5 | 2,165.89 | 2,163.0 | 2,257.4 | 2,367.0 | 2,461.8 | |
| 42 | Mo | 17,479.34 | 17,374.3 | 19,608.3 | 2,293.16 | 2,289.85 | 2,394.81 | 2,518.3 | 2,623.5 | |
| 43 | Tc | 18,367.1 | 18,250.8 | 20,619 | 2,424 | 2,420 | 2,538 | 2,674 | 2,792 | |
| 44 | Ru | 19,279.2 | 19,150.4 | 21,656.8 | 2,558.55 | 2,554.31 | 2,683.23 | 2,836.0 | 2,964.5 | |
| 45 | Rh | 20,216.1 | 20,073.7 | 22,723.6 | 2,696.74 | 2,692.05 | 2,834.41 | 3,001.3 | 3,143.8 | |
| 46 | Pd | 21,177.1 | 21,020.1 | 23,818.7 | 2,838.61 | 2,833.29 | 2,990.22 | 3,171.79 | 3,328.7 | |
| 47 | Ag | 22,162.92 | 21,990.3 | 24,942.4 | 2,984.31 | 2,978.21 | 3,150.94 | 3,347.81 | 3,519.59 | |
| 48 | Cd | 23,173.6 | 22,984.1 | 26,095.5 | 3,133.73 | 3,126.91 | 3,316.57 | 3,528.12 | 3,716.86 | |
| 49 | In | 24,209.7 | 24,002.0 | 27,275.9 | 3,286.94 | 3,279.29 | 3,487.21 | 3,713.81 | 3,920.81 | |
| 50 | Sn | 25,271.3 | 25,044.0 | 28,486.0 | 3,443.98 | 3,435.42 | 3,662.80 | 3,904.86 | 4,131.12 | |
| 51 | Sb | 26,359.1 | 26,110.8 | 29,725.6 | 3,604.72 | 3,595.32 | 3,843.57 | 4,100.78 | 4,347.79 | |
| 52 | Te | 27,472.3 | 27,201.7 | 30,995.7 | 3,769.33 | 3,758.8 | 4,029.58 | 4,301.7 | 4,570.9 | |
| 53 | I | 28,612.0 | 28,317.2 | 32,294.7 | 3,937.65 | 3,926.04 | 4,220.72 | 4,507.5 | 4,800.9 | |
| 54 | Xe | 29,779 | 29,458 | 33,624 | 4,109.9 | — | — | — | — | |
| 55 | Cs | 30,972.8 | 30,625.1 | 34,986.9 | 4,286.5 | 4,272.2 | 4,619.8 | 4,935.9 | 5,280.4 | |
| 56 | Ba | 32,193.6 | 31,817.1 | 36,378.2 | 4,466.26 | 4,450.90 | 4,827.53 | 5,156.5 | 5,531.1 | |
| 57 | La | 33,441.8 | 33,034.1 | 37,801.0 | 4,650.97 | 4,634.23 | 5,042.1 | 5,383.5 | 5,788.5 | 833 |
| 58 | Ce | 34,719.7 | 34,278.9 | 39,257.3 | 4,840.2 | 4,823.0 | 5,262.2 | 5,613.4 | 6,052 | 883 |
| 59 | Pr | 36,026.3 | 35,550.2 | 40,748.2 | 5,033.7 | 5,013.5 | 5,488.9 | 5,850 | 6,322.1 | 929 |
| 60 | Nd | 37,361.0 | 36,847.4 | 42,271.3 | 5,230.4 | 5,207.7 | 5,721.6 | 6,089.4 | 6,602.1 | 978 |
| 61 | Pm | 38,724.7 | 38,171.2 | 43,826 | 5,432.5 | 5,407.8 | 5,961 | 6,339 | 6,892 | — |
| 62 | Sm | 40,118.1 | 39,522.4 | 45,413 | 5,636.1 | 5,609.0 | 6,205.1 | 6,586 | 7,178 | 1,081 |
| Z | Element | Kα1 | Kα2 | Kβ1 | Lα1 | Lα2 | Lβ1 | Lβ2 | Lγ1 | Mα1 |
| 63 | Eu | 41,542.2 | 40,901.9 | 47,037.9 | 5,845.7 | 5,816.6 | 6,456.4 | 6,843.2 | 7,480.3 | 1,131 |
| 64 | Gd | 42,996.2 | 42,308.9 | 48,697 | 6,057.2 | 6,025.0 | 6,713.2 | 7,102.8 | 7,785.8 | 1,185 |
| 65 | Tb | 44,481.6 | 43,744.1 | 50,382 | 6,272.8 | 6,238.0 | 6,978 | 7,366.7 | 8,102 | 1,240 |
| 66 | Dy | 45,998.4 | 45,207.8 | 52,119 | 6,495.2 | 6,457.7 | 7,247.7 | 7,635.7 | 8,418.8 | 1,293 |
| 67 | Ho | 47,546.7 | 46,699.7 | 53,877 | 6,719.8 | 6,679.5 | 7,525.3 | 7,911 | 8,747 | 1,348 |
| 68 | Er | 49,127.7 | 48,221.1 | 55,681 | 6,948.7 | 6,905.0 | 7,810.9 | 8,189.0 | 9,089 | 1,406 |
| 69 | Tm | 50,741.6 | 49,772.6 | 57,517 | 7,179.9 | 7,133.1 | 8,101 | 8,468 | 9,426 | 1,462 |
| 70 | Yb | 52,388.9 | 51,354.0 | 59,370 | 7,415.6 | 7,367.3 | 8,401.8 | 8,758.8 | 9,780.1 | 1,521.4 |
| 71 | Lu | 54,069.8 | 52,965.0 | 61,283 | 7,655.5 | 7,604.9 | 8,709.0 | 9,048.9 | 10,143.4 | 1,581.3 |
| 72 | Hf | 55,790.2 | 54,611.4 | 63,234 | 7,899.0 | 7,844.6 | 9,022.7 | 9,347.3 | 10,515.8 | 1,644.6 |
| 73 | Ta | 57,532 | 56,277 | 65,223 | 8,146.1 | 8,087.9 | 9,343.1 | 9,651.8 | 10,895.2 | 1,710 |
| 74 | W | 59,318.24 | 57,981.7 | 67,244.3 | 8,397.6 | 8,335.2 | 9,672.35 | 9,961.5 | 11,285.9 | 1,775.4 |
| 75 | Re | 61,140.3 | 59,717.9 | 69,310 | 8,652.5 | 8,586.2 | 10,010.0 | 10,275.2 | 11,685.4 | 1,842.5 |
| 76 | Os | 63,000.5 | 61,486.7 | 71,413 | 8,911.7 | 8,841.0 | 10,355.3 | 10,598.5 | 12,095.3 | 1,910.2 |
| 77 | Ir | 64,895.6 | 63,286.7 | 73,560.8 | 9,175.1 | 9,099.5 | 10,708.3 | 10,920.3 | 12,512.6 | 1,979.9 |
| 78 | Pt | 66,832 | 65,112 | 75,748 | 9,442.3 | 9,361.8 | 11,070.7 | 11,250.5 | 12,942.0 | 2,050.5 |
| 79 | Au | 68,803.7 | 66,989.5 | 77,984 | 9,713.3 | 9,628.0 | 11,442.3 | 11,584.7 | 13,381.7 | 2,122.9 |
| 80 | Hg | 70,819 | 68,895 | 80,253 | 9,988.8 | 9,897.6 | 11,822.6 | 11,924.1 | 13,830.1 | 2,195.3 |
| 81 | Tl | 72,871.5 | 70,831.9 | 82,576 | 10,268.5 | 10,172.8 | 12,213.3 | 12,271.5 | 14,291.5 | 2,270.6 |
| 82 | Pb | 74,969.4 | 72,804.2 | 84,936 | 10,551.5 | 10,449.5 | 12,613.7 | 12,622.6 | 14,764.4 | 2,345.5 |
| 83 | Bi | 77,107.9 | 74,814.8 | 87,343 | 10,838.8 | 10,730.91 | 13,023.5 | 12,979.9 | 15,247.7 | 2,422.6 |
| 84 | Po | 79,290 | 76,862 | 89,800 | 11,130.8 | 11,015.8 | 13,447 | 13,340.4 | 15,744 | — |
| 85 | At | 81,520 | 78,950 | 92,300 | 11,426.8 | 11,304.8 | 13,876 | — | 16,251 | — |
| 86 | Rn | 83,780 | 81,070 | 94,870 | 11,727.0 | 11,597.9 | 14,316 | — | 16,770 | — |
| 87 | Fr | 86,100 | 83,230 | 97,470 | 12,031.3 | 11,895.0 | 14,770 | 14,450 | 17,303 | — |
| 88 | Ra | 88,470 | 85,430 | 100,130 | 12,339.7 | 12,196.2 | 15,235.8 | 14,841.4 | 17,849 | — |
| 89 | Ac | 90,884 | 87,670 | 102,850 | 12,652.0 | 12,500.8 | 15,713 | — | 18,408 | — |
| 90 | Th | 93,350 | 89,953 | 105,609 | 12,968.7 | 12,809.6 | 16,202.2 | 15,623.7 | 18,982.5 | 2,996.1 |
| 91 | Pa | 95,868 | 92,287 | 108,427 | 13,290.7 | 13,122.2 | 16,702 | 16,024 | 19,568 | 3,082.3 |
| 92 | U | 98,439 | 94,665 | 111,300 | 13,614.7 | 13,438.8 | 17,220.0 | 16,428.3 | 20,167.1 | 3,170.8 |
| 93 | Np | — | — | — | 13,944.1 | 13,759.7 | 17,750.2 | 16,840.0 | 20,784.8 | — |
| 94 | Pu | — | — | — | 14,278.6 | 14,084.2 | 18,293.7 | 17,255.3 | 21,417.3 | — |
| 95 | Am | — | — | — | 14,617.2 | 14,411.9 | 18,852.0 | 17,676.5 | 22,065.2 | — |
4.2. Atomic binding energies
X-Ray Data Booklet Table 1-1. Electron binding energies, in electron volts, for the elements in their natural forms. https://xdb.lbl.gov/Section1/Table_1-1.pdf
| Z | Element | K 1s | L1 2s | L2 2p1/2 | L3 2p3/2 | M1 3s | M2 3p1/2 | M3 3p3/2 | M4 3d3/2 | M5 3d5/2 | N1 4s | N2 4p1/2 | N3 4p3/2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | H | 13.6 | |||||||||||
| 2 | He | 24.6* | |||||||||||
| 3 | Li | 54.7* | |||||||||||
| 4 | Be | 111.5* | |||||||||||
| 5 | B | 188* | |||||||||||
| 6 | C | 284.2* | |||||||||||
| 7 | N | 409.9* | 37.3* | ||||||||||
| 8 | O | 543.1* | 41.6* | ||||||||||
| 9 | F | 696.7* | |||||||||||
| 10 | Ne | 870.2* | 48.5* | 21.7* | 21.6* | ||||||||
| 11 | Na | 1070.8† | 63.5† | 30.65 | 30.81 | ||||||||
| 12 | Mg | 1303.0† | 88.7 | 49.78 | 49.50 | ||||||||
| 13 | Al | 1559.6 | 117.8 | 72.95 | 72.55 | ||||||||
| 14 | Si | 1839 | 149.7*b | 99.82 | 99.42 | ||||||||
| 15 | P | 2145.5 | 189* | 136* | 135* | ||||||||
| 16 | S | 2472 | 230.9 | 163.6* | 162.5* | ||||||||
| 17 | Cl | 2822.4 | 270* | 202* | 200* | ||||||||
| 18 | Ar | 3205.9* | 326.3* | 250.6† | 248.4* | 29.3* | 15.9* | 15.7* | |||||
| 19 | K | 3608.4* | 378.6* | 297.3* | 294.6* | 34.8* | 18.3* | 18.3* | |||||
| 20 | Ca | 4038.5* | 438.4† | 349.7† | 346.2† | 44.3 | † | 25.4† | 25.4† | ||||
| 21 | Sc | 4492 | 498.0* | 403.6* | 398.7* | 51.1* | 28.3* | 28.3* | |||||
| 22 | Ti | 4966 | 560.9† | 460.2† | 453.8† | 58.7† | 32.6† | 32.6† | |||||
| 23 | V | 5465 | 626.7† | 519.8† | 512.1† | 66.3† | 37.2† | 37.2† | |||||
| 24 | Cr | 5989 | 696.0† | 583.8† | 574.1† | 74.1† | 42.2† | 42.2† | |||||
| 25 | Mn | 6539 | 769.1† | 649.9† | 638.7† | 82.3† | 47.2† | 47.2† | |||||
| 26 | Fe | 7112 | 844.6† | 719.9† | 706.8† | 91.3† | 52.7† | 52.7† | |||||
| 27 | Co | 7709 | 925.1† | 793.2† | 778.1† | 101.0† | 58.9† | 59.9† | |||||
| 28 | Ni | 8333 | 1008.6† | 870.0† | 852.7† | 110.8† | 68.0† | 66.2† | |||||
| 29 | Cu | 8979 | 1096.7† | 952.3† | 932.7 | 122.5† | 77.3† | 75.1† | |||||
| 30 | Zn | 9659 | 1196.2* | 1044.9* | 1021.8* | 139.8* | 91.4* | 88.6* | 10.2* | 10.1* | |||
| 31 | Ga | 10367 | 1299.0*b | 1143.2† | 1116.4† | 159.5† | 103.5† | 100.0† | 18.7† | 18.7† | |||
| 32 | Ge | 11103 | 1414.6*b | 1248.1*b | 1217.0*b | 180.1* | 124.9* | 120.8* | 29.8 | 29.2 | |||
| 33 | As | 11867 | 1527.0*b | 1359.1*b | 1323.6*b | 204.7* | 146.2* | 141.2* | 41.7* | 41.7* | |||
| 34 | Se | 12658 | 1652.0*b | 1474.3*b | 1433.9*b | 229.6* | 166.5* | 160.7* | 55.5* | 54.6* | |||
| 35 | Br | 13474 | 1782* | 1596* | 1550* | 257* | 189* | 182* | 70* | 69* | |||
| 36 | Kr | 14326 | 1921 | 1730.9* | 1678.4* | 292.8* | 222.2* | 214.4 | 95.0* | 93.8* | 27.5* | 14.1* | 14.1* |
| 37 | Rb | 15200 | 2065 | 1864 | 1804 | 326.7* | 248.7* | 239.1* | 113.0* | 112* | 30.5* | 16.3* | 15.3* |
| 38 | Sr | 16105 | 2216 | 2007 | 1940 | 358.7† | 280.3† | 270.0† | 136.0† | 134.2† | 38.9† | 21.3 | 20.1† |
| 39 | Y | 17038 | 2373 | 2156 | 2080 | 392.0*b | 310.6* | 298.8* | 157.7† | 155.8† | 43.8* | 24.4* | 23.1* |
| 40 | Zr | 17998 | 2532 | 2307 | 2223 | 430.3† | 343.5† | 329.8† | 181.1† | 178.8† | 50.6† | 28.5† | 27.1† |
| 41 | Nb | 18986 | 2698 | 2465 | 2371 | 466.6† | 376.1† | 360.6† | 205.0† | 202.3† | 56.4† | 32.6† | 30.8† |
| 42 | Mo | 20000 | 2866 | 2625 | 2520 | 506.3† | 411.6† | 394.0† | 231.1† | 227.9† | 63.2† | 37.6† | 35.5† |
| 43 | Tc | 21044 | 3043 | 2793 | 2677 | 544* | 447.6 | 417.7 | 257.6 | 253.9* | 69.5* | 42.3* | 39.9* |
| 44 | Ru | 22117 | 3224 | 2967 | 2838 | 586.1* | 483.5† | 461.4† | 284.2† | 280.0† | 75.0† | 46.3† | 43.2† |
| 45 | Rh | 23220 | 3412 | 3146 | 3004 | 628.1† | 521.3† | 496.5† | 311.9† | 307.2† | 81.4*b | 50.5† | 47.3† |
| 46 | Pd | 24350 | 3604 | 3330 | 3173 | 671.6† | 559.9† | 532.3† | 340.5† | 335.2† | 87.1*b | 55.7†a | 50.9† |
| 47 | Ag | 25514 | 3806 | 3524 | 3351 | 719.0† | 603.8† | 573.0† | 374.0† | 368.3 | 97.0† | 63.7† | 58.3† |
| Z | Element | K 1s | L1 2s | L2 2p1/2 | L3 2p3/2 | M1 3s | M2 3p1/2 | M3 3p3/2 | M4 3d3/2 | M5 3d5/2 | N 4s | N2 4p1/2 | N3 4p3/2 |
| 48 | Cd | 26711 | 4018 | 3727 | 3538 | 772.0† | 652.6† | 618.4† | 411.9† | 405.2† | 109.8† | 63.9†a | 63.9†a |
| 49 | In | 27940 | 4238 | 3938 | 3730 | 827.2† | 703.2† | 665.3† | 451.4† | 443.9† | 122.9† | 73.5†a | 73.5†a |
| 50 | Sn | 29200 | 4465 | 4156 | 3929 | 884.7† | 756.5† | 714.6† | 493.2† | 484.9† | 137.1† | 83.6†a | 83.6†a |
| 51 | Sb | 30491 | 4698 | 4380 | 4132 | 946† | 812.7† | 766.4† | 537.5† | 528.2† | 153.2† | 95.6†a | 95.6†a |
| 52 | Te | 31814 | 4939 | 4612 | 4341 | 1006† | 870.8† | 820.0† | 583.4† | 573.0† | 169.4† | 103.3†a | 103.3†a |
| 53 | I | 33169 | 5188 | 4852 | 4557 | 1072* | 931* | 875* | 630.8 | 619.3 | 186* | 123* | 123* |
| 54 | Xe | 34561 | 5453 | 5107 | 4786 | 1148.7* | 1002.1* | 940.6* | 689.0* | 676.4* | 213.2* | 146.7 | 145.5* |
| 55 | Cs | 35985 | 5714 | 5359 | 5012 | 1211*b | 1071* | 1003* | 740.5* | 726.6* | 232.3* | 172.4* | 161.3* |
| 56 | Ba | 37441 | 5989 | 5624 | 5247 | 1293*b | 1137*b | 1063*b | 795.7† | 780.5* | 253.5† | 192 | 178.6† |
| 57 | La | 38925 | 6266 | 5891 | 5483 | 1362*b | 1209*b | 1128*b | 853* | 836* | 274.7* | 205.8 | 196.0* |
| 58 | Ce | 40443 | 6549 | 6164 | 5723 | 1436*b | 1274*b | 1187*b | 902.4* | 883.8* | 291.0* | 223.2 | 206.5* |
| 59 | Pr | 41991 | 6835 | 6440 | 5964 | 1511 | 1337 | 1242 | 948.3* | 928.8* | 304.5 | 236.3 | 217.6 |
| 60 | Nd | 43569 | 7126 | 6722 | 6208 | 1575 | 1403 | 1297 | 1003.3* | 980.4* | 319.2* | 243.3 | 224.6 |
| 61 | Pm | 45184 | 7428 | 7013 | 6459 | --- | 1471 | 1357 | 1052 | 1027 | --- | 242 | 242 |
| 62 | Sm | 46834 | 7737 | 7312 | 6716 | 1723 | 1541 | 1420 | 1110.9* | 1083.4* | 347.2* | 265.6 | 247.4 |
| 63 | Eu | 48519 | 8052 | 7617 | 6977 | 1800 | 1614 | 1481 | 1158.6* | 1127.5* | 360 | 284 | 257 |
| 64 | Gd | 50239 | 8376 | 7930 | 7243 | 1881 | 1688 | 1544 | 1221.9* | 1189.6* | 378.6* | 286 | 271 |
| 65 | Tb | 51996 | 8708 | 8252 | 7514 | 1968 | 1768 | 1611 | 1276.9* | 1241.1* | 396.0* | 322.4* | 284.1* |
| 66 | Dy | 53789 | 9046 | 8581 | 7790 | 2047 | 1842 | 1676 | 1333 | 1292.6* | 414.2* | 333.5* | 293.2* |
| 67 | Ho | 55618 | 9394 | 8918 | 8071 | 2128 | 1923 | 1741 | 1392 | 1351 | 432.4* | 343.5 | 308.2* |
| 68 | Er | 57486 | 9751 | 9264 | 8358 | 2207 | 2006 | 1812 | 1453 | 1409 | 449.8* | 366.2 | 320.2* |
| 69 | Tm | 59390 | 10116 | 9617 | 8648 | 2307 | 2090 | 1885 | 1515 | 1468 | 470.9* | 385.9* | 332.6* |
| 70 | Yb | 61332 | 10486 | 9978 | 8944 | 2398 | 2173 | 1950 | 1576 | 1528 | 480.5* | 388.7* | 339.7* |
| Z | Element | N4 4d3/2 | N5 4d5/2 | N6 4f5/2 | N7 4f7/2 | O1 5s | O2 5p1/2 | O3 5p3/2 | O4 5d3/2 | O5 5d5/2 | P1 6s | P2 6p1/2 | P3 6p3/2 |
| 48 | Cd | 11.7† | l0.7† | ||||||||||
| 49 | In | 17.7† | 16.9† | ||||||||||
| 50 | Sn | 24.9† | 23.9† | ||||||||||
| 51 | Sb | 33.3† | 32.1† | ||||||||||
| 52 | Te | 41.9† | 40.4† | ||||||||||
| 53 | I | 50.6 | 48.9 | ||||||||||
| 54 | Xe | 69.5* | 67.5* | --- | --- | 23.3* | 13.4* | 12.1* | |||||
| 55 | Cs | 79.8* | 77.5* | --- | --- | 22.7 | 14.2* | 12.1* | |||||
| 56 | Ba | 92.6† | 89.9† | --- | --- | 30.3† | 17.0† | 14.8† | |||||
| 57 | La | 105.3* | 102.5* | --- | --- | 34.3* | 19.3* | 16.8* | |||||
| 58 | Ce | 109* | --- | 0.1 | 0.1 | 37.8 | 19.8* | 17.0* | |||||
| 59 | Pr | 115.1* | 115.1* | 2.0 | 2.0 | 37.4 | 22.3 | 22.3 | |||||
| 60 | Nd | 120.5* | 120.5* | 1.5 | 1.5 | 37.5 | 21.1 | 21.1 | |||||
| 61 | Pm | 120 | 120 | --- | --- | --- | --- | --- | |||||
| 62 | Sm | 129 | 129 | 5.2 | 5.2 | 37.4 | 21.3 | 21.3 | |||||
| 63 | Eu | 133 | 127.7* | 0 | 0 | 32 | 22 | 22 | |||||
| 64 | Gd | --- | 142.6* | 8.6* | 8.6* | 36 | 28 | 21 | |||||
| 65 | Tb | 150.5* | 150.5* | 7.7* | 2.4* | 45.6* | 28.7* | 22.6* | |||||
| 66 | Dy | 153.6* | 153.6* | 8.0* | 4.3* | 49.9* | 26.3 | 26.3 | |||||
| 67 | Ho | 160* | 160* | 8.6* | 5.2* | 49.3* | 30.8* | 24.1* | |||||
| 68 | Er | 167.6* | 167.6* | --- | 4.7* | 50.6* | 31.4* | 24.7* | |||||
| 69 | Tm | 175.5* | 175.5* | --- | 4.6 | 54.7* | 31.8* | 25.0* | |||||
| 70 | Yb | 191.2* | 182.4* | 2.5* | 1.3* | 52.0* | 30.3* | 24.1* | |||||
| Z | Element | K 1s | L1 2s | L2 2p1/2 | L3 2p3/2 | M1 3s | M2 3p1/2 | M3 3p3/2 | M4 3d3/2 | M5 3d5/2 | N1 4s | N2 4p1/2 | N3 4p3/2 |
| 71 | Lu | 63314 | 10870 | 10349 | 9244 | 2491 | 2264 | 2024 | 1639 | 1589 | 506.8* | 412.4* | 359.2* |
| 72 | Hf | 65351 | 11271 | 10739 | 9561 | 2601 | 2365 | 2108 | 1716 | 1662 | 538* | 438.2† | 380.7† |
| 73 | Ta | 67416 | 11682 | 11136 | 9881 | 2708 | 2469 | 2194 | 1793 | 1735 | 563.4† | 463.4† | 400.9† |
| 74 | W | 69525 | 12100 | 11544 | 10207 | 2820 | 2575 | 2281 | 1872 | 1809 | 594.1† | 490.4† | 423.6† |
| 75 | Re | 71676 | 12527 | 11959 | 10535 | 2932 | 2682 | 2367 | 1949 | 1883 | 625.4† | 518.7† | 446.8† |
| 76 | Os | 73871 | 12968 | 12385 | 10871 | 3049 | 2792 | 2457 | 2031 | 1960 | 658.2† | 549.1† | 470.7† |
| 77 | Ir | 76111 | 13419 | 12824 | 11215 | 3174 | 2909 | 2551 | 2116 | 2040 | 691.1† | 577.8† | 495.8† |
| 78 | Pt | 78395 | 13880 | 13273 | 11564 | 3296 | 3027 | 2645 | 2202 | 2122 | 725.4† | 609.1† | 519.4† |
| 79 | Au | 80725 | 14353 | 13734 | 11919 | 3425 | 3148 | 2743 | 2291 | 2206 | 762.1† | 642.7† | 546.3† |
| 80 | Hg | 83102 | 14839 | 14209 | 12284 | 3562 | 3279 | 2847 | 2385 | 2295 | 802.2† | 680.2† | 576.6† |
| 81 | Tl | 85530 | 15347 | 14698 | 12658 | 3704 | 3416 | 2957 | 2485 | 2389 | 846.2† | 720.5† | 609.5† |
| 82 | Pb | 88005 | 15861 | 15200 | 13035 | 3851 | 3554 | 3066 | 2586 | 2484 | 891.8† | 761.9† | 643.5† |
| 83 | Bi | 90524 | 16388 | 15711 | 13419 | 3999 | 3696 | 3177 | 2688 | 2580 | 939† | 805.2† | 678.8† |
| 84 | Po | 93105 | 16939 | 16244 | 13814 | 4149 | 3854 | 3302 | 2798 | 2683 | 995* | 851* | 705* |
| 85 | At | 95730 | 17493 | 16785 | 14214 | 4317 | 4008 | 3426 | 2909 | 2787 | 1042* | 886* | 740* |
| 86 | Rn | 98404 | 18049 | 17337 | 14619 | 4482 | 4159 | 3538 | 3022 | 2892 | 1097* | 929* | 768* |
| 87 | Fr | 101137 | 18639 | 17907 | 15031 | 4652 | 4327 | 3663 | 3136 | 3000 | 1153* | 980* | 810* |
| 88 | Ra | 103922 | 19237 | 18484 | 15444 | 4822 | 4490 | 3792 | 3248 | 3105 | 1208* | 1058 | 879* |
| 89 | Ac | 106755 | 19840 | 19083 | 15871 | 5002 | 4656 | 3909 | 3370 | 3219 | 1269* | 1080* | 890* |
| 90 | Th | 109651 | 20472 | 19693 | 16300 | 5182 | 4830 | 4046 | 3491 | 3332 | 1330* | 1168* | 966.4† |
| 91 | Pa | 112601 | 21105 | 20314 | 16733 | 5367 | 5001 | 4174 | 3611 | 3442 | 1387* | 1224* | 1007* |
| 92 | U | 115606 | 21757 | 20948 | 17166 | 5548 | 5182 | 4303 | 3728 | 3552 | 1439*b | 1271*b | 1043† |
| Z | Element | N4 4d3/2 | N5 4d5/2 | N6 4f5/2 | N7 4f7/2 | O1 5s | O2 5p1/2 | O3 5p3/2 | O4 5d3/2 | O5 5d5/2 | P1 6s | P2 6p1/2 | P3 6p3/2 |
| 71 | Lu | 206.1* | 196.3* | 8.9* | 7.5* | 57.3* | 33.6* | 26.7* | |||||
| 72 | Hf | 220.0† | 211.5† | 15.9† | 14.2† | 64.2† | 38* | 29.9† | |||||
| 73 | Ta | 237.9† | 226.4† | 23.5† | 21.6† | 69.7† | 42.2* | 32.7† | |||||
| 74 | W | 255.9† | 243.5† | 33.6* | 31.4† | 75.6† | 45.3*b | 36.8† | |||||
| 75 | Re | 273.9† | 260.5† | 42.9* | 40.5* | 83† | 45.6* | 34.6*b | |||||
| 76 | Os | 293.1† | 278.5† | 53.4† | 50.7† | 84* | 58* | 44.5† | |||||
| 77 | Ir | 311.9† | 296.3† | 63.8† | 60.8† | 95.2*b | 63.0*b | 48.0† | |||||
| 78 | Pt | 331.6† | 314.6† | 74.5† | 71.2† | 101.7*b | 65.3*b | 51.7† | |||||
| 79 | Au | 353.2† | 335.1† | 87.6† | 84.0 | 107.2*b | 74.2† | 57.2† | |||||
| 80 | Hg | 378.2† | 358.8† | 104.0† | 99.9† | 127† | 83.1† | 64.5† | 9.6† | 7.8† | |||
| 81 | Tl | 405.7† | 385.0† | 122.2† | 117.8† | 136.0*b | 94.6† | 73.5† | 14.7† | 12.5† | |||
| 82 | Pb | 434.3† | 412.2† | 141.7† | 136.9† | 147*b | 106.4† | 83.3† | 20.7† | 18.1† | |||
| 83 | Bi | 464.0† | 440.1† | 162.3† | 157.0† | 159.3*b | 119.0† | 92.6† | 26.9† | 23.8† | |||
| 84 | Po | 500* | 473* | 184* | 184* | 177* | 132* | 104* | 31* | 31* | |||
| 85 | At | 533* | 507 | 210* | 210* | 195* | 148* | 115* | 40* | 40* | |||
| 86 | Rn | 567* | 541* | 238* | 238* | 214* | 164* | 127* | 48* | 48* | 26 | ||
| 87 | Fr | 603* | 577* | 268* | 268* | 234* | 182* | 140* | 58* | 58* | 34 | 15 | 15 |
| 88 | Ra | 636* | 603* | 299* | 299* | 254* | 200* | 153* | 68* | 68* | 44 | 19 | 19 |
| 89 | Ac | 675* | 639* | 319* | 319* | 272* | 215* | 167* | 80* | 80* | --- | --- | --- |
| 90 | Th | 712.1† | 675.2† | 342.4† | 333.1† | 290*a | 229*a | 182*a | 92.5† | 85.4† | 41.4† | 24.5† | 16.6† |
| 91 | Pa | 743* | 708* | 371* | 360* | 310* | 232* | 232* | 94* | 94* | --- | --- | --- |
| 92 | U | 778.3† | 736.2† | 388.2* | 377.4† | 321*ab | 257*ab | 192*ab | 102.8† | 94.2† | 43.9† | 26.8† | 16.8† |
4.3. X-ray fluorescence line intensities
Ref:
X-Ray Data Booklet Table 1-3. Photon energies and relative intensities of K-, L-, and M-shell lines shown in Fig. 1-1, arranged by increasing energy. An intensity of 100 is assigned to the strongest line in each shell for each element.
| Energy [eV] | Z | Element | Line | Intensity |
|---|---|---|---|---|
| 54.3 | 3 | Li | Kα1,2 | 150 |
| 108.5 | 4 | Be | Kα1,2 | 150 |
| 183.3 | 5 | B | Kα1,2 | 151 |
| 277 | 6 | C | Kα1,2 | 147 |
| 348.3 | 21 | Sc | Ll | 21 |
| 392.4 | 7 | N | Kα1,2 | 150 |
| 395.3 | 22 | Ti | Ll | 46 |
| 395.4 | 21 | Sc | Lα1,2 | 111 |
| 399.6 | 21 | Sc | Lβ1 | 77 |
| 446.5 | 23 | V | Ll | 28 |
| 452.2 | 22 | Ti | Lα1,2 | 111 |
| 458.4 | 22 | Ti | Lβ1 | 79 |
| 500.3 | 24 | Cr | Ll | 17 |
| 511.3 | 23 | V | Lα1,2 | 111 |
| 519.2 | 23 | V | Lβ1 | 80 |
| 524.9 | 8 | O | Kα1,2 | 151 |
| 556.3 | 25 | Mn | Ll | 15 |
| 572.8 | 24 | Cr | Lα1,2 | 111 |
| 582.8 | 24 | Cr | Lβ1 | 79 |
| 615.2 | 26 | Fe | Ll | 10 |
| 637.4 | 25 | Mn | Lα1,2 | 111 |
| 648.8 | 25 | Mn | Lβ1 | 77 |
| 676.8 | 9 | F | Kα1,2 | 148 |
| 677.8 | 27 | Co | Ll | 10 |
| 705.0 | 26 | Fe | Lα1,2 | 111 |
| 718.5 | 26 | Fe | Lβ1 | 66 |
| 742.7 | 28 | Ni | Ll | 9 |
| 776.2 | 27 | Co | Lα1,2 | 111 |
| 791.4 | 27 | Co | Lβ1 | 76 |
| 811.1 | 29 | Cu | Ll | 8 |
| 833 | 57 | La | Mα1 | 100 |
| 848.6 | 10 | Ne | Kα1,2 | 150 |
| 851.5 | 28 | Ni | Lα1,2 | 111 |
| 868.8 | 28 | Ni | Lβ1 | 68 |
| 883 | 58 | Ce | Mα1 | 100 |
| 884 | 30 | Zn | Ll | 7 |
| 929.2 | 59 | Pr | Mα1 | 100 |
| 929.7 | 29 | Cu | Lα1,2 | 111 |
| 949.8 | 29 | Cu | Lβ1 | 65 |
| 957.2 | 31 | Ga | Ll | 7 |
| 978 | 60 | Nd | Mα1 | 100 |
| 1011.7 | 30 | Zn | Lα1,2 | 111 |
| 1034.7 | 30 | Zn | Lβ1 | 65 |
| 1036.2 | 32 | Ge | Ll | 6 |
| 1041.0 | 11 | Na | Kα1,2 | 150 |
| 1081 | 62 | Sm | Mα1 | 100 |
| 1097.9 | 31 | Ga | Lα1,2 | 111 |
| 1120 | 33 | As | Ll | 6 |
| 1124.8 | 31 | Ga | Lβ1 | 66 |
| 1131 | 63 | Eu | Mα1 | 100 |
| 1185 | 64 | Gd | Mα1 | 100 |
| 1188.0 | 32 | Ge | Lα1,2 | 111 |
| 1204.4 | 34 | Se | Ll | 6 |
| 1218.5 | 32 | Ge | Lβ1 | 60 |
| 1240 | 65 | Tb | Mα1 | 100 |
| 1253.6 | 12 | Mg | Kα1,2 | 150 |
| 1282.0 | 33 | As | Lα1,2 | 111 |
| 1293 | 66 | Dy | Mα1 | 100 |
| 1293.5 | 35 | Br | Ll | 5 |
| 1317.0 | 33 | As | Lβ1 | 60 |
| 1348 | 67 | Ho | Mα1 | 100 |
| 1379.1 | 34 | Se | Lα1,2 | 111 |
| 1386 | 36 | Kr | Ll | 5 |
| 1406 | 68 | Er | Mα1 | 100 |
| 1419.2 | 34 | Se | Lβ1 | 59 |
| 1462 | 69 | Tm | Mα1 | 100 |
| 1480.4 | 35 | Br | Lα1,2 | 111 |
| 1482.4 | 37 | Rb | Ll | 5 |
| 1486.3 | 13 | Al | Kα2 | 50 |
| 1486.7 | 13 | Al | Kα1 | 100 |
| 1521.4 | 70 | Yb | Mα1 | 100 |
| 1525.9 | 35 | Br | Lβ1 | 59 |
| 1557.4 | 13 | Al | Kβ1 | 1 |
| 1581.3 | 71 | Lu | Mα1 | 100 |
| 1582.2 | 38 | Sr | Ll | 5 |
| 1586.0 | 36 | Kr | Lα1,2 | 111 |
| 1636.6 | 36 | Kr | Lβ1 | 57 |
| 1644.6 | 72 | Hf | Mα1 | 100 |
| 1685.4 | 39 | Y | Ll | 5 |
| 1692.6 | 37 | Rb | Lα2 | 11 |
| 1694.1 | 37 | Rb | Lα1 | 100 |
| 1709.6 | 73 | Ta | Mα1 | 100 |
| 1739.4 | 14 | Si | Kα2 | 50 |
| 1740.0 | 14 | Si | Kα1 | 100 |
| 1752.2 | 37 | Rb | Lβ1 | 58 |
| 1775.4 | 74 | W | Mα1 | 100 |
| 1792.0 | 40 | Zr | Ll | 5 |
| 1804.7 | 38 | Sr | Lα2 | 11 |
| 1806.6 | 38 | Sr | Lα1 | 100 |
| 1835.9 | 14 | Si | Kβ1 | 2 |
| 1842.5 | 75 | Re | Mα1 | 100 |
| 1871.7 | 38 | Sr | Lβ1 | 58 |
| 1902.2 | 41 | Nb | Ll | 5 |
| 1910.2 | 76 | Os | Mα1 | 100 |
| 1920.5 | 39 | Y | Lα2 | 11 |
| 1922.6 | 39 | Y | Lα1 | 100 |
| 1979.9 | 77 | Ir | Mα1 | 100 |
| 1995.8 | 39 | Y | Lβ1 | 57 |
| 2012.7 | 15 | P | Kα2 | 50 |
| 2013.7 | 15 | P | Kα1 | 100 |
| 2015.7 | 42 | Mo | Ll | 5 |
| 2039.9 | 40 | Zr | Lα2 | 11 |
| 2042.4 | 40 | Zr | Lα1 | 100 |
| 2050.5 | 78 | Pt | Mα1 | 100 |
| 2122 | 43 | Tc | Ll | 5 |
| 2122.9 | 79 | Au | Mα1 | 100 |
| 2124.4 | 40 | Zr | Lβ1 | 54 |
| 2139.1 | 15 | P | Kβ1 | 3 |
| 2163.0 | 41 | Nb | Lα2 | 11 |
| 2165.9 | 41 | Nb | Lα1 | 100 |
| 2195.3 | 80 | Hg | Mα1 | 100 |
| 2219.4 | 40 | Zr | Lβ2,15 | 1 |
| 2252.8 | 44 | Ru | Ll | 4 |
| 2257.4 | 41 | Nb | Lβ1 | 52 |
| 2270.6 | 81 | Tl | Mα1 | 100 |
| 2289.8 | 42 | Mo | Lα2 | 11 |
| 2293.2 | 42 | Mo | Lα1 | 100 |
| 2302.7 | 40 | Zr | Lγ1 | 2 |
| 2306.6 | 16 | S | Kα2 | 50 |
| 2307.8 | 16 | S | Kα1 | 100 |
| 2345.5 | 82 | Pb | Mα1 | 100 |
| 2367.0 | 41 | Nb | Lβ2,15 | 3 |
| 2376.5 | 45 | Rh | Ll | 4 |
| 2394.8 | 42 | Mo | Lβ1 | 53 |
| 2420 | 43 | Tc | Lα2 | 11 |
| 2422.6 | 83 | Bi | Mα1 | 100 |
| 2424 | 43 | Tc | Lα1 | 100 |
| 2461.8 | 41 | Nb | Lγ1 | 2 |
| 2464.0 | 16 | S | Kβ1 | 5 |
| 2503.4 | 46 | Pd | Ll | 4 |
| 2518.3 | 42 | Mo | Lβ2,15 | 5 |
| 2538 | 43 | Tc | Lβ1 | 54 |
| 2554.3 | 44 | Ru | Lα2 | 11 |
| 2558.6 | 44 | Ru | Lα1 | 100 |
| 2620.8 | 17 | Cl | Kα2 | 50 |
| 2622.4 | 17 | Cl | Kα1 | 100 |
| 2623.5 | 42 | Mo | Lγ1 | 3 |
| 2633.7 | 47 | Ag | Ll | 4 |
| 2674 | 43 | Tc | Lβ2,15 | 7 |
| 2683.2 | 44 | Ru | Lβ1 | 54 |
| 2692.0 | 45 | Rh | Lα2 | 11 |
| 2696.7 | 45 | Rh | Lα1 | 100 |
| 2767.4 | 48 | Cd | Ll | 4 |
| 2792 | 43 | Tc | Lγ1 | 3 |
| 2815.6 | 17 | Cl | Kβ1 | 6 |
| 2833.3 | 46 | Pd | Lα2 | 11 |
| 2834.4 | 45 | Rh | Lβ1 | 52 |
| 2836.0 | 44 | Ru | Lβ2,15 | 10 |
| 2838.6 | 46 | Pd | Lα1 | 100 |
| 2904.4 | 49 | In | Ll | 4 |
| 2955.6 | 18 | Ar | Kα2 | 50 |
| 2957.7 | 18 | Ar | Kα1 | 100 |
| 2964.5 | 44 | Ru | Lγ1 | 4 |
| 2978.2 | 47 | Ag | Lα2 | 11 |
| 2984.3 | 47 | Ag | Lα1 | 100 |
| 2990.2 | 46 | Pd | Lβ1 | 53 |
| 2996.1 | 90 | Th | Mα1 | 100 |
| 3001.3 | 45 | Rh | Lβ2,15 | 10 |
| 3045.0 | 50 | Sn | Ll | 4 |
| 3126.9 | 48 | Cd | Lα2 | 11 |
| 3133.7 | 48 | Cd | Lα1 | 100 |
| 3143.8 | 45 | Rh | Lγ1 | 5 |
| 3150.9 | 47 | Ag | Lβ1 | 56 |
| 3170.8 | 92 | U | Mα1 | 100 |
| 3171.8 | 46 | Pd | Lβ2,15 | 12 |
| 3188.6 | 51 | Sb | Ll | 4 |
| 3190.5 | 18 | Ar | Kβ1,3 | 10 |
| 3279.3 | 49 | In | Lα2 | 11 |
| 3286.9 | 49 | In | Lα1 | 100 |
| 3311.1 | 19 | K | Kα2 | 50 |
| 3313.8 | 19 | K | Kα1 | 100 |
| 3316.6 | 48 | Cd | Lβ1 | 58 |
| 3328.7 | 46 | Pd | Lγ1 | 6 |
| 3335.6 | 52 | Te | Ll | 4 |
| 3347.8 | 47 | Ag | Lβ2,15 | 13 |
| 3435.4 | 50 | Sn | Lα2 | 11 |
| 3444.0 | 50 | Sn | Lα1 | 100 |
| 3485.0 | 53 | I | Ll | 4 |
| 3487.2 | 49 | In | Lβ1 | 58 |
| 3519.6 | 47 | Ag | Lγ1 | 6 |
| 3528.1 | 48 | Cd | Lβ2,15 | 15 |
| 3589.6 | 19 | K | Kβ1,3 | 11 |
| 3595.3 | 51 | Sb | Lα2 | 11 |
| 3604.7 | 51 | Sb | Lα1 | 100 |
| 3636 | 54 | Xe | Ll | 4 |
| 3662.8 | 50 | Sn | Lβ1 | 60 |
| 3688.1 | 20 | Ca | Kα2 | 50 |
| 3691.7 | 20 | Ca | Kα1 | 100 |
| 3713.8 | 49 | In | Lβ2,15 | 15 |
| 3716.9 | 48 | Cd | Lγ1 | 6 |
| 3758.8 | 52 | Te | Lα2 | 11 |
| 3769.3 | 52 | Te | Lα1 | 100 |
| 3795.0 | 55 | Cs | Ll | 4 |
| 3843.6 | 51 | Sb | Lβ1 | 61 |
| 3904.9 | 50 | Sn | Lβ2,15 | 16 |
| 3920.8 | 49 | In | Lγ1 | 6 |
| 3926.0 | 53 | I | Lα2 | 11 |
| 3937.6 | 53 | I | Lα1 | 100 |
| 3954.1 | 56 | Ba | Ll | 4 |
| 4012.7 | 20 | Ca | Kβ1,3 | 13 |
| 4029.6 | 52 | Te | Lβ1 | 61 |
| 4086.1 | 21 | Sc | Kα2 | 50 |
| 4090.6 | 21 | Sc | Kα1 | 100 |
| 4093 | 54 | Xe | Lα2 | 11 |
| 4100.8 | 51 | Sb | Lβ2,15 | 17 |
| 4109.9 | 54 | Xe | Lα1 | 100 |
| 4124 | 57 | La | Ll | 4 |
| 4131.1 | 50 | Sn | Lγ1 | 7 |
| 4220.7 | 53 | I | Lβ1 | 61 |
| 4272.2 | 55 | Cs | Lα2 | 11 |
| 4286.5 | 55 | Cs | Lα1 | 100 |
| 4287.5 | 58 | Ce | Ll | 4 |
| 4301.7 | 52 | Te | Lβ2,15 | 18 |
| 4347.8 | 51 | Sb | Lγ1 | 8 |
| 4414 | 54 | Xe | Lβ1 | 60 |
| 4450.9 | 56 | Ba | Lα2 | 11 |
| 4453.2 | 59 | Pr | Ll | 4 |
| 4460.5 | 21 | Sc | Kβ1,3 | 15 |
| 4466.3 | 56 | Ba | Lα1 | 100 |
| 4504.9 | 22 | Ti | Kα2 | 50 |
| 4507.5 | 53 | I | Lβ2,15 | 19 |
| 4510.8 | 22 | Ti | Kα1 | 100 |
| 4570.9 | 52 | Te | Lγ1 | 8 |
| 4619.8 | 55 | Cs | Lβ1 | 61 |
| 4633.0 | 60 | Nd | Ll | 4 |
| 4634.2 | 57 | La | Lα2 | 11 |
| 4651.0 | 57 | La | Lα1 | 100 |
| 4714 | 54 | Xe | Lβ2,15 | 20 |
| 4800.9 | 53 | I | Lγ1 | 8 |
| 4809 | 61 | Pm | Ll | 4 |
| 4823.0 | 58 | Ce | Lα2 | 11 |
| 4827.5 | 56 | Ba | Lβ1 | 60 |
| 4840.2 | 58 | Ce | Lα1 | 100 |
| 4931.8 | 22 | Ti | Kβ1,3 | 15 |
| 4935.9 | 55 | Cs | Lβ2,15 | 20 |
| 4944.6 | 23 | V | Kα2 | 50 |
| 4952.2 | 23 | V | Kα1 | 100 |
| 4994.5 | 62 | Sm | Ll | 4 |
| 5013.5 | 59 | Pr | Lα2 | 11 |
| 5033.7 | 59 | Pr | Lα1 | 100 |
| 5034 | 54 | Xe | Lγ1 | 8 |
| 5042.1 | 57 | La | Lβ1 | 60 |
| 5156.5 | 56 | Ba | Lβ2,15 | 20 |
| 5177.2 | 63 | Eu | Ll | 4 |
| 5207.7 | 60 | Nd | Lα2 | 11 |
| 5230.4 | 60 | Nd | Lα1 | 100 |
| 5262.2 | 58 | Ce | Lβ1 | 61 |
| 5280.4 | 55 | Cs | Lγ1 | 8 |
| 5362.1 | 64 | Gd | Ll | 4 |
| 5383.5 | 57 | La | Lβ2,15 | 21 |
| 5405.5 | 24 | Cr | Kα2 | 50 |
| 5408 | 61 | Pm | Lα2 | 11 |
| 5414.7 | 24 | Cr | Kα1 | 100 |
| 5427.3 | 23 | V | Kβ1,3 | 15 |
| 5432 | 61 | Pm | Lα1 | 100 |
| 5488.9 | 59 | Pr | Lβ1 | 61 |
| 5531.1 | 56 | Ba | Lγ1 | 9 |
| 5546.7 | 65 | Tb | Ll | 4 |
| 5609.0 | 62 | Sm | Lα2 | 11 |
| 5613.4 | 58 | Ce | Lβ2,15 | 21 |
| 5636.1 | 62 | Sm | Lα1 | 100 |
| 5721.6 | 60 | Nd | Lβ1 | 60 |
| 5743.1 | 66 | Dy | Ll | 4 |
| 5788.5 | 57 | La | Lγ1 | 9 |
| 5816.6 | 63 | Eu | Lα2 | 11 |
| 5845.7 | 63 | Eu | Lα1 | 100 |
| 5850 | 59 | Pr | Lβ2,15 | 21 |
| 5887.6 | 25 | Mn | Kα2 | 50 |
| 5898.8 | 25 | Mn | Kα1 | 100 |
| 5943.4 | 67 | Ho | Ll | 4 |
| 5946.7 | 24 | Cr | Kβ1,3 | 15 |
| 5961 | 61 | Pm | Lβ1 | 61 |
| 6025.0 | 64 | Gd | Lα2 | 11 |
| 6052 | 58 | Ce | Lγ1 | 9 |
| 6057.2 | 64 | Gd | Lα1 | 100 |
| 6089.4 | 60 | Nd | Lβ2,15 | 21 |
| 6152 | 68 | Er | Ll | 4 |
| 6205.1 | 62 | Sm | Lβ1 | 61 |
| 6238.0 | 65 | Tb | Lα2 | 11 |
| 6272.8 | 65 | Tb | Lα1 | 100 |
| 6322.1 | 59 | Pr | Lγ1 | 9 |
| 6339 | 61 | Pm | Lβ2 | 21 |
| 6341.9 | 69 | Tm | Ll | 4 |
| 6390.8 | 26 | Fe | Kα2 | 50 |
| 6403.8 | 26 | Fe | Kα1 | 100 |
| 6456.4 | 63 | Eu | Lβ1 | 62 |
| 6457.7 | 66 | Dy | Lα2 | 11 |
| 6490.4 | 25 | Mn | Kβ1,3 | 17 |
| 6495.2 | 66 | Dy | Lα1 | 100 |
| 6545.5 | 70 | Yb | Ll | 4 |
| 6587.0 | 62 | Sm | Lβ2,15 | 21 |
| 6602.1 | 60 | Nd | Lγ1 | 10 |
| 6679.5 | 67 | Ho | Lα2 | 11 |
| 6713.2 | 64 | Gd | Lβ1 | 62 |
| 6719.8 | 67 | Ho | Lα1 | 100 |
| 6752.8 | 71 | Lu | Ll | 4 |
| 6843.2 | 63 | Eu | Lβ2,15 | 21 |
| 6892 | 61 | Pm | Lγ1 | 10 |
| 6905.0 | 68 | Er | Lα2 | 11 |
| 6915.3 | 27 | Co | Kα2 | 51 |
| 6930.3 | 27 | Co | Kα1 | 100 |
| 6948.7 | 68 | Er | Lα1 | 100 |
| 6959.6 | 72 | Hf | Ll | 5 |
| 6978 | 65 | Tb | Lβ1 | 61 |
| 7058.0 | 26 | Fe | Kβ1,3 | 17 |
| 7102.8 | 64 | Gd | Lβ2,15 | 21 |
| 7133.1 | 69 | Tm | Lα2 | 11 |
| 7173.1 | 73 | Ta | Ll | 5 |
| 7178.0 | 62 | Sm | Lγ1 | 10 |
| 7179.9 | 69 | Tm | Lα1 | 100 |
| 7247.7 | 66 | Dy | Lβ1 | 62 |
| 7366.7 | 65 | Tb | Lβ2,15 | 21 |
| 7367.3 | 70 | Yb | Lα2 | 11 |
| 7387.8 | 74 | W | Ll | 5 |
| 7415.6 | 70 | Yb | Lα1 | 100 |
| 7460.9 | 28 | Ni | Kα2 | 51 |
| 7478.2 | 28 | Ni | Kα1 | 100 |
| 7480.3 | 63 | Eu | Lγ1 | 10 |
| 7525.3 | 67 | Ho | Lβ1 | 64 |
| 7603.6 | 75 | Re | Ll | 5 |
| 7604.9 | 71 | Lu | Lα2 | 11 |
| 7635.7 | 66 | Dy | Lβ2 | 20 |
| 7649.4 | 27 | Co | Kβ1,3 | 17 |
| 7655.5 | 71 | Lu | Lα1 | 100 |
| 7785.8 | 64 | Gd | Lγ1 | 11 |
| 7810.9 | 68 | Er | Lβ1 | 64 |
| 7822.2 | 76 | Os | Ll | 5 |
| 7844.6 | 72 | Hf | Lα2 | 11 |
| 7899.0 | 72 | Hf | Lα1 | 100 |
| 7911 | 67 | Ho | Lβ2,15 | 20 |
| 8027.8 | 29 | Cu | Kα2 | 51 |
| 8045.8 | 77 | Ir | Ll | 5 |
| 8047.8 | 29 | Cu | Kα1 | 100 |
| 8087.9 | 73 | Ta | Lα2 | 11 |
| 8101 | 69 | Tm | Lβ1 | 64 |
| 8102 | 65 | Tb | Lγ1 | 11 |
| 8146.1 | 73 | Ta | Lα1 | 100 |
| 8189.0 | 68 | Er | Lβ2,15 | 20 |
| 8264.7 | 28 | Ni | Kβ1,3 | 17 |
| 8268 | 78 | Pt | Ll | 5 |
| 8335.2 | 74 | W | Lα2 | 11 |
| 8397.6 | 74 | W | Lα1 | 100 |
| 8401.8 | 70 | Yb | Lβ1 | 65 |
| 8418.8 | 66 | Dy | Lγ1 | 11 |
| 8468 | 69 | Tm | Lβ2,15 | 20 |
| 8493.9 | 79 | Au | Ll | 5 |
| 8586.2 | 75 | Re | Lα2 | 11 |
| 8615.8 | 30 | Zn | Kα2 | 51 |
| 8638.9 | 30 | Zn | Kα1 | 100 |
| 8652.5 | 75 | Re | Lα1 | 100 |
| 8709.0 | 71 | Lu | Lβ1 | 66 |
| 8721.0 | 80 | Hg | Ll | 5 |
| 8747 | 67 | Ho | Lγ1 | 11 |
| 8758.8 | 70 | Yb | Lβ2,15 | 20 |
| 8841.0 | 76 | Os | Lα2 | 11 |
| 8905.3 | 29 | Cu | Kβ1,3 | 17 |
| 8911.7 | 76 | Os | Lα1 | 100 |
| 8953.2 | 81 | Tl | Ll | 6 |
| 9022.7 | 72 | Hf | Lβ1 | 67 |
| 9048.9 | 71 | Lu | Lβ2 | 19 |
| 9089 | 68 | Er | Lγ1 | 11 |
| 9099.5 | 77 | Ir | Lα2 | 11 |
| 9175.1 | 77 | Ir | Lα1 | 100 |
| 9184.5 | 82 | Pb | Ll | 6 |
| 9224.8 | 31 | Ga | Kα2 | 51 |
| 9251.7 | 31 | Ga | Kα1 | 100 |
| 9343.1 | 73 | Ta | Lβ1 | 67 |
| 9347.3 | 72 | Hf | Lβ2 | 20 |
| 9361.8 | 78 | Pt | Lα2 | 11 |
| 9420.4 | 83 | Bi | Ll | 6 |
| 9426 | 69 | Tm | Lγ1 | 12 |
| 9442.3 | 78 | Pt | Lα1 | 100 |
| 9572.0 | 30 | Zn | Kβ1,3 | 17 |
| 9628.0 | 79 | Au | Lα2 | 11 |
| 9651.8 | 73 | Ta | Lβ2 | 20 |
| 9672.4 | 74 | W | Lβ1 | 67 |
| 9713.3 | 79 | Au | Lα1 | 100 |
| 9780.1 | 70 | Yb | Lγ1 | 12 |
| 9855.3 | 32 | Ge | Kα2 | 51 |
| 9886.4 | 32 | Ge | Kα1 | 100 |
| 9897.6 | 80 | Hg | Lα2 | 11 |
| 9961.5 | 74 | W | Lβ2 | 21 |
| 9988.8 | 80 | Hg | Lα1 | 100 |
| 10010.0 | 75 | Re | Lβ1 | 66 |
| 10143.4 | 71 | Lu | Lγ1 | 12 |
| 10172.8 | 81 | Tl | Lα2 | 11 |
| 10260.3 | 31 | Ga | Kβ3 | 5 |
| 10264.2 | 31 | Ga | Kβ1 | 66 |
| 10268.5 | 81 | Tl | Lα1 | 100 |
| 10275.2 | 75 | Re | Lβ2 | 22 |
| 10355.3 | 76 | Os | Lβ1 | 67 |
| 10449.5 | 82 | Pb | Lα2 | 11 |
| 10508.0 | 33 | As | Kα2 | 51 |
| 10515.8 | 72 | Hf | Lγ1 | 12 |
| 10543.7 | 33 | As | Kα1 | 100 |
| 10551.5 | 82 | Pb | Lα1 | 100 |
| 10598.5 | 76 | Os | Lβ2 | 22 |
| 10708.3 | 77 | Ir | Lβ1 | 66 |
| 10730.9 | 83 | Bi | Lα2 | 11 |
| 10838.8 | 83 | Bi | Lα1 | 100 |
| 10895.2 | 73 | Ta | Lγ1 | 12 |
| 10920.3 | 77 | Ir | Lβ2 | 22 |
| 10978.0 | 32 | Ge | Kβ3 | 6 |
| 10982.1 | 32 | Ge | Kβ1 | 60 |
| 11070.7 | 78 | Pt | Lβ1 | 67 |
| 11118.6 | 90 | Th | Ll | 6 |
| 11181.4 | 34 | Se | Kα2 | 52 |
| 11222.4 | 34 | Se | Kα1 | 100 |
| 11250.5 | 78 | Pt | Lβ2 | 23 |
| 11285.9 | 74 | W | Lγ1 | 13 |
| 11442.3 | 79 | Au | Lβ1 | 67 |
| 11584.7 | 79 | Au | Lβ2 | 23 |
| 11618.3 | 92 | U | Ll | 7 |
| 11685.4 | 75 | Re | Lγ1 | 13 |
| 11720.3 | 33 | As | Kβ3 | 6 |
| 11726.2 | 33 | As | Kβ1 | 13 |
| 11822.6 | 80 | Hg | Lβ1 | 67 |
| 11864 | 33 | As | Kβ2 | 1 |
| 11877.6 | 35 | Br | Kα2 | 52 |
| 11924.1 | 80 | Hg | Lβ2 | 24 |
| 11924.2 | 35 | Br | Kα1 | 100 |
| 12095.3 | 76 | Os | Lγ1 | 13 |
| 12213.3 | 81 | Tl | Lβ1 | 67 |
| 12271.5 | 81 | Tl | Lβ2 | 25 |
| 12489.6 | 34 | Se | Kβ3 | 6 |
| 12495.9 | 34 | Se | Kβ1 | 13 |
| 12512.6 | 77 | Ir | Lγ1 | 13 |
| 12598 | 36 | Kr | Kα2 | 52 |
| 12613.7 | 82 | Pb | Lβ1 | 66 |
| 12622.6 | 82 | Pb | Lβ2 | 25 |
| 12649 | 36 | Kr | Kα1 | 100 |
| 12652 | 34 | Se | Kβ2 | 1 |
| 12809.6 | 90 | Th | Lα2 | 11 |
| 12942.0 | 78 | Pt | Lγ1 | 13 |
| 12968.7 | 90 | Th | Lα1 | 100 |
| 12979.9 | 83 | Bi | Lβ2 | 25 |
| 13023.5 | 83 | Bi | Lβ1 | 67 |
| 13284.5 | 35 | Br | Kβ3 | 7 |
| 13291.4 | 35 | Br | Kβ1 | 14 |
| 13335.8 | 37 | Rb | Kα2 | 52 |
| 13381.7 | 79 | Au | Lγ1 | 13 |
| 13395.3 | 37 | Rb | Kα1 | 100 |
| 13438.8 | 92 | U | Lα2 | 11 |
| 13469.5 | 35 | Br | Kβ2 | 1 |
| 13614.7 | 92 | U | Lα1 | 100 |
| 13830.1 | 80 | Hg | Lγ1 | 14 |
| 14097.9 | 38 | Sr | Kα2 | 52 |
| 14104 | 36 | Kr | Kβ3 | 7 |
| 14112 | 36 | Kr | Kβ1 | 14 |
| 14165.0 | 38 | Sr | Kα1 | 100 |
| 14291.5 | 81 | Tl | Lγ1 | 14 |
| 14315 | 36 | Kr | Kβ2 | 2 |
| 14764.4 | 82 | Pb | Lγ1 | 14 |
| 14882.9 | 39 | Y | Kα2 | 52 |
| 14951.7 | 37 | Rb | Kβ3 | 7 |
| 14958.4 | 39 | Y | Kα1 | 100 |
| 14961.3 | 37 | Rb | Kβ1 | 14 |
| 15185 | 37 | Rb | Kβ2 | 2 |
| 15247.7 | 83 | Bi | Lγ1 | 14 |
| 15623.7 | 90 | Th | Lβ2 | 26 |
| 15690.9 | 40 | Zr | Kα2 | 52 |
| 15775.1 | 40 | Zr | Kα1 | 100 |
| 15824.9 | 38 | Sr | Kβ3 | 7 |
| 15835.7 | 38 | Sr | Kβ1 | 14 |
| 16084.6 | 38 | Sr | Kβ2 | 3 |
| 16202.2 | 90 | Th | Lβ1 | 69 |
| 16428.3 | 92 | U | Lβ2 | 26 |
| 16521.0 | 41 | Nb | Kα2 | 52 |
| 16615.1 | 41 | Nb | Kα1 | 100 |
| 16725.8 | 39 | Y | Kβ3 | 8 |
| 16737.8 | 39 | Y | Kβ1 | 15 |
| 17015.4 | 39 | Y | Kβ2 | 3 |
| 17220.0 | 92 | U | Lβ1 | 61 |
| 17374.3 | 42 | Mo | Kα2 | 52 |
| 17479.3 | 42 | Mo | Kα1 | 100 |
| 17654 | 40 | Zr | Kβ3 | 8 |
| 17667.8 | 40 | Zr | Kβ1 | 15 |
| 17970 | 40 | Zr | Kβ2 | 3 |
| 18250.8 | 43 | Tc | Kα2 | 53 |
| 18367.1 | 43 | Tc | Kα1 | 100 |
| 18606.3 | 41 | Nb | Kβ3 | 8 |
| 18622.5 | 41 | Nb | Kβ1 | 15 |
| 18953 | 41 | Nb | Kβ2 | 3 |
| 18982.5 | 90 | Th | Lγ1 | 16 |
| 19150.4 | 44 | Ru | Kα2 | 53 |
| 19279.2 | 44 | Ru | Kα1 | 100 |
| 19590.3 | 42 | Mo | Kβ3 | 8 |
| 19608.3 | 42 | Mo | Kβ1 | 15 |
| 19965.2 | 42 | Mo | Kβ2 | 3 |
| 20073.7 | 45 | Rh | Kα2 | 53 |
| 20167.1 | 92 | U | Lγ1 | 15 |
| 20216.1 | 45 | Rh | Kα1 | 100 |
| 20599 | 43 | Tc | Kβ3 | 8 |
| 20619 | 43 | Tc | Kβ1 | 16 |
| 21005 | 43 | Tc | Kβ2 | 4 |
| 21020.1 | 46 | Pd | Kα2 | 53 |
| 21177.1 | 46 | Pd | Kα1 | 100 |
| 21634.6 | 44 | Ru | Kβ3 | 8 |
| 21656.8 | 44 | Ru | Kβ1 | 16 |
| 21990.3 | 47 | Ag | Kα2 | 53 |
| 22074 | 44 | Ru | Kβ2 | 4 |
| 22162.9 | 47 | Ag | Kα1 | 100 |
| 22698.9 | 45 | Rh | Kβ3 | 8 |
| 22723.6 | 45 | Rh | Kβ1 | 16 |
| 22984.1 | 48 | Cd | Kα2 | 53 |
| 23172.8 | 45 | Rh | Kβ2 | 4 |
| 23173.6 | 48 | Cd | Kα1 | 100 |
| 23791.1 | 46 | Pd | Kβ3 | 8 |
| 23818.7 | 46 | Pd | Kβ1 | 16 |
| 24002.0 | 49 | In | Kα2 | 53 |
| 24209.7 | 49 | In | Kα1 | 100 |
| 24299.1 | 46 | Pd | Kβ2 | 4 |
| 24911.5 | 47 | Ag | Kβ3 | 9 |
| 24942.4 | 47 | Ag | Kβ1 | 16 |
| 25044.0 | 50 | Sn | Kα2 | 53 |
| 25271.3 | 50 | Sn | Kα1 | 100 |
| 25456.4 | 47 | Ag | Kβ2 | 4 |
| 26061.2 | 48 | Cd | Kβ3 | 9 |
| 26095.5 | 48 | Cd | Kβ1 | 17 |
| 26110.8 | 51 | Sb | Kα2 | 54 |
| 26359.1 | 51 | Sb | Kα1 | 100 |
| 26643.8 | 48 | Cd | Kβ2 | 4 |
| 27201.7 | 52 | Te | Kα2 | 54 |
| 27237.7 | 49 | In | Kβ3 | 9 |
| 27275.9 | 49 | In | Kβ1 | 17 |
| 27472.3 | 52 | Te | Kα1 | 100 |
| 27860.8 | 49 | In | Kβ2 | 5 |
| 28317.2 | 53 | I | Kα2 | 54 |
| 28444.0 | 50 | Sn | Kβ3 | 9 |
| 28486.0 | 50 | Sn | Kβ1 | 17 |
| 28612.0 | 53 | I | Kα1 | 100 |
| 29109.3 | 50 | Sn | Kβ2 | 5 |
| 29458 | 54 | Xe | Kα2 | 54 |
| 29679.2 | 51 | Sb | Kβ3 | 9 |
| 29725.6 | 51 | Sb | Kβ1 | 18 |
| 29779 | 54 | Xe | Kα1 | 100 |
| 30389.5 | 51 | Sb | Kβ2 | 5 |
| 30625.1 | 55 | Cs | Kα2 | 54 |
| 30944.3 | 52 | Te | Kβ3 | 9 |
| 30972.8 | 55 | Cs | Kα1 | 100 |
| 30995.7 | 52 | Te | Kβ1 | 18 |
| 31700.4 | 52 | Te | Kβ2 | 5 |
| 31817.1 | 56 | Ba | Kα2 | 54 |
| 32193.6 | 56 | Ba | Kα1 | 100 |
| 32239.4 | 53 | I | Kβ3 | 9 |
| 32294.7 | 53 | I | Kβ1 | 18 |
| 33034.1 | 57 | La | Kα2 | 54 |
| 33042 | 53 | I | Kβ2 | 5 |
| 33441.8 | 57 | La | Kα1 | 100 |
| 33562 | 54 | Xe | Kβ3 | 9 |
| 33624 | 54 | Xe | Kβ1 | 18 |
| 34278.9 | 58 | Ce | Kα2 | 55 |
| 34415 | 54 | Xe | Kβ2 | 5 |
| 34719.7 | 58 | Ce | Kα1 | 100 |
| 34919.4 | 55 | Cs | Kβ3 | 9 |
| 34986.9 | 55 | Cs | Kβ1 | 18 |
| 35550.2 | 59 | Pr | Kα2 | 55 |
| 35822 | 55 | Cs | Kβ2 | 6 |
| 36026.3 | 59 | Pr | Kα1 | 100 |
| 36304.0 | 56 | Ba | Kβ3 | 10 |
| 36378.2 | 56 | Ba | Kβ1 | 18 |
| 36847.4 | 60 | Nd | Kα2 | 55 |
| 37257 | 56 | Ba | Kβ2 | 6 |
| 37361.0 | 60 | Nd | Kα1 | 100 |
| 37720.2 | 57 | La | Kβ3 | 10 |
| 37801.0 | 57 | La | Kβ1 | 19 |
| 38171.2 | 61 | Pm | Kα2 | 55 |
| 38724.7 | 61 | Pm | Kα1 | 100 |
| 38729.9 | 57 | La | Kβ2 | 6 |
| 39170.1 | 58 | Ce | Kβ3 | 10 |
| 39257.3 | 58 | Ce | Kβ1 | 19 |
| 39522.4 | 62 | Sm | Kα2 | 55 |
| 40118.1 | 62 | Sm | Kα1 | 100 |
| 40233 | 58 | Ce | Kβ2 | 6 |
| 40652.9 | 59 | Pr | Kβ3 | 10 |
| 40748.2 | 59 | Pr | Kβ1 | 19 |
| 40901.9 | 63 | Eu | Kα2 | 56 |
| 41542.2 | 63 | Eu | Kα1 | 100 |
| 41773 | 59 | Pr | Kβ2 | 6 |
| 42166.5 | 60 | Nd | Kβ3 | 10 |
| 42271.3 | 60 | Nd | Kβ1 | 19 |
| 42308.9 | 64 | Gd | Kα2 | 56 |
| 42996.2 | 64 | Gd | Kα1 | 100 |
| 43335 | 60 | Nd | Kβ2 | 6 |
| 43713 | 61 | Pm | Kβ3 | 10 |
| 43744.1 | 65 | Tb | Kα2 | 56 |
| 43826 | 61 | Pm | Kβ1 | 19 |
| 44481.6 | 65 | Tb | Kα1 | 100 |
| 44942 | 61 | Pm | Kβ2 | 6 |
| 45207.8 | 66 | Dy | Kα2 | 56 |
| 45289 | 62 | Sm | Kβ3 | 10 |
| 45413 | 62 | Sm | Kβ1 | 19 |
| 45998.4 | 66 | Dy | Kα1 | 100 |
| 46578 | 62 | Sm | Kβ2 | 6 |
| 46699.7 | 67 | Ho | Kα2 | 56 |
| 46903.6 | 63 | Eu | Kβ3 | 10 |
| 47037.9 | 63 | Eu | Kβ1 | 19 |
| 47546.7 | 67 | Ho | Kα1 | 100 |
| 48221.1 | 68 | Er | Kα2 | 56 |
| 48256 | 63 | Eu | Kβ2 | 6 |
| 48555 | 64 | Gd | Kβ3 | 10 |
| 48697 | 64 | Gd | Kβ1 | 20 |
| 49127.7 | 68 | Er | Kα1 | 100 |
| 49772.6 | 69 | Tm | Kα2 | 57 |
| 49959 | 64 | Gd | Kβ2 | 7 |
| 50229 | 65 | Tb | Kβ3 | 10 |
| 50382 | 65 | Tb | Kβ1 | 20 |
| 50741.6 | 69 | Tm | Kα1 | 100 |
| 51354.0 | 70 | Yb | Kα2 | 57 |
| 51698 | 65 | Tb | Kβ2 | 7 |
| 51957 | 66 | Dy | Kβ3 | 10 |
| 52119 | 66 | Dy | Kβ1 | 20 |
| 52388.9 | 70 | Yb | Kα1 | 100 |
| 52965.0 | 71 | Lu | Kα2 | 57 |
| 53476 | 66 | Dy | Kβ2 | 7 |
| 53711 | 67 | Ho | Kβ3 | 11 |
| 53877 | 67 | Ho | Kβ1 | 20 |
| 54069.8 | 71 | Lu | Kα1 | 100 |
| 54611.4 | 72 | Hf | Kα2 | 57 |
| 55293 | 67 | Ho | Kβ2 | 7 |
| 55494 | 68 | Er | Kβ3 | 11 |
| 55681 | 68 | Er | Kβ1 | 21 |
| 55790.2 | 72 | Hf | Kα1 | 100 |
| 56277 | 73 | Ta | Kα2 | 57 |
| 57210 | 68 | Er | Kβ2 | 7 |
| 57304 | 69 | Tm | Kβ3 | 11 |
| 57517 | 69 | Tm | Kβ1 | 21 |
| 57532 | 73 | Ta | Kα1 | 100 |
| 57981.7 | 74 | W | Kα2 | 58 |
| 59090 | 69 | Tm | Kβ2 | 7 |
| 59140 | 70 | Yb | Kβ3 | 11 |
| 59318.2 | 74 | W | Kα1 | 100 |
| 59370 | 70 | Yb | Kβ1 | 21 |
| 59717.9 | 75 | Re | Kα2 | 58 |
| 60980 | 70 | Yb | Kβ2 | 7 |
| 61050 | 71 | Lu | Kβ3 | 11 |
| 61140.3 | 75 | Re | Kα1 | 100 |
| 61283 | 71 | Lu | Kβ1 | 21 |
| 61486.7 | 76 | Os | Kα2 | 58 |
| 62970 | 71 | Lu | Kβ2 | 7 |
| 62980 | 72 | Hf | Kβ3 | 11 |
| 63000.5 | 76 | Os | Kα1 | 100 |
| 63234 | 72 | Hf | Kβ1 | 22 |
| 63286.7 | 77 | Ir | Kα2 | 58 |
| 64895.6 | 77 | Ir | Kα1 | 100 |
| 64948.8 | 73 | Ta | Kβ3 | 11 |
| 64980 | 72 | Hf | Kβ2 | 7 |
| 65112 | 78 | Pt | Kα2 | 58 |
| 65223 | 73 | Ta | Kβ1 | 22 |
| 66832 | 78 | Pt | Kα1 | 100 |
| 66951.4 | 74 | W | Kβ3 | 11 |
| 66989.5 | 79 | Au | Kα2 | 59 |
| 66990 | 73 | Ta | Kβ2 | 7 |
| 67244.3 | 74 | W | Kβ1 | 22 |
| 68803.7 | 79 | Au | Kα1 | 100 |
| 68895 | 80 | Hg | Kα2 | 59 |
| 68994 | 75 | Re | Kβ3 | 12 |
| 69067 | 74 | W | Kβ2 | 8 |
| 69310 | 75 | Re | Kβ1 | 22 |
| 70819 | 80 | Hg | Kα1 | 100 |
| 70831.9 | 81 | Tl | Kα2 | 60 |
| 71077 | 76 | Os | Kβ3 | 12 |
| 71232 | 75 | Re | Kβ2 | 8 |
| 71413 | 76 | Os | Kβ1 | 23 |
| 72804.2 | 82 | Pb | Kα2 | 60 |
| 72871.5 | 81 | Tl | Kα1 | 100 |
| 73202.7 | 77 | Ir | Kβ3 | 12 |
| 73363 | 76 | Os | Kβ2 | 8 |
| 73560.8 | 77 | Ir | Kβ1 | 23 |
| 74814.8 | 83 | Bi | Kα2 | 60 |
| 74969.4 | 82 | Pb | Kα1 | 100 |
| 75368 | 78 | Pt | Kβ3 | 12 |
| 75575 | 77 | Ir | Kβ2 | 8 |
| 75748 | 78 | Pt | Kβ1 | 23 |
| 77107.9 | 83 | Bi | Kα1 | 100 |
| 77580 | 79 | Au | Kβ3 | 12 |
| 77850 | 78 | Pt | Kβ2 | 8 |
| 77984 | 79 | Au | Kβ1 | 23 |
| 79822 | 80 | Hg | Kβ3 | 12 |
| 80150 | 79 | Au | Kβ2 | 8 |
| 80253 | 80 | Hg | Kβ1 | 23 |
| 82118 | 81 | Tl | Kβ3 | 12 |
| 82515 | 80 | Hg | Kβ2 | 8 |
| 82576 | 81 | Tl | Kβ1 | 23 |
| 84450 | 82 | Pb | Kβ3 | 12 |
| 84910 | 81 | Tl | Kβ2 | 8 |
| 84936 | 82 | Pb | Kβ1 | 23 |
| 86834 | 83 | Bi | Kβ3 | 12 |
| 87320 | 82 | Pb | Kβ2 | 8 |
| 87343 | 83 | Bi | Kβ1 | 23 |
| 89830 | 83 | Bi | Kβ2 | 9 |
| 89953 | 90 | Th | Kα2 | 62 |
| 93350 | 90 | Th | Kα1 | 100 |
| 94665 | 92 | U | Kα2 | 62 |
| 98439 | 92 | U | Kα1 | 100 |
| 104831 | 90 | Th | Kβ3 | 12 |
| 105609 | 90 | Th | Kβ1 | 24 |
| 108640 | 90 | Th | Kβ2 | 9 |
| 110406 | 92 | U | Kβ3 | 13 |
| 111300 | 92 | U | Kβ1 | 24 |
| 114530 | 92 | U | Kβ2 | 9 |
4.4. Explanation of reconstruction
import ingrid / tos_helpers import ingridDatabase / [databaseRead, databaseDefinitions] import ggplotnim, nimhdf5, cligen proc main(file: string, head = 100, run = 0) = # 1. first plot events with more than 1 cluster using ToT as scale # 2. plot same events with clusters shown as separate # 3. plot cluster center (X), long axis, length, eccentricity, σ_T, σ_L, circle # of σ_T withH5(file, "r"): let fileInfo = getFileInfo(h5f) let run = if run == 0: fileInfo.runs[0] else: run let df = h5f.readAllDsets(run, chip = 3) echo df let septemDf = h5f.getSeptemDataFrame(run, allowedChips = @[3], ToT = true) echo septemDf var i = 0 for tup, subDf in groups(septemDf.group_by("eventNumber")): if i >= head: break if subDf.unique("cluster").len == 1: continue ggplot(subDf, aes("x", "y", color = "ToT")) + geom_point() + xlim(0, 256) + ylim(0, 256) + ggsave("/tmp/events/run_" & $run & "_event_" & $i & ".pdf") ggplot(subDf, aes("x", "y", color = "cluster", shape = "cluster")) + geom_point() + xlim(0, 256) + ylim(0, 256) + ggsave("/tmp/events/run_" & $run & "_event_" & $i & "_color_cluster.pdf") ggplot(subDf, aes("x", "y", color = "ToT", shape = "cluster")) + geom_point() + xlim(0, 256) + ylim(0, 256) + ggsave("/tmp/events/run_" & $run & "_event_" & $i & "_clustered.pdf") ## group again by cluster, ssDf ## - filter `df` to the correct event number (and cluster, uhh), event index? yes! ## - get center ## - get rotation angle ## - line through center & rot angle around center length - to max ## - inc i when isMainModule: dispatch main
4.5. Detector related
4.5.1. Water cooling
A short measurement of the flow rate of the water cooling system done at in the normal lab at the PI using a half open system (reservoir input open, output connected to cooling, cooling output into a reservoir), we measured:
1.6 L in 5:21 min
import unchained defUnit(L•min⁻¹) let vol = 1.6.Liter let time = 5.Minute + 21.Second echo "Flow rate: ", (vol / time).to(L•min⁻¹)
4.6. Data reconstruction
Data reconstruction of all CAST data can be done using
runAnalysisChain by:
cd $TPA/Analysis/ingrid ./runAnalysisChain -i ~/CastData/data \ --outpath ~/CastData/data \ --years 2017 --years 2018 \ --calib --back \ --reco
(where the paths must be correct of course!) if starting from the
already parsed raw data (i.e. H5 inputs). Otherwise --raw is also
needed.
Afterwards need to add the tracking information to the final H5 files by doing:
./cast_log_reader tracking \ -p ../resources/LogFiles/tracking-logs \ --startTime 2018/05/01 \ --endTime 2018/12/31 \ --h5out ~/CastData/data/DataRuns2018_Reco.h5 \ --dryRun
With the dryRun option you are only presented with what would be
written. Run without to actually add the data.
And the equivalent for the Run-2 data, adjusting the start and end
time as needed.
./cast_log_reader tracking \ -p ../resources/LogFiles/tracking-logs \ --startTime 2017/01/01 \ --endTime 2018/05/01 \ --h5out ~/CastData/data/DataRuns2017_Reco.h5 \ --dryRun
5. CDL measurements
To derive the background rate plots a likelihood method is used. Basically a likelihood distribution is built from 3 geometric properties of extracted pixel clusters:
- eccentricity
- length / transverse RMS
- fraction of pixels within transverse RMS
To define these distributions however a set of X-ray pure datasets is needed. In addition the geometric properties above are highly dependent on the X-ray's energy, see:
where the left plot compares the \(\ce{Mn}\) line (\(^{55}\ce{Fe}\) equivalent) to the \(\ce{Cu}\) line from \(\SI{0.9}{\kilo\volt}\) electrons and the right plot compres \(^{55}\ce{Fe}\) with typical cosmic background. Obvious that a single cut value will result in wildly different signal efficiencies and background rejections. Thus, take different distributions for different energies.
The distributions which the previous background rate plots were based on were obtained in 2014 with the Run-1 detector at the CAST Detector Lab (CDL). Using a different detector for this extremely sensitive part of the analysis chain will obviously introduce systematic errors. Thus, new calibration data was taken with the current Run-2 and Run-3 detector from 15-19 Feb 2019. A summary of the target filter combinations, applied HV and resulting pixel peak position is shown in 4 and the fluorescence lines these target filter combinations correspond to are listed in tab. 5.
| Run # | FADC? | Target | Filter | HV / kV | \(\langle\mu_{\text{peak}}\rangle\) | \(\Delta\mu\) |
|---|---|---|---|---|---|---|
| 315 | y | Mn | Cr | 12.0 | 223.89 | 8.79 |
| 319 | y | Cu | Ni | 15.0 | 347.77 | 8.49 |
| 320 | n | Cu | Ni | 15.0 | 323.23 | 21.81 |
| 323 | n | Mn | Cr | 12.0 | 224.78 | 8.92 |
| 325 | y | Ti | Ti | 9.0 | 176.51 | 1.22 |
| 326 | n | Ti | Ti | 9.0 | 173.20 | 2.20 |
| 328 | y | Ag | Ag | 6.0 | 117.23 | 2.02 |
| 329 | n | Ag | Ag | 6.0 | 118.66 | 1.21 |
| 332 | y | Al | Al | 4.0 | 55.36 | 1.26 |
| 333 | n | Al | Al | 4.0 | 54.79 | 2.33 |
| 335 | y | Cu | EPIC | 2.0 | 32.33 | 2.52 |
| 336 | n | Cu | EPIC | 2.0 | 33.95 | 0.67 |
| 337 | n | Cu | EPIC | 2.0 | 31.51 | 4.76 |
| 339 | y | Cu | EPIC | 0.9 | 25.00 | 0.79 |
| 340 | n | Cu | EPIC | 0.9 | 21.39 | 2.27 |
| 342 | y | C | EPIC | 0.6 | 18.04 | 1.46 |
| 343 | n | C | EPIC | 0.6 | 17.16 | 0.57 |
| 345 | y | Cu | Ni | 15.0 | 271.16 | 6.08 |
| 347 | y | Mn | Cr | 12.0 | 198.73 | 4.72 |
| 349 | y | Ti | Ti | 9.0 | 160.86 | 1.25 |
| 351 | y | Ag | Ag | 6.0 | 106.94 | 2.55 |
| Target | Filter | HV | line | Name in Marlin | Energy / keV |
|---|---|---|---|---|---|
| Cu | Ni | 15 | \(\ce{Cu}\) \(\text{K}_{\alpha}\) | A | 8.04 |
| Mn | Cr | 12 | \(\ce{Mn}\) \(\text{K}_{\alpha}\) | B | 5.89 |
| Ti | Ti | 9 | \(\ce{Ti}\) \(\text{K}_{\alpha}\) | C | 4.51 |
| Ag | Ag | 6 | \(\ce{Ag}\) \(\text{L}_{\alpha}\) | D | 2.98 |
| Al | Al | 4 | \(\ce{Al}\) \(\text{K}_{\alpha}\) | E | 1.49 |
| Cu | EPIC | 2 | \(\ce{Cu}\) \(\text{L}_{\alpha}\) | F | 0.930 |
| Cu | EPIC | 0.9 | \(\ce{O }\) \(\text{K}_{\alpha}\) | G | 0.525 |
| C | EPIC | 0.6 | \(\ce{C }\) \(\text{K}_{\alpha}\) | H | 0.277 |
For a reference of the X-ray fluorescence lines (for more exact values and \(\alpha_1\), \(\alpha_2\) values etc.) see: https://xdb.lbl.gov/Section1/Table_1-2.pdf.
The raw data is combined by target / filter combinations. To clean the data somewhat a few simple cuts are applied, as shown in tab. 15.
| Target | Filter | line | HV | length | rmsTmin | rmsTmax | eccentricity |
|---|---|---|---|---|---|---|---|
| Cu | Ni | \(\ce{Cu}\) \(\text{K}_{\alpha}\) | 15 | 0.1 | 1.0 | 1.3 | |
| Mn | Cr | \(\ce{Mn}\) \(\text{K}_{\alpha}\) | 12 | 0.1 | 1.0 | 1.3 | |
| Ti | Ti | \(\ce{Ti}\) \(\text{K}_{\alpha}\) | 9 | 0.1 | 1.0 | 1.3 | |
| Ag | Ag | \(\ce{Ag}\) \(\text{L}_{\alpha}\) | 6 | 6.0 | 0.1 | 1.0 | 1.4 |
| Al | Al | \(\ce{Al}\) \(\text{K}_{\alpha}\) | 4 | 0.1 | 1.1 | 2.0 | |
| Cu | EPIC | \(\ce{Cu}\) \(\text{L}_{\alpha}\) | 2 | 0.1 | 1.1 | 2.0 | |
| Cu | EPIC | \(\ce{O }\) \(\text{K}_{\alpha}\) | 0.9 | 0.1 | 1.1 | 2.0 | |
| C | EPIC | \(\ce{C }\) \(\text{K}_{\alpha}\) | 0.6 | 6.0 | 0.1 | 1.1 |
With these in place both to the pixel as well as charge spectra a mixture of gaussian / exponential gaussian functions is fitted.
Specifically the gaussian:
and exponential gaussian:
where the constant \(c\) is chosen such that the resulting function is continuous.
The functions fitted to the different spectra then depend on which fluorescence lines are visible. The full list of all combinations is shown in tab. 7 and 8.
| Target | Filter | line | HV | Fit function |
|---|---|---|---|---|
| Cu | Ni | \(\ce{Cu}\) \(\text{K}_{\alpha}\) | 15 | \(EG^{\mathrm{Cu,esc}}_{\mathrm{K}_{\alpha}}(a,b,N,\mu,\sigma) + EG^{\mathrm{Cu}}_{\mathrm{K}_{\alpha}}(a,b,N,\mu,\sigma)\) |
| Mn | Cr | \(\ce{Mn}\) \(\text{K}_{\alpha}\) | 12 | \(EG^{\mathrm{Mn,esc}}_{\mathrm{K}_{\alpha}}(a,b,N,\mu,\sigma) + EG^{\mathrm{Mn}}_{\mathrm{K}_{\alpha}}(a,b,N,\mu,\sigma)\) |
| Ti | Ti | \(\ce{Ti}\) \(\text{K}_{\alpha}\) | 9 | \(G^{\mathrm{Ti,esc}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma) + G^{\mathrm{Ti}}_{\mathrm{K}_{\beta}}(N,\mu,\sigma) + EG^{Ti}_{K_{\alpha}}(a,b,N,\mu,\sigma) + G^{Ti}_{K_{\beta}}(N,\mu,\sigma)\) |
| Ag | Ag | \(\ce{Ag}\) \(\text{L}_{\alpha}\) | 6 | \(EG^{\mathrm{Ag}}_{\mathrm{L}_{\alpha}}(a,b,N,\mu,\sigma) + G^{\mathrm{Ag}}_{\mathrm{L}_{\beta}}(N,\mu,\sigma)\) |
| Al | Al | \(\ce{Al}\) \(\text{K}_{\alpha}\) | 4 | \(EG^{\mathrm{Al}}_{\mathrm{K}_{\alpha}}(a,b,N,\mu,\sigma)\) |
| Cu | EPIC | \(\ce{Cu}\) \(\text{L}_{\alpha}\) | 2 | \(G^{\mathrm{Cu}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma)\) |
| Cu | EPIC | \(\ce{O }\) \(\text{K}_{\alpha}\) | 0.9 | \(G^{\mathrm{O}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma) + G^{\mathrm{C}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma) + G^{\mathrm{Fe,esc}}_{L_{\alpha,\beta}}(N,\mu,\sigma) + G^{\mathrm{Ni}}_{L_{\alpha,\beta}}(N,\mu,\sigma)\) |
| C | EPIC | \(\ce{C }\) \(\text{K}_{\alpha}\) | 0.6 | \(G^{\mathrm{C}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma) + G^{\mathrm{O}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma)\) |
| Target | Filter | line | HV | fit functions |
|---|---|---|---|---|
| Cu | Ni | \(\ce{Cu}\) \(\text{K}_{\alpha}\) | 15 | \(G^{\mathrm{Cu,esc}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma) + G^{\mathrm{Cu}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma)\) |
| Mn | Cr | \(\ce{Mn}\) \(\text{K}_{\alpha}\) | 12 | \(G^{\mathrm{Mn,esc}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma) + G^{\mathrm{Mn}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma)\) |
| Ti | Ti | \(\ce{Ti}\) \(\text{K}_{\alpha}\) | 9 | \(G^{\mathrm{Ti,esc}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma) + G^{\mathrm{Ti}}_{\mathrm{K}_{\beta}}(N,\mu,\sigma) + G^{\mathrm{Ti}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma) + G^{\mathrm{Ti}}_{\mathrm{K}_{\beta}}(N,\mu,\sigma)\) |
| Ag | Ag | \(\ce{Ag}\) \(\text{L}_{\alpha}\) | 6 | \(G^{\mathrm{Ag}}_{\mathrm{L}_{\alpha}}(N,\mu,\sigma) + G^{\mathrm{Ag}}_{\mathrm{L}_{\beta}}(N,\mu,\sigma)\) |
| Al | Al | \(\ce{Al}\) \(\text{K}_{\alpha}\) | 4 | \(G^{\mathrm{Al}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma)\) |
| Cu | EPIC | \(\ce{Cu}\) \(\text{L}_{\alpha}\) | 2 | \(G^{\mathrm{Cu}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma)\) |
| Cu | EPIC | \(\ce{O }\) \(\text{K}_{\alpha}\) | 0.9 | \(G^{\mathrm{O}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma) + G^{\mathrm{C}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma) + G^{\mathrm{Fe,esc}}_{L_{\alpha,\beta}}(N,\mu,\sigma) + G^{\mathrm{Ni}}_{\mathrm{L}_{\alpha,\beta}}(N,\mu,\sigma)\) |
| C | EPIC | \(\ce{C }\) \(\text{K}_{\alpha}\) | 0.6 | \(G^{\mathrm{C}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma) + G^{\mathrm{O}}_{\mathrm{K}_{\alpha}}(N,\mu,\sigma)\) |
The exact implementation in use for both the gaussian and exponential gaussian:
- Gauss: https://github.com/Vindaar/seqmath/blob/master/src/seqmath/smath.nim#L997-L1009
- exponential Gauss: https://github.com/Vindaar/TimepixAnalysis/blob/master/Analysis/ingrid/calibration.nim#L182-L194
The fitting was performed both with MPFit (Levenberg Marquardt C implementation) as a comparison, but mainly using NLopt. Specifically the gradient based "Method of Moving Asymptotes" algorithm was used (NLopt provides a large number of different minimization / maximization algorithms to choose from) to perform maximum likelihood estimation written in the form of a poisson distributed log likelihood \(\chi^2\):
where \(n_i\) is the number of events in bin \(i\) and \(y_i\) the model prediction of events in bin \(i\).
The required gradient was calculated simply using the symmetric derivative. Other algorithms and minimization functions were tried, but this proved to be the most reliable. See the implementation: https://github.com/Vindaar/TimepixAnalysis/blob/master/Analysis/ingrid/calibration.nim#L131-L162
The fits to all spectra are shown below.
The positions of the main peaks both in case of the pixel as well as in the charge case should be linear in terms of the energy. This we can see in fig. 41 and 42
Finally we can calculate the energy resolution from the peak position and the width of the peaks. It should roughly follow a \(1/E\) dependency. The plot is shown in fig. 43. We can see that the energy resolution is slightly better for the pixel spectra than for the charge spectra, which is mostly expected, because the charge values have an additional uncertainty due to the statistical fluctuation of the gas amplification. In both cases the resolution is better than \(\SI{10}{\percent}\) for above \(\SI{3}{\keV}\) and goes up to \(\sim\SI{30}{\percent}\) at the lowest measured energies.
5.1. Change CDL calculations to work run-by-run instead of by target/filter only
Points of note:
- we have established that the temperature variation is the main cause for the detector variations we see, both at CAST (and almost certainly, but not done direct plots of temperature vs gas gain) at the CDL
- the weather during CDL data taking was indeed very warm for
February and sunny (> 10°C during the day in Feb!):
- the variations of gas gain vs. run number show a significant
change during the week:
- the variations seen in the hit and charge spectra is much more
massive than thought:
All of this implies that we really should perform all the spectrum fits by run instead of by target & filter type. The latter doesn't work as we have to make drop certain runs completely to get decent looking data.
Note: the main 'difficulty' is the fact that we currently have a
hardcoded set of charges in the data for the likelihood reference
distribution inputs. Of course if we do it by run, the charges need
to be different by run. This however is useful, as it allows us to
fully get rid of the annoying hardcoded charges in the first
place. Instead we will write the charge bounds into ingridDatabase /
the calibration-cdl*.h5 file and read it from there by run!
[X]implement by run histograms of all InGrid properties intocdl_spectrum_creationbased on the cut data! -> these show clearly that the properties are fortunately not correlated with the gas gain! :rocket:
6. Implement vetos for likelihood
For some preliminary results regarding the veto power of the different detector features some reasonable cut values were chosen based on the different distributions. It is to be noted that these are not final and specifically are not based on a certain signal efficiency or similar! The main motivating factor for these values so far was having some numbers to write and test the implementation of the vetoes.
Relevant PR: https://github.com/Vindaar/TimepixAnalysis/pull/37 Contains both the veto code as well as the CDL code explained below for practical reasons.
6.1. FADC fall and rise time
IMPORTANT NOTE: For a continuation on this written during the writing process of my thesis (around ) see sec. 8.2. The below was written around 2019 for an SPSC update. Interestingly the distributions seen in these old plots cannot really be reproduced anymore by me. I don't quite understand what's going on yet, but it's of course possible that one of the many changes we made over the years fixed some issue there (maybe even just the pedestal from data calculation?).
UPDATE: As it turns out after having studied this all a bit more and looked into the implementation as well, the old FADC veto application not only used weird values (which may have been correct based on how we looked at the data back then, who knows), but way more importantly the implementation was broken! The FADC veto was never correctly applied!
Based on the fall and rise time distributions
the following cuts were chosen for fall time:
const cutFallLow = 400'u16 # in 1 GHz clock cycles const cutFallHigh = 600'u16 # in 1 GHz clock cycles
and for the rise time:
const cutRiseLow = 40'u16 # in 1 GHz clock cycles const cutRiseHigh = 130'u16 # in 1 GHz clock cycles
Application of this veto yields the following improvement for the gold region:
and over the whole chip:
That is, a marginal improvement. This is to be expected, if the interpretation of the fall and rise time distributions plots is such that the main peak visible for the calibration data actually correponds to well behaved X-rays whereas the tails correspond to background contamination, since this is already what the likelihood method is very efficient at. All "easily" cutable events have already been removed.
Given that all X-rays should correspond to a roughly spherical charge distribution entering the grid holes, the rise time distribution should, for a specific energy, be a peak around the perfectly spherical charge cloud with deviations based on the statistical nature of diffusion, which directly maps to the geometrical properties of the events seen on the InGrids, i.e. an event with larger deviation from the spherical case results in a longer / shorter rise time and also in a corresponding change in the eccentricity of said cluster. Although it has to be kept in mind that the FADC is sensitive to the axis orthogonal to the geometry as seen on the InGrid (so a streched rise time value does not necessarily correspond to a larger eccentricity in one event, but on average the same properties are seen in both methods).
6.2. Scinti vetoes
Similar to the FADC some cut values were chosen to act as a scintillator veto. Regarding the scintillators it is important to keep in mind that a real axion induced X-ray cannot ever trigger a scintillator. Thus, all events in which both the FADC triggered (i.e. our trigger to read out the scintillators in the first place and an event visible on the center chip) and a scintillator triggered are either a random coincidence or a physical coincidence. In the latter case we have a background event, which we want to cut away. Fortunately, the rate of random coincidences is very small, given the very short time scales under which physical coincidence can happen (\(\mathcal{O}(\SI{1.5}{\micro\second})\) as will be discussed below).
This can either be approximated by assuming a \(\cos^2\left(\theta\right)\) distribution for cosmics and taking into account the scintillator areas and rate of cosmics, or more easily by looking at a representative data run and considering the number of entries outside of the main distribution \(\numrange{0}{60}\) clock cycles. While we cannot be sure that events in the main peak are purely physical, we can be certain that above a certain threshold no physical coincidence can happen. So considering the region from \(\numrange{300}{4095}\) clock cycles @ \(\SI{40}{\mega\hertz}\) all events should be purely random.
Then we can estimate the rate of random events per second by considering the total open shutter time in which we can accept random triggers. The number of FADC tiggers minus the number of scintillator triggers in the main peak \numrange{0}{300} clock cycles:
is the number of possible instances, in which the scintillator can trigger. This gives us the time available for the scintillator to trigger \(t_{\text{shutter}}\):
The rate of random triggers can then be estimated to:
where \(N_{r, \text{scinti}}\) is just the real number of random triggers recorded in the given run.
- Total open shutter time: \(\SI{89.98}{\hour}\)
- Open shutter time w/ FADC triggers: \(\SI{5.62}{\hour}\)
Note that \(t_{\text{shutter}}\) is orders of magnitude smaller than the open shutter time with FADC triggers, due to us only being able to estimate from the 4095 clock cycles in which we can actually determine an individual trigger (and not even that technically. If there was a trigger at 4000 clock cycles before the FADC triggered and another at 500 clock cycles we will only be able to see the one at 500!), that is \(\SI{25}{\nano\second} \cdot 4095 = \SI{102.4}{\micro\second}\) for possibly up to \(\sim\SI{2.3}{\second}\) of open shutter!
| Scinti | \(N_{\text{FADC}}\) | \(N_{\text{main}}\) | \(N_{p, \text{scinti}}\) | \(t_{\text{shutter}}\) / s | \(N_{r, \text{scinti}}\) | \(n\) |
| SCS | 19640 | 412 | 19228 | 1.83 | 2 | 1.097 |
| SCL | 19640 | 6762 | 12878 | 1.22 | 79 | 64.67 |
At an estimated muon rate of
and a large veto scinti size of \(\sim\SI{0.33}{\meter\squared}\) this comes out to \(\SI{55.5}{\per\second}\), which is quite close to our estimation.
For the SCS the same estimation however yields a wildly unexpected result at \(\mathcal{O}(\SI{1}{\per\second})\), since the size of the SCS is \(\mathcal{O}(\SI{1}{\centi\meter})\). From the cosmic rate alone we would expect 0 events on average in the random case. Given the statistics of 2 events outside the main peak, the calculation is questionable though. In one of these two events the SCL saw a trigger 3 clock cycles away from SCS (341 vs. 338 clock cycles) which was most likely a muon traversing through both scintillators. Well.
Looking at the main peaks now:
Keep in mind that calibration data appearing in the two plots is due to contamination of calibration data sets with background events, essentially the random coincidences we talked about above, since the "background signal" can never be turned off. The low counts in the calibration distribution (so that is barely appears in the plot) is then mainly due to the extremely short total data taking duration, in which the shutter is open. Thus only very few background events are actually collected, because the \(^55\ce{Fe}\) source is a \(\mathcal{O}(\SI{15}{\mega\becquerel})\) source \(\sim \SI{40}{\centi\meter}\) from the detector window, but the detector has a dead time of \(\sim\SI{175}{\milli\second}\). This is an even more extreme case of the above, since the time for random events we consider here is only \(\SI{70}{clock\ cycles} = \SI{1.75}{\micro\second}\). Even for \(\mathcal{O}(1e5)\) events that amounts to less than a second. But given enough calibration in principle the signals visible in calibration dataset would reproduce the shape of the background dataset.
With the above in mind, we can safely say any trigger values below 300 clock cycles is reasonably surely related to a physical background event. These cuts
const scintLow = 0 const scintHigh = 300
result in the following background rate for the gold region and the whole chip, fig. 44:
Nevermind the whole chip fig. 45, which shows us some bug in our code, since we cannot veto events for which there physically cannot be a scintillator trigger (below \(\SI{1.3}{\kilo\electronvolt}\)). Let's just ignore that and investigate in due time… :) Good thing that's barely visible on the log plot!
6.2.1. Why are the scintillator counts so large in the first place?
Looking at the distributions of the scintillator counts above - and keeping in mind that the clock cycles correspond to a \(\SI{40}{\mega \hertz}\) clock - one might wonder why the values are so large in the first place.
This is easily explained by considering the gaseous properties at play here. First consider the SCS in fig. 46.
The first thing to highlight is where different times for two orthogonal muons come from. On average we expect a muon to deposit \(\sim\SI{2.67}{\keV\per\cm}\) of energy along its path through our Argon/Isobutane (97.7/2.3) gas mixture, resulting in \(\sim\SI{8}{\keV}\) deposited for an orthogonal muon. At the same time the FADC needs to collect about \(\sim\SI{1.3}{\keV}\) of charge equivalent before it can trigger.
Now if two muons have a different average ionization (since it's a statistical process), the amount of length equivalent that has to drift onto the grid to accumulate enough charge for the FADC will be different. This leads to a wider distribution of clock cycles.
Taking an average muon and the aforementioned trigger threshold, an equivalent of \(\SI{0.4875}{\cm}\) of track length has to be accumulated for the FADC to trigger. Given a drift velocity at our typical HV settings and gas mixture of \(\sim\SI{2}{\cm\per\micro\second}\) leads to an equivalent time of:
Given the clock frequency of \(\SI{40}{\mega\hertz}\) this amounts to:
The peak of the real distribution is rather at around 20 clock cycles. This is probably due to an inherent delay in the signal processing (I assume there will only really be offsets in delay, rather than unexpected non-linear behaviors?).
At around 60 clock cycles (= 1.5 µs) the whole track has drifted to the chip, assuming it is perfectly orthogonal. The size of the SiPM allows for shallow angles, which should explain the tail to ~ 70 clock cycles.
Thus, the edge at around 60 clock cycles must correspond to a deposited energy of around 1.3 keV (because the FADC triggered only after all the charge has drifted onto the grid).
The question then is why the distribution is almost flat (assuming the 20 ck peak is the 8 keV peak). This means that we have almost as many other orthogonal events with much lower energy.
Now consider the SCL in fig. 47.
In case of the SCL we see a much flatter distribution. This matches perfectly with the explanation above, except that the tracks on average come from above and drift to the readout plane parallel to the readout plane. Since the rate of cosmics is uniform along the detector volume we expect the same number of muons close to the readout plane as at a distance of \(\sim\SI{3}{\cm}\). The cut off then is again corresponding to the cathode end of the detector. A larger number of clock cycles would correspond to muons passing in front of the X-ray window.
6.3. Septem veto
Using the surrounding 6 InGrid as a veto is slightly more complicated. For a start, since we mostly restrict our analysis to the gold region (inner \(\SI{4.5}{\milli\meter}\) square of the chip), the septem board will not be of much help, because all events with their centers within the gold region either are obvious tracks (vetoed by the likelihood method) or do not extend onto the outer chips. However, one of the reasons we choose the gold region in the first place (aside from the axion image being centered within that region) is the stark increase in background towards the edges and especially corners of the chip.
Take the following heatmap fig. 48 of the cluster center positions, which illustrates it perfectly:
We can see that we have barely any background in the gold region ( \(\mathcal{O}(\SI{1e-5}{\cm^{-2}\second^{-1}\keV^{-1}})\), see fig. 49), whereas the background for the whole chip is between \(\SIrange{0.01}{0.0001}{cm^{-2}.s^{-1}.keV^{-1}}\) (ref fig. 50).
The reason for the visible increase is mostly that the events are not fully contained on the chip if they are close to the edges and especially in the corners. Cutting of from an eccentric cluster can lead to a more spherical cluster increasing the chance to look like an X-ray.
To use the surrounding chips as a veto then, works along the following way. First we generate an artifical event, which incorporates the active pixels not only from a single chip, but from all chips in a single coordinate system. For simplicity we assume that all chips are not separated by any spacing. So a real event like fig. 51 is reduced to a septemevent fig. 52:

The no spacing event displays are created with ./../../CastData/ExternCode/TimepixAnalysis/Tests/tpaPlusGgplot.nim (as of commit 16235d917325502a29eadc9c38d932a734d7b095 of TPA it produces the same plot as shown above). As can be seen the event number is \(\num{4}\). The data is from run \(\num{240}\) of the Run-3 dataset from the background data, i.e.: ./../../../../mnt/1TB/CAST/2018_2/DataRuns/Run_240_181021-14-54/. To generate the required file, simply:
./raw_data_manipulation /mnt/1TB/CAST/2018_2/DataRuns/Run_240_181021-14-54/ --out=run_240.h5 --nofadc ./reconstruction run_240.h5 ./reconstruction run_240.h5 --only_charge
In the same way as done for the FADC and scintillator vetoes the septem veto starts from all events, which pass the likelihood method for the center chip (either in the gold region or on the whole chip). For these events the discussed no spacing septemevents are built by collecting all active pixels corresponding to the event the cluster that passes the likelihood method belongs to. The resulting large event is processed in the exact same way that a normal single chip event is processed. Clusters are calculated from the whole event and geometric properties calculated for each cluster. Finally the energy of each cluster is calculated and the likelihood method applied to each.
The septem veto then demands that no cluster derived from such a septemevent may look like an X-ray. This is a pessimistic cut, since it's possible that we have an X-ray like event in the corner of the center chip, which turns out to belong to some track covering the surrounding chip. But at the same time we have a real X-ray on a different chip far away from this cluster. That real X-ray will pass the likelihood method. However, since we demand no cluster being X-ray like, this event will not be vetoed, despite the original cluster being now recognized as the background it really is.
This is done for simplicity in the implementation, since a smarter algorithm has to consider which cluster the original cluster that looked like an X-ray actually belongs to. This will be implemented in the future.
With this veto in mind, we get the following improved background rate fig 53 for the gold region and fig. 54 for the whole chip:
As expected we only really have an improvement outside of the gold region. This is also easily visible when considering the cluster centers of all those events on the whole chip, which pass both the likelihood method and the septem veto in fig. 55.
Note also that other than the FADC and scintillator vetoes, the septem veto works in all energy ranges, as it is not dependent on the FADC trigger.
6.3.1. TODO Septem veto rewrite
Talk about DBSCAN vs normal and create background rates
~/org/Mails/KlausUpdates/klaus_update_03_08_21/septemEvents_2017_logL_dbscan_eps_50.pdf
~/org/Mails/KlausUpdates/klaus_update_03_08_21/septemEvents_2017_logL_dbscan_eps_65_w_lines.pdf
(possibly) re-run with 65 and create background rate plot, this one is
a comparison of 65 w/ some 2017 or so background rate.
6.3.2. Additional veto using lines through cluster centers
By performing a check on the lines along the long axes of clusters, we can compute the distance between those lines and the original cluster centers of the cluster passing the logL cut.
Then, if that distance is small enough (maybe 3*RMS), we can veto those clusters, as it seems likely that the track is actually of the same origin, with just a relatively long distance without ionization.
Implemented in likelihood.nim now.
Example in fig. 56 that shows the veto working as intended.
6.3.3. DONE Investigate BAD clustering of the default clustering algorithm in some cases
For example 57 shows a case of the default clustering algorithm w/ 65 pix search radius, in which the clustering is utterly broken.
There are maybe 1 in 20 events that look like this!
NOTE: could this be due to some data ordering issues? I don't think so, but need to investigate that event.
TODO:
- extract the raw data of that cluster and run it through the simple cluster finder
UPDATE: The issue was rather straight
forward. It's an issue with the plotting only. We somehow overwrite
the aes for the coloring, which leads to a bunch of different
clusters 'being found'.
Why it exactly happens I'm not sure, but for now it doesn't matter too much.
UPDATE 2: As it turns out, it's a bug in the
code after all. The plotting works fine. The problem is us reusing the
septemFrame variable. We first use it to fill the pixels for the
clustering etc. and then reuse it to assign the cluster IDs. The
clustering works, but sometimes there are less pixels than originally
in the event, as they are part of no real cluster (less than min
number of pixels in a cluster). In this case there remain elements in
the septemFrame (just a seq[tuple]) that still contain ToT values.
6.3.4. TODO add logic for sparks checks
We might want to add a veto check that throws out events that contain sparks or highly ionizing events on an outer chip.
For example in fig. 58 we see a big spark on the 6th chip. In this case the few pixels on the central chip are quite likely some effect from that.
6.3.5. DONE Debug septem veto background rate
UPDATE: The summary of the whole mess below is as follows:
- the exact background rate from December cannot be reproduced
- there were multiple subtle bugs in the septem veto & the line veto
- there was a subtle bug in the mapping of septem pixels to single chip pixels (mainly affecting the line veto)
- the
crAllcase ininRegionwas broken, leading to the line veto effectively vetoing everything outside the gold region - probably more I forgot
Between the commits of sometime end of 2021 (reference commit:
9e841fa56091e0338e034503b916475f8bf145be
and now:
83445319bada0f9eef35c48527946c20ac21a5d0
there seems to have been some regression in the performance of the
septem veto & the line veto.
I'm still not 100% sure that the "old" commit referenced here does actually produce the correct result either.
The thing is for sure though:
The background rate as shown in fig:
and the clusters contained in the likelihood output files used in the
limit calculation, namely:
~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_all_chip_septem_dbscan.h5~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2018_all_chip_septem_dbscan.h5
show only about 9900 clusters over the whole data taking campaign.
This is not!! reproducible on the current code base!
I looked into the number of clusters passing the septem veto including line veto on the old and new code (by adding some file output). For the following command on the old code:
./likelihood /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 \ --h5out ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/lhood_2018_test_old2.h5 \ --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crGold --septemveto --plotSeptem
we get the following output file:
Run: 297 Passed indices before septem veto 19 Passed indices after septem veto 8 Run: 242 Passed indices before septem veto 9 Passed indices after septem veto 6 Run: 256 Passed indices before septem veto 9 Passed indices after septem veto 7 Run: 268 Passed indices before septem veto 4 Passed indices after septem veto 2 Run: 281 Passed indices before septem veto 16 Passed indices after septem veto 8 Run: 272 Passed indices before septem veto 18 Passed indices after septem veto 8 Run: 274 Passed indices before septem veto 14 Passed indices after septem veto 9 Run: 270 Passed indices before septem veto 8 Passed indices after septem veto 6 Run: 306 Passed indices before septem veto 2 Passed indices after septem veto 2 Run: 246 Passed indices before septem veto 2 Passed indices after septem veto 1 Run: 263 Passed indices before septem veto 6 Passed indices after septem veto 5 Run: 298 Passed indices before septem veto 11 Passed indices after septem veto 8 Run: 303 Passed indices before septem veto 7 Passed indices after septem veto 4 Run: 287 Passed indices before septem veto 2 Passed indices after septem veto 1 Run: 248 Passed indices before septem veto 5 Passed indices after septem veto 3 Run: 299 Passed indices before septem veto 3 Passed indices after septem veto 1 Run: 291 Passed indices before septem veto 9 Passed indices after septem veto 7 Run: 295 Passed indices before septem veto 6 Passed indices after septem veto 5 Run: 285 Passed indices before septem veto 6 Passed indices after septem veto 5 Run: 240 Passed indices before septem veto 3 Passed indices after septem veto 3 Run: 301 Passed indices before septem veto 13 Passed indices after septem veto 8 Run: 267 Passed indices before septem veto 1 Passed indices after septem veto 0 Run: 276 Passed indices before septem veto 26 Passed indices after septem veto 14 Run: 279 Passed indices before septem veto 10 Passed indices after septem veto 5 Run: 293 Passed indices before septem veto 10 Passed indices after septem veto 8 Run: 254 Passed indices before septem veto 6 Passed indices after septem veto 6 Run: 244 Passed indices before septem veto 5 Passed indices after septem veto 3 Run: 278 Passed indices before septem veto 7 Passed indices after septem veto 7 Run: 283 Passed indices before septem veto 17 Passed indices after septem veto 11 Run: 258 Passed indices before septem veto 7 Passed indices after septem veto 5 Run: 289 Passed indices before septem veto 8 Passed indices after septem veto 5 Run: 250 Passed indices before septem veto 8 Passed indices after septem veto 5 Run: 261 Passed indices before septem veto 20 Passed indices after septem veto 15 Run: 265 Passed indices before septem veto 15 Passed indices after septem veto 9
With the new code:
./likelihood /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 \ --h5out ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/lhood_2018_test_new2.h5 \ --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crGold --septemveto --lineveto --plotSeptem
(note the new additional --lineveto option!)
we get:
Run: 297 Passed indices before septem veto 19 Passed indices after septem veto 8 Run: 242 Passed indices before septem veto 9 Passed indices after septem veto 6 Run: 256 Passed indices before septem veto 9 Passed indices after septem veto 7 Run: 268 Passed indices before septem veto 4 Passed indices after septem veto 2 Run: 281 Passed indices before septem veto 16 Passed indices after septem veto 8 Run: 272 Passed indices before septem veto 18 Passed indices after septem veto 8 Run: 274 Passed indices before septem veto 14 Passed indices after septem veto 9 Run: 270 Passed indices before septem veto 8 Passed indices after septem veto 6 Run: 306 Passed indices before septem veto 2 Passed indices after septem veto 2 Run: 246 Passed indices before septem veto 2 Passed indices after septem veto 1 Run: 263 Passed indices before septem veto 6 Passed indices after septem veto 5 Run: 298 Passed indices before septem veto 11 Passed indices after septem veto 8 Run: 303 Passed indices before septem veto 7 Passed indices after septem veto 4 Run: 287 Passed indices before septem veto 2 Passed indices after septem veto 1 Run: 248 Passed indices before septem veto 5 Passed indices after septem veto 3 Run: 299 Passed indices before septem veto 3 Passed indices after septem veto 2 Run: 291 Passed indices before septem veto 9 Passed indices after septem veto 7 Run: 295 Passed indices before septem veto 6 Passed indices after septem veto 5 Run: 285 Passed indices before septem veto 6 Passed indices after septem veto 5 Run: 240 Passed indices before septem veto 3 Passed indices after septem veto 3 Run: 301 Passed indices before septem veto 13 Passed indices after septem veto 8 Run: 267 Passed indices before septem veto 1 Passed indices after septem veto 0 Run: 276 Passed indices before septem veto 26 Passed indices after septem veto 14 Run: 279 Passed indices before septem veto 10 Passed indices after septem veto 5 Run: 293 Passed indices before septem veto 10 Passed indices after septem veto 8 Run: 254 Passed indices before septem veto 6 Passed indices after septem veto 6 Run: 244 Passed indices before septem veto 5 Passed indices after septem veto 3 Run: 278 Passed indices before septem veto 7 Passed indices after septem veto 7 Run: 283 Passed indices before septem veto 17 Passed indices after septem veto 11 Run: 258 Passed indices before septem veto 7 Passed indices after septem veto 5 Run: 289 Passed indices before septem veto 8 Passed indices after septem veto 5 Run: 250 Passed indices before septem veto 8 Passed indices after septem veto 5 Run: 261 Passed indices before septem veto 20 Passed indices after septem veto 15 Run: 265 Passed indices before septem veto 15 Passed indices after septem veto 10
There are differences for runs 299 and 265:
# old Run: 299 Passed indices before septem veto 3 Passed indices after septem veto 1 # new Run: 299 Passed indices before septem veto 3 Passed indices after septem veto 2 # old Run: 265 Passed indices before septem veto 15 Passed indices after septem veto 9 # new Run: 265 Passed indices before septem veto 15 Passed indices after septem veto 10
So in each of these cases there is 1 more cluster passing in the new code base.
This is a start.
The file:
 contains all septem event displays of the old code base.
contains all septem event displays of the old code base.
Look at the events of runs 299 and 265 and whether they pass or not!
For the new code base the equivalent is:
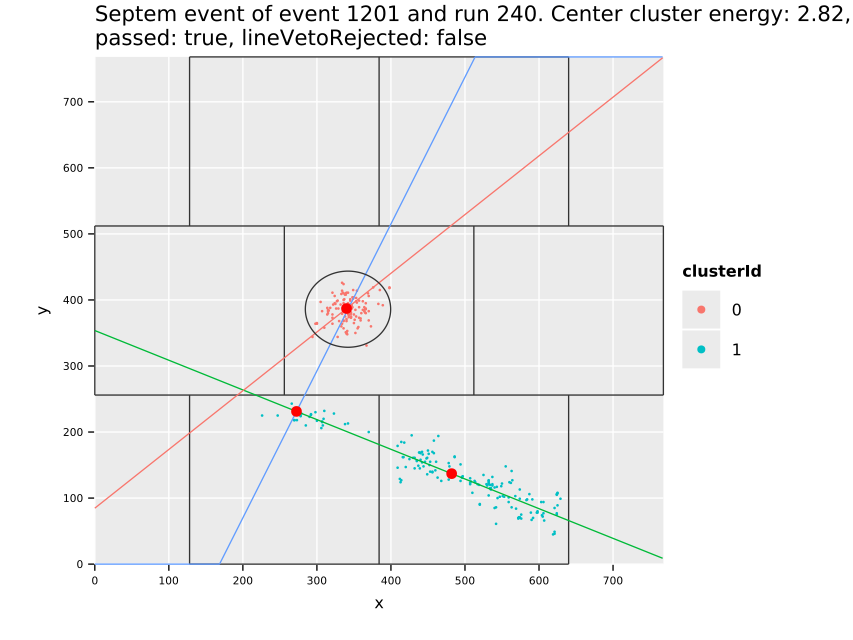
In particular of interest is the difference of run 299 event 6369.
Two things:
- both code bases actually reconstruct the center cluster as part of the cluster track to the left
- the old code doesn't know about the cluster in the top right and bottom left chips! Something is wrong in the old code about either the plotting (possible due to data assignment) or due to data reading!
However: looking at the passed and lineVetoRejected title
elements of each of these runs in the new plots, shows that we count
the same number of clusters as in the old code!! So something is
wrong about the exclusion logic!
UPDATE:
It turns out that the mismatch in removed indices must have been fixed
by the lineVetoRejected commit that I did. So this works as expected
now!
Next step: Do the same thing, but not only for the gold region, but for the whole chip! Will take a bit longer.
For now: Look at old code without line veto (commented out the line veto branch in old code): During cutting run 298 we got a KeyError from plotting:
tables.nim(233) raiseKeyError Error: unhandled exception: key not found: 128 [KeyError]
But we have the following data up to here:
Run: 297 Passed indices before septem veto 774 Passed indices after septem veto 14 Run: 242 Passed indices before septem veto 447 Passed indices after septem veto 13 Run: 256 Passed indices before septem veto 797 Passed indices after septem veto 13 Run: 268 Passed indices before septem veto 180 Passed indices after septem veto 4 Run: 281 Passed indices before septem veto 834 Passed indices after septem veto 17 Run: 272 Passed indices before septem veto 1176 Passed indices after septem veto 16 Run: 274 Passed indices before septem veto 1207 Passed indices after septem veto 15 Run: 270 Passed indices before septem veto 846 Passed indices after septem veto 10 Run: 306 Passed indices before septem veto 81 Passed indices after septem veto 2 Run: 246 Passed indices before septem veto 309 Passed indices after septem veto 1 Run: 263 Passed indices before septem veto 307 Passed indices after septem veto 6 Run: 298 Passed indices before septem veto 607
This is definitely enough to compare with the new code. Unfortunately it means we cannot look at the cluster positions right now. Need to rerun without plotting for that. First the equivalent for new code and then comparing events by event display.
The passing indices for the new code:
Run: 297 Passed indices before septem veto 774 Passed indices after septem veto 114 Run: 242 Passed indices before septem veto 447 Passed indices after septem veto 73 Run: 256 Passed indices before septem veto 797 Passed indices after septem veto 123 Run: 268 Passed indices before septem veto 180 Passed indices after septem veto 35 Run: 281 Passed indices before septem veto 834 Passed indices after septem veto 152 Run: 272 Passed indices before septem veto 1176 Passed indices after septem veto 195 Run: 274 Passed indices before septem veto 1207 Passed indices after septem veto 176 Run: 270 Passed indices before septem veto 846 Passed indices after septem veto 137 Run: 306 Passed indices before septem veto 81 Passed indices after septem veto 15 Run: 246 Passed indices before septem veto 309 Passed indices after septem veto 45 Run: 263 Passed indices before septem veto 307 Passed indices after septem veto 49 Run: 298 Passed indices before septem veto 607
Note: The same run 298 produces the same KeyError on the new code as well!
Looking into the comparison of run 268 for old and new code now. Plots as comparison:
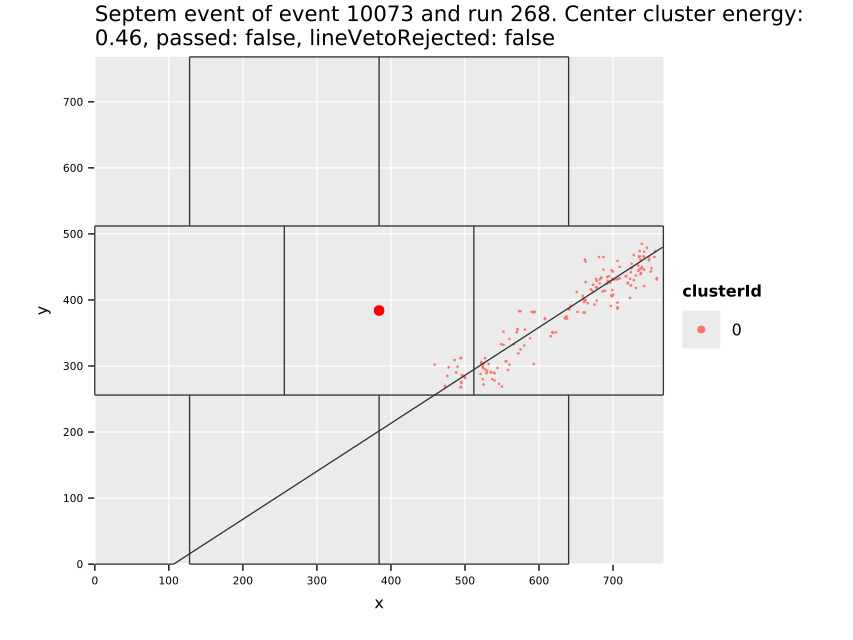
- ./../Figs/statusAndProgress/debugSeptemVeto/run_268_old_accidental_lineveto.pdf NOTE: file name adjusted after bug mentioned below found.
The reason for the difference is obvious quickly. Look at event 10542 in run 268 in both of these PDFs.
The reason the old code produces a background rate that is this much better, is plain and simply that it throws out events that it should not. So unfortunately it seems like a bug in the old code. :(
I still want to understand why that happens though. So check the old code explicitly for this event and see why it fails the logL cut suddenly.
UPDATE: The reason the old code produced such little background is
plainly that I messed up the passed = true part of the code when
commenting out the lineVeto stuff! Phew.
Checking again now with that fixed, if it reproduces the correct behavior. If so, will rerun the old code again with event displays looking at the passed indices. Indeed this fixed at least this event (10542) of the run. So rerun again now.
After the fix, we get these numbers for the passed indices:
Run: 297 Passed indices before septem veto 774 Passed indices after septem veto 141 Run: 242 Passed indices before septem veto 447 Passed indices after septem veto 86 Run: 256 Passed indices before septem veto 797 Passed indices after septem veto 150 Run: 268 Passed indices before septem veto 180 Passed indices after septem veto 42 Run: 281 Passed indices before septem veto 834 Passed indices after septem veto 178 Run: 272 Passed indices before septem veto 1176 Passed indices after septem veto 253 Run: 274 Passed indices before septem veto 1207 Passed indices after septem veto 218 Run: 270 Passed indices before septem veto 846 Passed indices after septem veto 175 Run: 306 Passed indices before septem veto 81 Passed indices after septem veto 18 Run: 246 Passed indices before septem veto 309 Passed indices after septem veto 56 Run: 263 Passed indices before septem veto 307 Passed indices after septem veto 56 Run: 298 Passed indices before septem veto 607
So comparing the numbers to the new code, we now actually get a more events in the old code!
Comparing the event displays again for run 268 (due to smaller number of events):
 (same file as above)
(same file as above)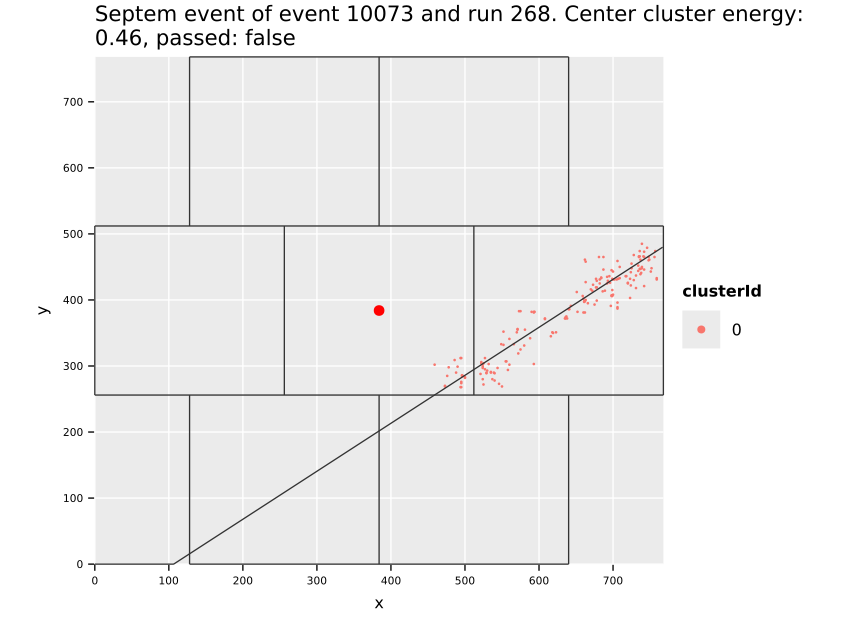
Look at event 16529 in this run 268.
The reason the old code removes less is the bug that was fixed
yesterday in the new code:
If there is a cluster on an outer chip, which passes the logL cut,
it causes the passed = true to be set!
So: From here, we'll rerun both the old and new code without plotting to generate output files that we can plot (background and clusters).
The resulting indices from the old code without lineveto:
Run: 297 Passed indices before septem veto 774 Passed indices after septem veto 141 Run: 242 Passed indices before septem veto 447 Passed indices after septem veto 86 Run: 256 Passed indices before septem veto 797 Passed indices after septem veto 150 Run: 268 Passed indices before septem veto 180 Passed indices after septem veto 42 Run: 281 Passed indices before septem veto 834 Passed indices after septem veto 178 Run: 272 Passed indices before septem veto 1176 Passed indices after septem veto 253 Run: 274 Passed indices before septem veto 1207 Passed indices after septem veto 218 Run: 270 Passed indices before septem veto 846 Passed indices after septem veto 175 Run: 306 Passed indices before septem veto 81 Passed indices after septem veto 18 Run: 246 Passed indices before septem veto 309 Passed indices after septem veto 56 Run: 263 Passed indices before septem veto 307 Passed indices after septem veto 56 Run: 298 Passed indices before septem veto 607 Passed indices after septem veto 128 Run: 303 Passed indices before septem veto 457 Passed indices after septem veto 91 Run: 287 Passed indices before septem veto 318 Passed indices after septem veto 69 Run: 248 Passed indices before septem veto 500 Passed indices after septem veto 98 Run: 299 Passed indices before septem veto 197 Passed indices after septem veto 36 Run: 291 Passed indices before septem veto 679 Passed indices after septem veto 124 Run: 295 Passed indices before septem veto 340 Passed indices after septem veto 64 Run: 285 Passed indices before septem veto 837 Passed indices after septem veto 177 Run: 240 Passed indices before septem veto 440 Passed indices after septem veto 91 Run: 301 Passed indices before septem veto 722 Passed indices after septem veto 150 Run: 267 Passed indices before septem veto 100 Passed indices after septem veto 24 Run: 276 Passed indices before septem veto 1842 Passed indices after septem veto 376 Run: 279 Passed indices before septem veto 889 Passed indices after septem veto 167 Run: 293 Passed indices before septem veto 941 Passed indices after septem veto 205 Run: 254 Passed indices before septem veto 499 Passed indices after septem veto 92 Run: 244 Passed indices before septem veto 319 Passed indices after septem veto 58 Run: 278 Passed indices before septem veto 320 Passed indices after septem veto 71 Run: 283 Passed indices before septem veto 1089 Passed indices after septem veto 212 Run: 258 Passed indices before septem veto 278 Passed indices after septem veto 62 Run: 289 Passed indices before septem veto 322 Passed indices after septem veto 62 Run: 250 Passed indices before septem veto 380 Passed indices after septem veto 72 Run: 261 Passed indices before septem veto 1095 Passed indices after septem veto 219 Run: 265 Passed indices before septem veto 916 Passed indices after septem veto 178
The clusters distributed on the chip:
The background rate:
Now redo the same with the new code.
The passed indices:
Run: 297 Passed indices before septem veto 774 Passed indices after septem veto 114 Run: 242 Passed indices before septem veto 447 Passed indices after septem veto 73 Run: 256 Passed indices before septem veto 797 Passed indices after septem veto 123 Run: 268 Passed indices before septem veto 180 Passed indices after septem veto 35 Run: 281 Passed indices before septem veto 834 Passed indices after septem veto 152 Run: 272 Passed indices before septem veto 1176 Passed indices after septem veto 195 Run: 274 Passed indices before septem veto 1207 Passed indices after septem veto 176 Run: 270 Passed indices before septem veto 846 Passed indices after septem veto 137 Run: 306 Passed indices before septem veto 81 Passed indices after septem veto 15 Run: 246 Passed indices before septem veto 309 Passed indices after septem veto 45 Run: 263 Passed indices before septem veto 307 Passed indices after septem veto 49 Run: 298 Passed indices before septem veto 607 Passed indices after septem veto 98 Run: 303 Passed indices before septem veto 457 Passed indices after septem veto 73 Run: 287 Passed indices before septem veto 318 Passed indices after septem veto 62 Run: 248 Passed indices before septem veto 500 Passed indices after septem veto 80 Run: 299 Passed indices before septem veto 197 Passed indices after septem veto 32 Run: 291 Passed indices before septem veto 679 Passed indices after septem veto 97 Run: 295 Passed indices before septem veto 340 Passed indices after septem veto 58 Run: 285 Passed indices before septem veto 837 Passed indices after septem veto 133 Run: 240 Passed indices before septem veto 440 Passed indices after septem veto 78 Run: 301 Passed indices before septem veto 722 Passed indices after septem veto 120 Run: 267 Passed indices before septem veto 100 Passed indices after septem veto 17 Run: 276 Passed indices before septem veto 1842 Passed indices after septem veto 296 Run: 279 Passed indices before septem veto 889 Passed indices after septem veto 134 Run: 293 Passed indices before septem veto 941 Passed indices after septem veto 166 Run: 254 Passed indices before septem veto 499 Passed indices after septem veto 79 Run: 244 Passed indices before septem veto 319 Passed indices after septem veto 50 Run: 278 Passed indices before septem veto 320 Passed indices after septem veto 61 Run: 283 Passed indices before septem veto 1089 Passed indices after septem veto 166 Run: 258 Passed indices before septem veto 278 Passed indices after septem veto 55 Run: 289 Passed indices before septem veto 322 Passed indices after septem veto 48 Run: 250 Passed indices before septem veto 380 Passed indices after septem veto 56 Run: 261 Passed indices before septem veto 1095 Passed indices after septem veto 178 Run: 265 Passed indices before septem veto 916 Passed indices after septem veto 141
The cluster distribution is found in:
And the background rate:
Comparing these two background rates, we see that the background is lower than with the old code!
This is much more visible when comparing all the clusters. We do indeed have almost 1000 clusters less in this case!
The next step is to also apply the likelihood cut on both the 2017 and
2018 data & also use the line cut to see if we can actually reproduce
the following background rate:
.
First though, we check if we can find the exact files & command to reproduce that file:
Looking into the zsh history:
: 1640019890:0;hdfview /tmp/lhood_2018_septemveto.h5. : 1640019897:0;hdfview /tmp/lhood_2017_septemveto.h5 : 1640019986:0;./likelihood /mnt/1TB/CAST/2017/DataRuns2017_Reco.h5 --h5out /tmp/lhood_2017_septemveto_testing.h5 --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 --cdlYear=2018 --region=crGold --septemveto : 1640020217:0;hdfview /tmp/lhood_2017_septemveto_testing.h5 : 1640021131:0;nim r tests/tgroups.nim : 1640021511:0;./plotBackgroundRate /tmp/lhood_2017_septemveto.h5 /tmp/lhood_2018_septemveto.h5 --title="GridPix background rate based on 2017/18 data at CAST" : 1640022228:0;./likelihood /mnt/1TB/CAST/2018/DataRuns2018_Reco.h5 --h5out /tmp/lhood_2018_septemveto.h5 --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 --cdlYear=2018 --region=crGold --septemveto : 1640022305:0;hdfview /mnt/1TB/CAST/2018/DataRuns2018_Reco.h5 : 1640022317:0;cd /mnt/1TB/CAST : 1640022328:0;rm DataRuns2018_Reco.h5 : 1640022390:0;rm /tmp/lhood_2018_septemveto.h5 : 1640022467:0;nim r hello.nim : 1640022605:0;mkdir examples : 1640022725:0;nim r hello_README.nim : 1640023030:0;hdfview /tmp/lhood_2018_septemveto.h5 : 1640025908:0;./plotBackgroundRate /tmp/lhood_2017_septemveto.h5 /tmp/lhood_2018_septemveto.h5 : 1640028916:0;./plotBackgroundRate /tmp/lhood_2017_septemveto.h5 /tmp/lhood_2018_septemveto.h5 --separateFiles : 1640028919:0;evince plots/background_rate_2017_2018_show2014_false_separate_true.pdf : 1640032332:0;./plotBackgroundRate /tmp/lhood_2017_septemveto.h5 /tmp/lhood_2018_septemveto.h5 --combName bla --combYear 2018 : 1640033107:0;dragon background_rate_2017_2018_show2014_false_separate_false. : 1640034992:0;dragon background_rate_2017_2018_show2014_false_separate_false.pdf : 1640035057:0;mv background_rate_2017_2018_septemveto_gold_minorTicks.pdf : 1640035065:0;mv background_rate_2017_2018_show2014_false_separate_false.pdf background_rate_2017_2018_septemveto_gold_minorTicks.pdf : 1640035087:0;evince background_rate_2017_2018_show2014_false_separate_false.pdf : 1640035110:0;mv background_rate_2017_2018_show2014_false_separate_false.pdf background_rate_2017_2018_septemveto_gold_12ticks.pdf : 1640035121:0;dragon background_rate_2017_2018_septemveto_gold_12ticks.pdf background_rate_2017_2018_septemveto_gold_minorTicks.pdf : 1640035181:0;evince background_rate_2017_2018_septemveto_gold_minorTicks.pdf : 1640085508:0;./plotBackgroundRate /tmp/lhood_run3_sigEff_65.h5 ../../resources/LikelihoodFiles/lhood_2018_no_tracking.h5 --separateFiles : 1640085515:0;./plotBackgroundRate /tmp/lhood_2017_septemveto.h5 /tmp/lhood_2018_septemveto.h5 --combName bla --combYear 2018 --title "GridPix background rate based on CAST data in 2017/18" --useTeX : 1640088525:0;cp background_rate_2017_2018_septemveto_gold_12ticks.pdf ~/org/Figs/statusAndProgress/backgroundRates/
This is a bit fishy:
- The 12 ticks background rate was definitely created in the call at
1640085515(second to last line) - the input files
/tmp/lhood_2017_septemveto.h5and/tmp/lhood_2018_septemveto.h5can be found to be created further above at1640022228(or 2018), but not for 2017. At1640019986we created the file with _testing suffix. - the 2018 file is removed before the
plotBackgroundRatecall
This probably implies the order is a bit weird / some history is
missing, as things were done asynchronously from different shells?
The last reference to a lhood_2017_septemveto.h5 is actually from
much earlier, namely:
: 1635345634:0;./likelihood /mnt/1TB/CAST/2017/DataRuns2017_Reco.h5 --h5out /tmp/lhood_2017_septemveto.h5 --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 --cdlYear=2018 --region=--septemveto
Furthermore, the lhood*.h5 files used here do not exist anymore (no
reboot since the plots I think). There are such files in /tmp/ at
the time of writing this (), but the files do
not produce the same background rate.
What we can check, is what the date is the plot was created to narrow down the state of the code we were running:
From the plotting call above, the timestamp is
(decode-time (seconds-to-time 1640085515)) ;; C-u C-x C-e to insert result into buffer (35 18 12 21 12 2021 2 nil 3600)
So it was created on the 21st of December 2021.
The last commit before this date was:
185e9eceab204d2b400ed787bbd02ecf986af983 [geometry] fix severe pitch conversion bug
from Dec 14.
It is possible of course that the pitch conversion was precisely the reason for the wrong background? But at the same time we don't know what local state we ran with, i.e. whether there were local changes etc.
As a final thing, let's at least check whether the lhood*.h5 files
used back then were created only for the gold region or the full chip.
Going by the zsh history above, the argument was always --crGold.
IMPORTANT: A big takeaways from all this is that we really need the git hash in the output of the likelihood H5 files and the vetoes & clustering algorithm settings used!
Thus, as a final test, let's rerun the code with the "old" code as we used (which was a commit from Jan 14) and see if we get the same result including the line veto, but only for the gold region.
Old code, gold region w/ line veto:
./likelihood /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 \ --h5out ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crGold_old_septemveto_lineveto.h5 \ --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crGold --septemveto
After this we'll run 2017 as well.
./likelihood /mnt/1TB/CAST/2017/DataRuns2017_Reco.h5 \ --h5out ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2017_crGold_old_septemveto_lineveto.h5 \ --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crGold --septemveto
Using these output files to generate a background rate
./plotBackgroundRate ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2017_crGold_old_septemveto_lineveto.h5 \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crGold_old_septemveto_lineveto.h5 \ --combName 2017/18 --combYear 2018 --region crGold
results in:
So, also not the background rate we got in December.
As a final check, I'd checkout the code from the commit mentioned above in December and see what happens if we do the same as this.
A theory might be the pitch conversion bug: in the commit from Dec 14, we only fixed it in one out of two places!
Running now:
./likelihood /mnt/1TB/CAST/2017/DataRuns2017_Reco.h5 --h5out \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2017_crGold_dec_14_2021_septemveto_lineveto.h5 \ --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crGold --septemveto
ok, great. That code doesn't even run properly…
Tried another commit, which has the same issue.
At this point it's likely that something fishy was going on there.
As a sanity check, try again the current code with the line veto and gold only. The files:
~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2017_crGold_new_septemveto_lineveto.h5~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crGold_new_septemveto_lineveto.h5
giving the following background rate:
Comparing this background to the one using the old code actually shows a very nice improvement all across the board and in particular in the Argon peak at 3 keV.
The shape is similar to the 12ticks plot from December last
year. Just a bit higher in the very low energy range.
As a final check, I'll now recreate the cluster maps for old & new code. For the old code without the line cut and for the new one with the line cut. The files:
~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2017_crAll_new_septemveto_lineveto.h5~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crAll_new_septemveto_lineveto.h5
Gives the following background clusters over the whole chip:
So: we have about 3500 clusters more than with the files that we use for the limit calculation!
Two questions:
- what are these clusters? Why are they not removed?
- is the background rate in the gold region also higher?
The background rate in the gold region is:
It's even worse then. Not only do we have more clusters in over the whole chip than our best case scenario (that we cannot reproduce), but also our background rate is worse if computed from the full chip logL file than from the gold region only. The only difference between these two cases should be the lineveto, as the "in region check" happens in the gold region in one and in the whole chip in the other case.
Let's extract clusters from each of the likelihood files and then see which clusters appear in what context.
UPDATE:
While writing the code to extract the cluster information, I
remembered to check the inRegion procedure for the crAll case:
func inRegion*(centerX, centerY: float, region: ChipRegion): bool {.inline.} = # ... of crAll: # simply always return good result = true
This is the reason there are more clusters in the crAll case than
a) we expect and more importantly b) the background rate is different
from crAll to crGold!
Of course not all coordinates are valid for crAll!! Only those
that are actually on the freaking chip.
It effectively meant that with the change of crAll on the "in region
check" for the line veto, the veto never did anything!
Let's change that and re-run the code again… :(
The output files:
~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2017_crAll_new_septemveto_lineveto_fixed_inRegion.h5~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crAll_new_septemveto_lineveto_fixed_inRegion.h5
The background clusters of the version with fixed inRegion is:
We can see that finally we have a "good" number (i.e. expected number) of clusters again. ~9500 clusters is similar to the number we get from the files we use as input for the limit calculation at this point.
Looking at the background rate in the gold region for these files:
We can see that this background rate is still (!!!) higher than in
the direct crGold case.
Need to pick up the extractCluster tool again and compare the actual
clusters used in each of these two cases.
While we can in principle plot the clusters that pass directly, it
won't be very helpful by themselves. Better if we print out the
clusters of a single run that survive in the crAll case within the
gold region and do the same with the direct crGold file. Then just
get the event numbers and look at the plots using the --plotSeptem
option of likelihood.nim.
Ideally we should refactor out the drawing logic to a standalone tool
that is imported in likelihood, but all the additional information
is so tightly coupled to the veto logic, that it'd get ugly.
First call it for run 261 (relatively long, should give enough
mismatches between the files) on the crGold file:
cd ~/CastData/ExternCode/TimepixAnalysis/Tools/. ./extractClusterInfo -f ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crGold_new_septemveto_lineveto.h5 \ --region crGold --run 261
And now the same for the crAll file:
extractClusterInfo -f ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crAll_new_septemveto_lineveto_fixed_inRegion.h5 \
--region crGold --run 261
There we have it. One event is more in the crAll case. Namely event
14867 of run 261.
Let's look at it, call likelihood with --plotSeptem option.
./likelihood /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 --h5out \ /tmp/test_noworries.h5 --altCdlFile \ /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crAll --septemveto --lineveto --plotSeptem
The event is the following:
What the hell? How does not that pass in case we only look at crGold?…
Let's create the plots for that case…
Run same command as above with --region crGold.
Great, even making sure the correct region is used in
inRegionOfInterest in likelihood.nim, this event suddenly does
pass, even if we run it just with crGold…
Guess it's time to rerun the likelihood again, but this time only
on the gold region….
./likelihood /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 --h5out \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crGold_new_septemveto_lineveto_fixed_inRegion.h5 \ --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crGold --septemveto --lineveto
First, let's look at the same run 261 of the new output file:
extractClusterInfo -f ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crGold_new_septemveto_lineveto_fixed_inRegion.h5 \
--region crGold --run 261
And indeed, now the same event is found here, 14867…
I assume the background rate will now be the same as in the crAll case
cut to crGold?
First checked the clusters in the gold region again:
basti at void in ~/CastData/ExternCode/TimepixAnalysis/Tools ツ ./extractClusterInfo -f ../resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crGold_new_septemveto_lineveto_fixed_inRegion.h5 --region crGold > crGold.txt basti at void in ~/CastData/ExternCode/TimepixAnalysis/Tools ツ ./extractClusterInfo -f ../resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crAll_new_septemveto_lineveto_fixed_inRegion.h5 --region crGold > crAll.txt basti at void in ~/CastData/ExternCode/TimepixAnalysis/Tools ツ diff crGold.txt crAll.txt 12a13 > (run: 263, event: 22755, cX: 9.488187499999999, cY: 6.219125) 27d27 < INFO: no events left in run number 267 for chip 3 56a57 > (run: 256, event: 30527, cX: 7.846476683937824, cY: 9.488069948186528) 108a110 > (run: 297, event: 62781, cX: 9.433578431372547, cY: 8.849428104575162) 128a131 > (run: 283, event: 94631, cX: 9.469747838616716, cY: 9.37306195965418) 202c205 < Found 200 clusters in region: crGold --- > Found 204 clusters in region: crGold
So they are still different by 4 events. Let's look at these….
Run 263.
The obvious thing looking at the coordinates of these clusters is that they are all very close to 9.5 in one coordinate. That is the cutoff of the gold region (4.5 to 9.5 mm). Does the filtering go weird somewhere?
The event in run 263 is:
Looking at the title, we can see that the issue is the line veto. It
seems like for these close clusters, somehow they are interpreted as
"outside" the region of interest and thus they veto themselves.
From the debug output of likelihood:
Cluster center: 23.5681875 and 20.299125 line veto?? false at energy ? 5.032889059014033 with log 5.60286264130613 and ut 11.10000000000002 for cluster: 0 f or run 263 and event 22755
Computing the cluster center from the given coordinates:
23.5681875 - 14 = 9.5681875 20.299125 - 14 = 6.299125
which is obviously outside the 9.5 region…
But the coordinates reported above were cX: 9.488187499999999, cY:
6.219125
So something is once again amiss. Are the septem coordinates simply
not computed correctly? One pixel off?
I have an idea what might be going on. Possibly the pixels reported by TOS start at 1 instead of 0. That would mean the pixel ⇒ Septem pixel conversion is off by 1 / 2 pixels.
Check with printXYDataset, by just printing one run:
printXyDataset -f /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 --run 263 --chip 3 --dset "x" --reco
So, no. The pixel information does indeed start at 0…
Need to check where the center cluster position is computed in
likelihood then.
Or rather, first let's check what applyPitchConversion actually does
in these cases:
const NPIX = 256 const PITCH = 0.055 let TimepixSize = NPIX * PITCH func applyPitchConversion*[T: (float | SomeInteger)](x, y: T, npix: int): (float, float) = ## template which returns the converted positions on a Timepix ## pixel position --> position from center in mm ((float(npix) - float(x) - 0.5) * PITCH, (float(y) + 0.5) * PITCH) # first find boundary of gold region let s84 = applyPitchConversion(84, 127, NPIX) echo s84 # what's max echo applyPitchConversion(0, 0, NPIX) echo applyPitchConversion(255, 255, NPIX) let center84 = applyPitchConversion(256 + 84, 127, NPIX * 3) echo center84 echo "Convert to center: ", center84[0] - TimepixSize
So, from the code snippet above we learned the following:
- either the pixel pitch is not exactly 0.055 μm
- or the size of the Timepix is not 14 mm
I think the former is more likely, the real size is larger. Using that
size, TimepixSize for the computation of the pixel position on the
center chip that corresponds to just inside of the gold region
(pixel 84; the computation is the same as the withSeptem template!)
So, once we fix that in likelihood, it should be finally correct.
Rerunning with crGold to verify that the above event 22755 does
indeed now pass.
As we can see, the event does not pass now, as it shouldn't.
Final check, run likelihood on full crGold and compare output of
clusters with extractClusterInfo.
extractClusterInfo -f ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crGold_new_septemveto_lineveto_fixed_inRegion_fixed_timepixSize.h5 \
--region crGold --short
Indeed, we get the same number of clusters as in the crAll case
now. Yay.
That final file is:
~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crGold_new_septemveto_lineveto_fixed_inRegion_fixed_timepixSize.h5
After that, we need to check how many clusters we get in the new
code using the line veto for the whole chip. We should hopefully end
up with < 10000 clusters over the whole center chip. For that we at
least have the likelihood files in the resources directory as a reference.
6.3.6. TODO Create background cluster plot from H5 files used for limit as comparison
6.3.7. TODO Implement clustering & veto & git hash as attributes from likelihood!
6.4. Estimating the random coincidence rate of the septem & line veto [/]
UPDATE: : We reran the code today after fixing the issues with the septem veto (clustering with real spacing instead of without and rotation angle for septem geometry / normal) and the numbers are changed a bit. See ./../../phd/thesis.html for the currently up to date numbers. The resulting files are in ./../../phd/resources/estimateRandomCoinc/ produced ./../journal.html.
[ ]NEED to explain that eccentricity line veto cutoff is not used, but tested. Also NEED to obviously give the numbers for both setups.
[ ]NAME THE ABSOLUTE EFFICIENCIES OF EACH SETUP[ ]IMPORTANT: The random coincidence we calculate here changes not only the dead time for the tracking time, but also for the background rate! As such we need to regulate both![ ]REWRITE THIS! -> Important parts are that background rates are only interesting if one understands the associated efficiencies. So need to explain that. This part should become :noexport:, but a shortened simpler version of this should remain.
One potential issue with the septem and line veto is that the shutter times we ran with at CAST are very long (\(> \SI{2}{s}\)), but only the center chip is triggered by the FADC. This means that the outer chips can record cluster data that is not correlated to what the center chip sees. When applying one of these two vetoes the chance for random coincidence might be non negligible.
In order to estimate this we can create fake events from real clusters on the center chip and clusters for the outer chips using different events. This way we bootstrap a larger number of events than otherwise available and knowing that the geometric data cannot be correlated. Any vetoing in these cases therefore must be a random coincidence.
As the likelihood tool already uses effectively an index to map the
cluster indices for each chip to their respective event number, we've
implemented this there (--estimateRandomCoinc) by rewriting the
index.
It is a good idea to also run it together with the --plotseptem
option to actually see some events and verify with your own eyes that
the events are actually "correct" (i.e. not the original ones). You
will note that there are many events that "clearly" look as if the
bootstrapping is not working correctly, because they look way too much
as if they are "obviously correlated". To give yourself a better sense
that this is indeed just coincidence, you can run the tool with the
--estFixedEvents option, which bootstraps events using a fixed
cluster in the center for each run. Checking out the event displays of
those is a convincing affair that unfortunately random coincidences
are even convincing to our own eyes.
likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/lhood_2018_crAll_80eff_septem_fake.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --septemveto --estimateRandomCoinc
which writes the file /tmp/septem_fake_veto.txt, which for this case
is found
./../resources/septem_veto_random_coincidences/septem_veto_before_after_fake_events_septem_old.txt
(note: updated numbers from latest state of code is the same file
without _old suffix)
Mean value of and fraction (from script in next section): File: /home/basti/org/resources/septemvetorandomcoincidences/septemvetobeforeafterfakeeventsseptem.txt Mean output = 1674.705882352941 Fraction of events left = 0.8373529411764704
From this file the method seems to remove typically a bit more than 300 out of 2000 bootstrapped fake events. This seems to imply a random coincidence rate of about 17% (or effectively a reduction of further 17% in efficiency / 17% increase in dead time).
Of course this does not even include the line veto, which will drop it further. Before we combine both of them, let's run it with the line veto alone:
likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/lhood_2018_crAll_80eff_line_fake.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --lineveto --estimateRandomCoinc
this results in: ./../resources/septem_veto_random_coincidences/septem_veto_before_after_fake_events_line.txt
Mean value of: File: /home/basti/org/resources/septemvetorandomcoincidences/septemvetobeforeafterfakeeventsline.txt Mean output = 1708.382352941177 Fraction of events left = 0.8541911764705882
And finally both together:
likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/lhood_2018_crAll_80eff_septem_line_fake.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --septemveto --lineveto --estimateRandomCoinc
which generated the following output: ./../resources/septem_veto_random_coincidences/septem_veto_before_after_fake_events_septem_line.txt
Mean value of: File: /home/basti/org/resources/septemvetorandomcoincidences/septemvetobeforeafterfakeeventsseptemline.txt Mean output = 1573.676470588235 Fraction of events left = 0.7868382352941178
This comes out to a fraction of 78.68% of the events left after running the vetoes on our bootstrapped fake events. Combining it with a software efficiency of ε = 80% the total combined efficiency then would be \(ε_\text{total} = 0.8 · 0.7868 = 0.629\), so about 63%.
Finally now let's prepare some event displays for the case of using
different center clusters and using the same ones. We run the
likelihood tool with the --plotSeptem option and stop the program
after we have enough plots.
In this context note the energy cut range for the --plotseptem
option (by default set to 5 keV), adjustable by the
PLOT_SEPTEM_E_CUTOFF environment variable.
First with different center clusters:
PLOT_SEPTEM_E_CUTOFF=10.0 likelihood \ -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/dummy.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --septemveto --lineveto --estimateRandomCoinc --plotseptem
which are wrapped up using pdfunite and stored in:
./Figs/background/estimateSeptemVetoRandomCoinc/fake_events_septem_line_veto_all_outer_events.pdf
and now with fixed clusters:
PLOT_SEPTEM_E_CUTOFF=10.0 likelihood \ -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/dummy.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --septemveto --lineveto --estimateRandomCoinc --estFixedEvent --plotseptem
(Note that the cluster that is chosen can be set using
SEPTEM_FAKE_FIXED_CLUSTER to a different index, by default it just
uses 5).
These events are here:
./Figs/background/estimateSeptemVetoRandomCoinc/fake_events_fixed_cluster_septem_line_veto_all_outer_events.pdf
Combining different options of the line veto and the eccentricity cut
for the line veto, as well as applying both the septem and the line
veto for real data as well as fake bootstrapped data we can make an
informed decision about the settings to use. At the same time get an
understanding for the real dead time we
introduce. Fig. 60
shows precisely such data. We can see that the fraction that passes
the veto setups (y axis) drops the further we go towards a low
eccentricity cut (x axis). For the real data (Real suffix in the
legend) the drop is faster than for fake boostrapped data (Fake suffix
in the legend) however, which means that we can use the lowest
eccentricity cut as we like (effectively disabling the cut at
\(ε_\text{cut} = 1.0\)). The exact choice between the purple / green
pair (line veto including all clusters, even the one containing the
original cluster) and the turquoise / blue pair (septem veto + line
veto with only those clusters that do not contain the original; those
are covered by the septem veto) is not entirely clear. Both will be
investigated for their effect on the expected limit. The important
point is that the fake data allows us to estimate the random
coincidence rate, which needs to be treated as an additional dead time
during background and solar tracking time. A lower background may or
may not be beneficial, compared to a higher dead time.
6.4.1. TODO Rewrite the whole estimation to a proper program [/]
IMPORTANT
That program should call likelihood alone, and likelihood needs to
be rewritten such that it outputs the septem random coincidence (or
real removal) into the H5 output file. Maybe just add a type that
stores the information which we serialize.
With the serialized info about the veto settings we can then
reconstruct in code what is what.
Or possibly better if the output is written to a separate file such that we don't store all the cluster data.
Anyhow, then rewrite the code snippet in the section below that prints the information about the random coincidence rates and creates the plot.
6.4.2. Run a whole bunch more cases
The below is running now . Still running as of , damn this is slow.
[X]INVESTIGATE PERFORMANCE AFTER IT'S DONE[ ]We should be able to run ~4 (depending on choice even more) in parallel, no?
import shell, strutils, os #let vals = @[1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9, 2.0] #let vals = @[1.0, 1.1] let vals = @[1.0, 1.1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9, 2.0] #let vetoes = @["--lineveto", "--lineveto --estimateRandomCoinc"] let vetoes = @["--septemveto --lineveto", "--septemveto --lineveto --estimateRandomCoinc"] ## XXX: ADD CODE DIFFERENTIATING SEPTEM + LINE & LINE ONLY IN NAMES AS WELL! #const lineVeto = "lvRegular" const lineVeto = "lvRegularNoHLC" let cmd = """ LINE_VETO_KIND=$# \ ECC_LINE_VETO_CUT=$# \ USE_REAL_LAYOUT=true \ likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /t/lhood_2018_crAll_80eff_septem_line_ecc_cutoff_$#_$#_real_layout$#.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 $# """ proc toName(veto: string): string = (if "estimateRandomCoinc" in veto: "_fake_events" else: "") for val in vals: for veto in vetoes: let final = cmd % [ lineVeto, $val, $val, lineVeto, toName(veto), $veto ] let (res, err) = shellVerbose: one: cd /tmp ($final) writeFile("/tmp/logL_output_septem_line_ecc_cutoff_$#_$#_real_layout$#.txt" % [$val, lineVeto, toName(veto)], res) let outpath = "/home/basti/org/resources/septem_veto_random_coincidences/autoGen/" let outfile = "septem_veto_before_after_septem_line_ecc_cutoff_$#_$#_real_layout$#.txt" % [$val, lineVeto, toName(veto)] copyFile("/tmp/septem_veto_before_after.txt", outpath / outfile) removeFile("/tmp/septem_veto_before_after.txt") # remove file to not append more and more to file
It has finally finished some time before . Holy moly how slow.
We will keep the generated lhood_* and logL_output_* files in
./../resources/septem_veto_random_coincidences/autoGen/ together
with the septem_veto_befor_after_* files.
See the code in one of the next sections for the 'analysis' of this dataset.
[X]RERUN THE ABOVE AFTER LINE VETO BUGFIX & PERF IMPROVEMENTS[ ]Rerun everything in check for thesis final.
6.4.3. Number of events removed in real usage
[ ]MAYBE EXTEND CODE SNIPPET ABOVE TO ALLOW CHOOSING BETWEEN εcut ANALYSIS AND REAL FRACTIONS
As a reference let's quickly run the code also for the normal use case where we don't do any bootstrapping:
likelihood \ -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/dummy_real.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --septemveto
which results in ./../resources/septem_veto_random_coincidences/septem_veto_before_after_only_septem.txt
Next the line veto alone:
likelihood \ -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/dummy_real.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --lineveto
which results in: ./../resources/septem_veto_random_coincidences/septem_veto_before_after_only_line.txt
And finally both together:
likelihood \ -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/dummy_real_2.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --septemveto --lineveto
and this finally yields:
./../resources/septem_veto_random_coincidences/septem_veto_before_after_septem_line.txt
And further for reference let's compute the fake rate when only using the septem veto (as we have no eccentricity dependence, hence a single value):
likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/lhood_2018_crAll_80eff_septem_real_layout.h5 \ --region crAll \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --septemveto \ --estimateRandomCoinc
Run the line veto with new features:
- real septemboard layout
- eccentricity cut off for tracks participating (ecc > 1.6)
LINE_VETO_KIND=lvRegularNoHLC \ ECC_LINE_VETO_CUT=1.6 \ USE_REAL_LAYOUT=true \ likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/lhood_2018_crAll_80eff_line_ecc_cutof_1.6_real_layout.h5 \ --region crAll \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --lineveto
[ ]WE SHOULD REALLY LOOK INTO RUNNING THE LINE VETO ONLY USING DIFFERENT ε CUTOFFS! -> Then compare the real application with the fake bootstrap application and see if there is a sweet spot in terms of S/N.
Let's calculate the fraction in all cases:
import strutils let files = @["/home/basti/org/resources/septem_veto_random_coincidences/septem_veto_before_after_only_septem.txt", "/home/basti/org/resources/septem_veto_random_coincidences/septem_veto_before_after_only_line.txt", "/home/basti/org/resources/septem_veto_random_coincidences/septem_veto_before_after_septem_line.txt", "/home/basti/org/resources/septem_veto_random_coincidences/septem_veto_before_after_fake_events_septem.txt", "/home/basti/org/resources/septem_veto_random_coincidences/septem_veto_before_after_fake_events_line.txt", "/home/basti/org/resources/septem_veto_random_coincidences/septem_veto_before_after_fake_events_septem_line.txt"] proc parseFile(fname: string): float = var lines = fname.readFile.strip.splitLines() var line = 0 var numRuns = 0 var outputs = 0 # if file has more than 68 lines, remove everything before, as that means # those were from a previous run if lines.len > 68: lines = lines[^68 .. ^1] doAssert lines.len == 68 while line < lines.len: if lines[line].len == 0: break # parse input # `Septem events before: 1069 (L,F) = (false, false)` let input = lines[line].split(':')[1].strip.split()[0].parseInt # parse output # `Septem events after fake cut: 137` inc line let output = lines[line].split(':')[1].strip.parseInt result += output.float / input.float outputs += output inc numRuns inc line echo "\tMean output = ", outputs.float / numRuns.float result = result / numRuns.float # first the predefined files: for f in files: echo "File: ", f echo "\tFraction of events left = ", parseFile(f) # now all files in our eccentricity cut run directory const path = "/home/basti/org/resources/septem_veto_random_coincidences/autoGen/" import std / [os, parseutils, strutils] import ggplotnim proc parseEccentricityCutoff(f: string): float = let str = "ecc_cutoff_" let startIdx = find(f, str) + str.len var res = "" let stopIdx = parseUntil(f, res, until = "_", start = startIdx) echo res result = parseFloat(res) proc determineType(f: string): string = ## I'm sorry for this. :) if "only_line_ecc" in f: result.add "Line" elif "septem_line_ecc" in f: result.add "SeptemLine" else: doAssert false, "What? " & $f if "lvRegularNoHLC" in f: result.add "lvRegularNoHLC" elif "lvRegular" in f: result.add "lvRegular" else: # also lvRegularNoHLC, could use else above, but clearer this way. Files result.add "lvRegularNoHLC" # without veto kind are older, therefore no HLC if "_fake_events.txt" in f: result.add "Fake" else: result.add "Real" var df = newDataFrame() # walk all files and determine the type for f in walkFiles(path / "septem_veto_before_after*.txt"): echo "File: ", f let frac = parseFile(f) let eccCut = parseEccentricityCutoff(f) let typ = determineType(f) echo "\tFraction of events left = ", frac df.add toDf({"Type" : typ, "ε_cut" : eccCut, "FractionPass" : frac}) df.writeCsv("/home/basti/org/resources/septem_line_random_coincidences_ecc_cut.csv", precision = 8) ggplot(df, aes("ε_cut", "FractionPass", color = "Type")) + geom_point() + ggtitle("Fraction of events passing line veto based on ε cutoff") + margin(right = 9) + ggsave("Figs/background/estimateSeptemVetoRandomCoinc/fraction_passing_line_veto_ecc_cut.pdf", width = 800, height = 480) #ggsave("/tmp/fraction_passing_line_veto_ecc_cut.pdf", width = 800, height = 480) ## XXX: we probably don't need the following plot for the real data, as the eccentricity ## cut does not cause anything to get worse at lower values. Real improvement better than ## fake coincidence rate. #df = df.spread("Type", "FractionPass").mutate(f{float: "Ratio" ~ `Real` / `Fake`}) #ggplot(df, aes("ε_cut", "Ratio")) + # geom_point() + # ggtitle("Ratio of fraction of events passing line veto real/fake based on ε cutoff") + # #ggsave("Figs/background/estimateSeptemVetoRandomCoinc/ratio_real_fake_fraction_passing_line_veto_ecc_cut.pdf") # ggsave("/tmp/ratio_real_fake_fraction_passing_line_veto_ecc_cut.pdf")
(about the first set of files) So about 14.8% in the only septem case and 9.9% in the septem + line veto case.
[ ]MOVE BELOW TO PROPER THESIS PART!
(about the ε cut)
- Investigate significantly lower fake event fraction passing
UPDATE:
The numbers visible in the plot are MUCH LOWER than what we had previously after implementing the line veto alone!!
Let's run with the equivalent of the old parameters:
LINE_VETO_KIND=lvRegular \ ECC_LINE_VETO_CUT=1.0 \ USE_REAL_LAYOUT=false \ likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /t/lhood_2018_crAll_80eff_line_ecc_cutof_1.0_tight_layout_lvRegular.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 --lineveto --estimateRandomCoinc
-> As it turns out this was a bug in our logic that decides which cluster is of interest to the line veto. We accidentally always deemed it interesting, if the original cluster was on its own… Fixed now.
6.5. On the line veto without septem veto
When dealing with the line veto without the septem veto there are multiple questions that come up of course.
First of all, what is the cluster that you're actually targeting with our 'line'? The original cluster (OC) that passed lnL or a hypothetical larger cluster that was found during the septem event reconstruction (HLC).
Assuming the former, the next question is whether we want to allow an HLC to veto our OC? In a naive implementation this is precisely what's happening, because in the regular use case of septem veto + line veto, the line veto would never have any effect anymore, as an HLC would almost certainly be vetoed by the septem veto! But without the septem veto, this decision is fully up to the line veto and the question becomes relevant. (we will implement a switch, maybe based on an environment variable or flag)
In the latter case the tricky part is mainly just identifying the correct cluster which to test for in order to find its center. However, this needs to be implemented to avoid the HLC in the above mentioned case. With it done, we then have 3 different ways to do the line veto:
- 'regular' line veto. Every cluster checks the line to the center cluster. Without septem veto this includes HLC checking OC.
- 'regular without HLC' line veto: Lines check the OC, but the HLC is explicitly not considered.
- 'checking the HLC' line veto: In this case all clusters check the center of the HLC.
Thoughts on LvCheckHLC:
- The radii around the new HLC become so large that in practice this won't be a very good idea I think!
- The
lineVetoRejectedpart of the title seems to be "true" in too many cases. What's going on here? See:for example "2882 and run 297" on page 31. Like huh? My first guess is that the distance calculation is off somehow? Similar page 33 and probably many more. Even worse is page 34: "event 30 and run 297"! -> Yeah, as it turns out the problem was just that our
inRegionOfInterestcheck had become outdated due to our change of [ ]Select example events for each of the 'line veto kinds' to demonstrate their differences.
OC: Original Cluster (passing lnL cut on center chip)
HCL: Hypothetical Large Cluster (new cluster that OC is part of after
septemboard reco)
Regular:
is an example event in which we see the "regular" line veto without
using the septem veto. Things to note:
- the black circle shows the 'radius' of the OC, not the HLC
- the OC is actually part of a HLC
- because of this and because the HLC is a nice track, the event is vetoed, not by the green track, but by the HLC itself!
This wouldn't be a problem if we also used the septem veto, as this
event would already be removed due to the septem veto!
(More plots:  )
)
Regular no HLC:
The reference cluster to check for is still the regular OC with the
same radius. And again the OC is part of an HLC. However, in contrast
to the 'regular' case, this event is not vetoed. The green and purple
clusters simply don't point at the black circle and the HLC itself is
not considered here. This defines the 'regular no HLC' veto.
is just an example of an event that proves the method works & a nice
example of a cluster barely hitting the radius of the OC.
On the other hand though this is also a good example for why we should
have an eccentricity cut on those clusters that we use to check for
lines! The green cluster in this second event is not even remotely
eccentric enough and indeed is actually part of the orange track!
(More plots:
 )
)
Check HLC cluster:
Is an example event where we can see how ridiculous the "check HLC"
veto kind can become. There is a very large cluster that the OC is
actually part of (in red). But because of that the radius is SO
LARGE that it even encapsulates a whole other cluster (that
technically should ideally be part of the 'lower' of the tracks!).
For this reason I don't think this method is particularly useful. In
other events of course it looks more reasonable, but still. There
probably isn't a good way to make this work reliably. In any case
though, for events that are significant in size, they would almost
certainly never pass any lnL cuts anyhow.
(More plots:
 )
)
The following is a broken event. THe purple cluster is not used for line veto. Why? /t/problemevent12435run297.pdf
[X]Implement a cutoff for the eccentricity that a cluster must have in order to partake in the line veto. Currently this can only be set via an environment variable (ECC_LINE_VETO_CUT). A good value is around the 1.4 - 1.6 range I think (anything that rules out most X-ray like clusters!)
6.5.1. Note on real septemboard spacing being important extended
is an example event that shows we need to introduce the correct chip
spacing for the line veto. For the septem veto it's not very
important, because the distance is way more important than the angle
of how things match up. But for the line veto it's essential, as can
be seen in that example (note that it uses lvRegularNoHLC and no
septem veto, i.e. that's why the veto is false, despite the purple HLC of
course "hitting" the original cluster)
-> This has been implemented now. Activated (for now) via an
environment variable USE_REAL_LAYOUT.
An example event for the spacing & the eccentricity cutoff is:
file:///home/basti/org/Figs/statusAndProgress/exampleEvents/example_event_with_line_spacing_and_ecc_cutoff.pdf
which was generated using:
LINE_VETO_KIND=lvRegularNoHLC \ ECC_LINE_VETO_CUT=1.6 \ USE_REAL_LAYOUT=true \ likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/lhood_2018_crAll_80eff_line.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --lineveto --plotseptem
and then just extract it from the /plots/septemEvents
directory. Note the definition of the environment variables like this!
6.5.2. Outdated: Estimation using subset of outer ring events
The text here was written when we were still bootstrapping events only from the subset of event numbers that actually have a cluster that passes lnL on the center chip. This subset is of course biased even on the outer chip. Assuming that center clusters often come with activity on the outer chips, means there are less events representing those cases where there isn't even any activity in the center. This over represents activity on the outer chip.
likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/lhood_2018_crAll_80eff_septem_fake.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --septemveto --estimateRandomCoinc
which writes the file /tmp/septem_fake_veto.txt, which for this case
is found
./../resources/septem_veto_random_coincidences/estimates_septem_veto_random_coincidences.txt
Mean value of: 1522.61764706.
From this file the method seems to remove typically a bit less than 500 out of 2000 bootstrapped fake events. This seems to imply a random coincidence rate of almost 25% (or effectively a reduction of further 25% in efficiency / 25% increase in dead time). Pretty scary stuff.
Of course this does not even include the line veto, which will drop it further. Let's run that:
likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/lhood_2018_crAll_80eff_septem_line_fake.h5 \ --region crAll --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --septemveto --lineveto --estimateRandomCoinc
which generated the following output: ./../resources/septem_veto_random_coincidences/estimates_septem_line_veto_random_coincidences.txt
Mean value of: 1373.70588235.
This comes out to a fraction of 68.68% of the events left after running the vetoes on our bootstrapped fake events. Combining it with a software efficiency of ε = 80% the total combined efficiency then would be \(ε_\text{total} = 0.8 · 0.6868 = 0.5494\), so about 55%.
7. Application of CDL data to analysis
Relevant PR: https://github.com/Vindaar/TimepixAnalysis/pull/37
All calculations and plots above so far are done with the CDL data obtained in 2014. This imposes many uncertainties on those results and is one of the reasons the vetoes explained above were only implemented so far, but are not very refined yet. Before these shortcomings are adressed then, the new CDL data should be used as the basis for the likelihood method.
The idea behind using the CDL data as reference spectra is quite simple. One starts with the full spectrum of each target / filter combination of the data. From this two different "datasets" are created:
7.1. CDL calibration file
The CDL calibration file simply contains all reconstructed clusters from the CDL runs sorted by target / filter combinations.
The only addition to that is the calculation of the likelihood value dataset. For an explanation on this, see the 7.3 section below.
7.2. X-ray reference file
This file contains our reference spectra stored as histograms. We take each target / filter combination from the above file. Then we apply the following cuts:
- cluster center in silver region (circle around chip center with \(\SI{4.5}{\mm}\) radius)
- cut on transverse RMS, see below
- cut on length, see below
- cut on min number of pixels, at least 3
- cut on total charge, see below
where the latter 4 cuts depend on the energy. The full table is shown in tab. 14.
NOTE: Due to a bug in the implementation of the total charge calculation the charge values here are actually off by about a factor of 2! New values have yet to be calculated by redoing the CDL charge reconstruction and fits.
| Target | Filter | HV / \si{\kV} | Qmin / \(e^-\) | Qmax / \(e^-\) | length / mm | rmsT,min | rmsT,max |
|---|---|---|---|---|---|---|---|
| Cu | Ni | 15 | \num{5.9e5} | \num{1.0e6} | 7.0 | 0.1 | 1.1 |
| Mn | Cr | 12 | \num{3.5e5} | \num{6.0e5} | 7.0 | 0.1 | 1.1 |
| Ti | Ti | 9 | \num{2.9e5} | \num{5.5e5} | 7.0 | 0.1 | 1.1 |
| Ag | Ag | 6 | \num{2.0e5} | \num{4.0e5} | 7.0 | 0.1 | 1.1 |
| Al | Al | 4 | \num{5.9e4} | \num{2.1e5} | 7.0 | 0.1 | 1.1 |
| Cu | EPIC | 2 | \num{1.3e5} | \num{7.0e4} | 7.0 | 0.1 | 1.1 |
| Cu | EPIC | 0.9 | \num{3.0e4} | \num{8.0e4} | 7.0 | 0.1 | 1.1 |
| C | EPIC | 0.6 | \num{ 0.0} | \num{5.0e4} | 6.0 |
| Target | Filter | HV / \si{\kV} | Qcenter / \(e^-\) | Qsigma / \(e^-\) | length / mm | rmsT,min | rmsT,max |
|---|---|---|---|---|---|---|---|
| Cu | Ni | 15 | \num{6.63e5} | \num{7.12e4} | 7.0 | 0.1 | 1.1 |
| Mn | Cr | 12 | \num{4.92e5} | \num{5.96e4} | 7.0 | 0.1 | 1.1 |
| Ti | Ti | 9 | \num{4.38e5} | \num{6.26e4} | 7.0 | 0.1 | 1.1 |
| Ag | Ag | 6 | \num{2.90e5} | \num{4.65e4} | 7.0 | 0.1 | 1.1 |
| Al | Al | 4 | \num{1.34e5} | \num{2.33e4} | 7.0 | 0.1 | 1.1 |
| Cu | EPIC | 2 | \num{7.76e4} | \num{2.87e4} | 7.0 | 0.1 | 1.1 |
| Cu | EPIC | 0.9 | \num{4.17e4} | \num{1.42e4} | 7.0 | 0.1 | 1.1 |
| C | EPIC | 0.6 | \num{ 0.0} | \num{1.31e4} | 6.0 |
After these cuts are applied and all clusters are thrown out, which do not pass these cuts, histograms are calculated for all properties according to the binnings shown in tab. 11.
| name | bins | min | max |
|---|---|---|---|
| skewnessLongitudinal | 100 | \num{-5.05} | \num{4.85} |
| skewnessTransverse | 100 | \num{-5.05} | \num{4.85} |
| rmsTransverse | 150 | \num{-0.0166667} | \num{4.95} |
| eccentricity | 150 | \num{0.97} | \num{9.91} |
| hits | 250 | \num{-0.5} | \num{497.5} |
| kurtosisLongitudinal | 100 | \num{-5.05} | \num{4.85} |
| kurtosisTransverse | 100 | \num{-5.05} | \num{4.85} |
| length | 200 | \num{-0.05} | \num{19.85} |
| width | 100 | \num{-0.05} | \num{9.85} |
| rmsLongitudinal | 150 | \num{-0.0166667} | \num{4.95} |
| lengthDivRmsTrans | 150 | \num{-0.1} | \num{29.7} |
| rotationAngle | 100 | \num{-0.015708} | \num{3.09447} |
| energyFromCharge | 100 | \num{-0.05} | \num{9.85} |
| likelihood | 200 | \num{-40.125} | \num{9.625} |
| fractionInTransverseRms | 100 | \num{-0.005} | \num{0.985} |
| totalCharge | 200 | \num{-6250} | \num{2.48125e+06} |
7.3. Calculation of likelihood values
With both the calibration CDL file present and the X-ray reference file present, we can complete the process required to use the new CDL data for the analysis by calculating the likelihood values for all clusters found in the calibration CDL file.
This works according to the following idea:
- choose the correct energy bin for a cluster (its energy is calculated from the total charge) according to tab. 12 and get its X-ray reference histogram
- calculate log likelihood value for the clusters eccentricity under the reference spectrum
- add logL value for length / RMS transverse
- add logL value for fraction in transverse RMS
- invert value
where the likelihood is just calculated according to (ref: https://github.com/Vindaar/seqmath/blob/master/src/seqmath/smath.nim#L845-L867)
proc likelihood(hist: seq[float], val: float, bin_edges: seq[float]): float = let ind = bin_edges.lowerBound(val).int if ind < hist.len: result = hist[ind].float / hist.sum.float else: result = 0 proc logLikelihood(hist: seq[float], val: float, bin_edges: seq[float]): float = let lhood = likelihood(hist, val, bin_edges) if lhood <= 0: result = NegInf else: result = ln(lhood)
| Target | Filter | HV / \si{\kV} | min Energy / \si{\keV} | max Energy / \si{\keV} |
|---|---|---|---|---|
| Cu | Ni | 15 | 6.9 | ∞ |
| Mn | Cr | 12 | 4.9 | 6.9 |
| Ti | Ti | 9 | 3.2 | 4.9 |
| Ag | Ag | 6 | 2.1 | 3.2 |
| Al | Al | 4 | 1.2 | 2.1 |
| Cu | EPIC | 2 | 0.7 | 1.2 |
| Cu | EPIC | 0.9 | 0.4 | 0.7 |
| C | EPIC | 0.6 | 0.0 | 0.4 |
The result is a likelihood dataset for each target filter combination. This is now the foundation to determine the cut values on the logL values we wish to use for one energy bin. But for that we do not wish to use the raw likelihood dataset obviously. Instead we apply both the cuts previously mentioned which are used to generate the X-ray reference spectra tab. 14 and in addition some more cuts, which filter out some more unphysical single clusters, see tab. 15.
All clusters, which pass these combined cuts cuts are added to our likelihood distriution for each target filter combination.
These are then binned to 200 bins in a range from \(\numrange{0.0}{30.0}\) for the logL values. Finally the cut value is determine by demanding an \(\SI{80}{\percent}\) software efficiency. Our assumption is that for each target filter combination the distribution created by the listed cuts is essentially "background free". Then the signal efficiency is simply the ratio of accepted values divided by the total number of entries in the histogram. The actual calculation is:
proc determineCutValue(hist: seq[float], efficiency: float): int = var cur_eff = 0.0 last_eff = 0.0 let hist_sum = hist.sum.float while cur_eff < efficiency: inc result last_eff = cur_eff cur_eff = hist[0..result].sum.float / hist_sum
where the input is the described cleaned likelihood histogram and the result is the bin index corresponding to an \(\SI{80}{\percent}\) signal efficiency below the index (based on the fact that we accept all values smaller than the logL value corresponding to that index in the likelihood distribution). Relevant code: https://github.com/Vindaar/TimepixAnalysis/blob/master/Analysis/ingrid/likelihood.nim#L200-L211
With these in place the usage of the 2019 CDL data is done.
The background rate for the gold region comparing the 2014 CDL data with the 2019 CDL data then is shown in fig. 62
As can be seen the behavior of the background rate for the 2019 CDL data is somewhat smoother, while roughly the same background rate is recovered.
7.4. 2014 CDL Dataset description
The original calibration CDL file used by Christoph (only converted to H5 from the original ROOT file) is found at:
./../../CastData/ExternCode/TimepixAnalysis/resources/calibration-cdl.h5
and the X-ray reference file:
./../../CastData/ExternCode/TimepixAnalysis/resources/XrayReferenceDataSet.h5
Using the ./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/cdl_spectrum_creation.nim tool we can recreate this file from the raw data.
However, in order to do that, we need access to the raw data of the 2014 CDL runs and know which of those runs belongs to which target filter combination.
Fortunately, we have access to both the .xlsx file describing the different runs during the CDL data taking: ./../../CastData/ExternCode/TimepixAnalysis/resources/CDL-Apr14-D03-W0063-Runlist_stick.xlsx or https://github.com/Vindaar/TimepixAnalysis/blob/master/resources/CDL-Apr14-D03-W0063-Runlist_stick.xlsx.
By looking both at the "Comment" column and the "Run ok" column as well as the names of the runs, we can gleam first insights into which runs are used.
A different way to look at it (a good cross check) is to use Christoph's actual folders from his computer. A copy is found in: ./../../../../mnt/1TB/CAST/CDL-reference/ and each subdirectory for each target filter combination contains symbolic links to the used runs (from which we can determine the run number):
cd /mnt/1TB/CAST/CDL-reference for f in *kV; do ls $f/reco/*.root; done
Based on these two approaches the accepted runs were sorted by their target filter combination and live here: ./../../../../mnt/1TB/CAST/2014_15/CDL_Runs_raw/
cd /mnt/1TB/CAST/2014_15/CDL_Runs_raw #for f in *kV; do ls -lh $f; done tree -d -L 2
These directories can then be easily used to recreate the
calibration-cdl.h5 file and XrayReferenceFile.h5.
Finally, in order to actually create the files using
cdl_spectrum_creation.nim, we need a table that contains each run
number and the corresponding target filter kind, akin to the following
file for the 2019 CDL data:
https://github.com/Vindaar/TimepixAnalysis/blob/master/resources/cdl_runs_2019.org
Let's create such a file from the above mentioned directories. The
important thing is to use the exact same layout as the
cdl_runs_2019.org file so that we don't have to change the parsing
depending on the year of CDL data.
UPDATE: Indeed, Hendrik already created such a file and sent it to me. It now lives at ./../../CastData/ExternCode/TimepixAnalysis/resources/cdl_runs_2014.html
The code here will remain as a way to generate that file (although it's not actually done).
import os, sequtils, strutils, strformat import ingrid / cdl_spectrum_creation const path = "/mnt/1TB/CAST/2014_15/CDL_Runs_raw" proc readDir(path: string): seq[string] = ## reads a calibration-cdl-* directory and returns a sequence of correctly ## formatted lines for the resulting Org table for (pc, path) in walkDir(path): echo path var lines: seq[string] for (pc, path) in walkDir(path): case pc of pcDir: let dirName = extractFilename path if dirName.startsWith "calibration-cdl": lines.add readDir(path) else: discard
Now we need to generate a H5 file that contains all calibration runs, which we can use as a base.
So first let's create links for all runs:
cd /mnt/1TB/CAST/2014_15/CDL_Runs_raw mkdir all_cdl_runs cd all_cdl_runs # generate symbolc links to all runs for dir in ../calibration-cdl-apr2014-*; do for f in $dir/*; do ln -s $f `basename $f`; done; done
And now run through raw + reco:
raw_data_manipulation all_cdl_runs --runType xray --out calibration-cdl-apr2014_raw.h5 --ignoreRunList reconstruction calibration-cdl-apr2014_raw.h5 --out calibration-cdl-apr2014_reco.h5 reconstruction calibration-cdl-apr2014_reco.h5 --only_charge reconstruction calibration-cdl-apr2014_reco.h5 --only_gas_gain reconstruction calibration-cdl-apr2014_reco.h5 --only_energy_from_e
Now we're done with our input file for the CDL creation.
cdl_spectrum_creation calibration-cdl-apr2014_reco.h5 --cutcdl cdl_spectrum_creation calibration-cdl-apr2014_reco.h5 --genCdlFile --year=2014 cdl_spectrum_creation calibration-cdl-apr2014_reco.h5 --genRefFile --year=2014
And that's it.
7.5. Comment on confusion between CDL / Ref file & cuts
This section is simply a comment on the relation between the CDL data file, the X-ray reference file and the different cuts, because every time I don't look at this for a while I end up confused again.
Files:
- calibration CDL data file /
calibration-cdl.h5/cdlFile - Xray reference file /
XrayReferenceFile.h5/refFile
Cuts:
- X-ray cleaning cuts /
getXrayCleaningCuts, tab. 15 - CDL reference cuts /
getEnergyBinMinMaxVals201*, tab. 14.
Usage in likelihood.nim:
buildLogLHist: receives bothcdlFileandrefFile. However:refFileis only used to callcalcLikelihoodDatasetas a fallback in the case where thecdlFilegiven does not yet havelogLvalues computed (which only happens when the CDL file is first generated fromcdl_spectrum_generation.nim). ThebuildLogLHistprocedure builds two sequences:- the logL values of all clusters of the CDL data for one target/filter combination which pass both sets of cuts mentioned above.
- the corresponding energy values of these clusters.
calcCutValueTab: receives bothcdlFileandrefFile. Computes the cut values used for each target/filter combination (or once morphing is implemented for each energy). The procedure usesbuildLogLHistto get all valid clusters (that pass both of the above mentioned cuts!) and computes the histogram of those values. These are then the logL distributions from which a cut value is computed by looking for the ε (default 80%) value in the CDF (cumulative distribution function).calcLogLikelihood: receives bothcdlFileandrefFile. It computes the actual logL values of each cluster in the input file to which the logL cut is to be applied. CallscalcLikelihoodDatasetsinternally, which actually uses therefFileas well aswriteLogLDsetAttributeswriteLogLDsetAttributes: takes bothcdlFileandrefFileand simply adds the names of usedcdlFileandrefFileto the input H5 file.calcLikelihoodDataset: only takes therefFile. Computes the logL value for each cluster in the input H5 file.calcLikelihoodForEvent: takes indirectly therefFile(as data fromcalcLikelihoodDataset). Computes the logL value for each cluster explicitly.filterClustersByLogL: takes bothcdlFileandrefFile. Performs the application of the logL cuts. Mainly callscalcCutValueTaband uses it to perform the filtering (plus additional vetoes etc.)
All of this implies the following:
- The
refFileis only used to compute thelogLvalues for each cluster. That's what's meant by reference distribution. It only considers the CDL cuts, i.e. cuts to clean out unlikely to be X-rays from the set of CDL data by filtering to the peaks of the CDL data. - The
cdlFileis used to compute the logL distributions and their cut values for each target/filter combination. It uses both sets of cuts. The logL distributions, which are used to determine the ε efficiency are from thecdlFile!
| File | Uses X-ray cleaning cuts | Uses CDL reference cuts | Purpose |
|---|---|---|---|
| refFile | false | true | used to compute the logL values of each cluster |
| cdlFile | true | true | used to compute the logL distributions, which are then used |
| to compute the cut values given a certain signal efficiency | |||
| to be used to decide if a given input cluster is Xray like or not |
In a sense we have the following branching situation:
- CDL raw data:
- -> CDL cuts -> binning by pre determined bin edges -> X-ray reference spectra. Used to compute logL values of each cluster, because each spectrum (for each observable) is used to determine the likelihood value of each property.
-> CDL cuts + X-ray cleaning cuts -> gather logL values of all clusters passing these cuts (logL values are also computed using reference spectra above!) and bin by:
- 200 bins in (0, 30) (logL value)
to get the logL distributions. Look at CDF of logL distributions to determine cut values requiring a specific signal efficiency (by default 80%).
7.5.1. What does this imply for section 22 on CDL morphing?
It means the morphing we computed in cdlMorphing.nim in practice
does not actually have to be applied to the studied distributions, but
rather to those with the CDL cuts applied!
This is a bit annoying to be honest.
To make this a bit more palateable, let's extract the buildLogLHist
procedure into its own module, so that we can easier check what the
data looks like in comparison to the distributions we looked at
before.
NOTE: Ok, my brain is starting to digest what this really implies. Namely it means that the interpolation has to be done in 2 stages.
- interpolate the X-ray reference spectra in the same as we have implemented for the CDL morphing and compute each clusters logL value based on that interpolation.
- Perform not an interpolation on the logL input variables (eccentricity, …) but on the final logL distributions.
In a sense one could do either of these independently. Number 2 seems
easier to implement, because it applies the interpolation logic to the
logL histograms in buildLogLHisto directly and computes the cut
values from each interpolated distribution.
Then in another step we can later perform the interpolation of the
logL variable distributions as done while talking about the CDL
morphing in the first place. For that we have to modify
calcLikelihoodForEvent (and parents of course) to not receive a
tuple[ecc, ldivRms, fracRms: Table[string: histTuple]] but rather a
more abstract interpolator type that stores internally a number
(~1000) different, morphed distributions for each energy and picks the
correct one when asked in the call to logLikelihood.
The thing that makes number 1 so annoying is that it means the logL
dataset not only for the actual datafiles, but also for the
calibration-cdl.h5 file need to be recomputed.
7.5.2. Applying interpolation to logL distributions
Before we apply any kind of interpolation, it seems important to
visualize what the logL distributions actually look like again.
See the discussion in sec. 22.5.
7.6. Explanation of CDL datasets for Klaus
This section contains an explanation I wrote for Klaus trying to clarify what the difference between all the different datasets and cuts is.
7.6.1. Explanation for Klaus: CDL data and reference spectra
One starts from the raw data taken in the CAST detector lab. After selecting the data runs that contain useful information we are left with what we will call "raw CDL data" in the following.
This raw CDL data is stored in a file called calibration-cdl.h5, a
HDF5 file that went through the general TPA pipeline so that all
clusters are selected, geometric properties and the energy for each
cluster are computed.
From this file we compute the so called "X-ray reference spectra". These reference spectra define the likelihood reference distributions for each observable:
- eccentricity
- cluster length / transverse RMS
- fraction of pixels in a circle of radius 'transverse RMS' around the cluster center
These spectra are stored in the XrayReferenceFile.h5 file.
This file is generated as follows:
- take the
calibration-cdl.h5file - apply the cuts of tab. 14 to filter clusters passing these cuts
- compute histograms of the remaining clusters according to predefined bin ranges and bin widths (based on Christoph's work)
| Target | Filter | HV / \si{\kV} | Qcenter / \(e^-\) | Qsigma / \(e^-\) | length / mm | rmsT,min | rmsT,max |
|---|---|---|---|---|---|---|---|
| Cu | Ni | 15 | \num{6.63e5} | \num{7.12e4} | 7.0 | 0.1 | 1.1 |
| Mn | Cr | 12 | \num{4.92e5} | \num{5.96e4} | 7.0 | 0.1 | 1.1 |
| Ti | Ti | 9 | \num{4.38e5} | \num{6.26e4} | 7.0 | 0.1 | 1.1 |
| Ag | Ag | 6 | \num{2.90e5} | \num{4.65e4} | 7.0 | 0.1 | 1.1 |
| Al | Al | 4 | \num{1.34e5} | \num{2.33e4} | 7.0 | 0.1 | 1.1 |
| Cu | EPIC | 2 | \num{7.76e4} | \num{2.87e4} | 7.0 | 0.1 | 1.1 |
| Cu | EPIC | 0.9 | \num{4.17e4} | \num{1.42e4} | 7.0 | 0.1 | 1.1 |
| C | EPIC | 0.6 | \num{ 0.0} | \num{1.31e4} | 6.0 |
These are the spectra we have looked at when talking about the CDL morphing.
This file however is not used to derive the actual logL distributions and therefore not to determine the cut values on said distributions.
Instead, to compute the logL distributions we take the
calibration-cdl.h5 file again.
This is now done as follows:
- make sure the
calibration-cdl.h5file already has logL values for each cluster computed. If not, use theXrayReferenceFile.h5to compute logL values for each cluster. - apply the cuts of tab. 14 to the clusters
(now we have selected the same clusters as contained in
XrayReferenceFile.h5) - in addition apply the cuts of tab. 15 to further remove clusters that could be background events in the raw CDL data. Note that some of these cuts overlap with the previous cuts. Essentially it's a slightly stricter cut on the transverse RMS and an additional cut on the cluster eccentricity.
- gather the logL values of all remaining clusters
- compute a histogram given:
- 200 bins in the range (0, 30) of logL values
The resulting distribution is the logL distribution that is then
used to compute a cut value for a specified signal efficiency by
scanning the CDF for the corresponding value.
| Target | Filter | line | HV | length | rmsTmin | rmsTmax | eccentricity |
|---|---|---|---|---|---|---|---|
| Cu | Ni | \(\ce{Cu}\) \(\text{K}_{\alpha}\) | 15 | 0.1 | 1.0 | 1.3 | |
| Mn | Cr | \(\ce{Mn}\) \(\text{K}_{\alpha}\) | 12 | 0.1 | 1.0 | 1.3 | |
| Ti | Ti | \(\ce{Ti}\) \(\text{K}_{\alpha}\) | 9 | 0.1 | 1.0 | 1.3 | |
| Ag | Ag | \(\ce{Ag}\) \(\text{L}_{\alpha}\) | 6 | 6.0 | 0.1 | 1.0 | 1.4 |
| Al | Al | \(\ce{Al}\) \(\text{K}_{\alpha}\) | 4 | 0.1 | 1.1 | 2.0 | |
| Cu | EPIC | \(\ce{Cu}\) \(\text{L}_{\alpha}\) | 2 | 0.1 | 1.1 | 2.0 | |
| Cu | EPIC | \(\ce{O }\) \(\text{K}_{\alpha}\) | 0.9 | 0.1 | 1.1 | 2.0 | |
| C | EPIC | \(\ce{C }\) \(\text{K}_{\alpha}\) | 0.6 | 6.0 | 0.1 | 1.1 |
These logL distributions are shown in fig. 63.
So in the end linear interpolation had to be implemented in 2 different places:
- between the different distributions of the reference spectra for all three logL variables
- between the different logL distributions
7.6.2. Aside: Fun bug
The following plot cost me a few hours of debugging:
7.7. Extraction of CDL data to CSV
For Tobi I wrote a mini script ./../../CastData/ExternCode/TimepixAnalysis/Tools/cdlH5ToCsv.nim, which extracts the CDL data (after the CDL cuts are applied, i.e. "cleaning cuts") and stores them in CSV files.
These are the datasets as they are created in
cdl_spectrum_creation.nim in cutAndWrite after:
let passIdx = cutOnProperties(h5f, grp, cut.cutTo, ("rmsTransverse", cut.minRms, cut.maxRms), ("length", 0.0, cut.maxLength), ("hits", cut.minPix, Inf), ("eccentricity", 0.0, cut.maxEccentricity))
8. FADC
For FADC info see the thesis.
FADC manual: https://archive.org/details/manualzilla-id-5646050/
and
8.1. Pedestal [/]
Initially the pedestal data was used from the single pedestal run we took before the first CAST data taking.
The below was initially written for the thesis.
[ ]INSERT PLOTS OF COMPARISON OF OLD PEDESTAL AND NEW PEDESTAL!!!
8.2. Rise and fall times of data
Let's look at the rise and fall times of FADC data comparing the 55Fe data with background data to understand where one might put cuts. In sec. 6.1 we already looked at this years ago, but for the thesis we need new plots that are reproducible and verify the cuts we use make sense (hint: they don't).
The following is just a small script to generate plots comparing these.
import nimhdf5, ggplotnim import std / [strutils, os, sequtils, sets, strformat] import ingrid / [tos_helpers, ingrid_types] import ingrid / calibration / [calib_fitting, calib_plotting] import ingrid / calibration proc plotFallTimeRiseTime(df: DataFrame, suffix: string, riseTimeHigh: float) = ## Given a full run of FADC data, create the ## Note: it may be sensible to compute a truncated mean instead # local copy filtered to maximum allowed rise time let df = df.filter(f{`riseTime` <= riseTimeHigh}) proc plotDset(dset: string) = let dfCalib = df.filter(f{`Type` == "⁵⁵Fe"}) echo "============================== ", dset, " ==============================" echo "Percentiles:" echo "\t 1-th: ", dfCalib[dset, float].percentile(1) echo "\t 5-th: ", dfCalib[dset, float].percentile(5) echo "\t50-th: ", dfCalib[dset, float].percentile(50) echo "\t mean: ", dfCalib[dset, float].mean echo "\t95-th: ", dfCalib[dset, float].percentile(95) echo "\t99-th: ", dfCalib[dset, float].percentile(99) ggplot(df, aes(dset, fill = "Type")) + geom_histogram(position = "identity", bins = 100, hdKind = hdOutline, alpha = 0.7) + ggtitle(&"FADC signal {dset} in ⁵⁵Fe vs background data in $#" % suffix) + xlab(dset & " [ns]") + ggsave(&"Figs/statusAndProgress/FADC/fadc_{dset}_signal_vs_background_$#.pdf" % suffix) ggplot(df, aes(dset, fill = "Type")) + geom_density(normalize = true, alpha = 0.7, adjust = 2.0) + ggtitle(&"FADC signal {dset} in ⁵⁵Fe vs background data in $#" % suffix) + xlab(dset & " [ns]") + ggsave(&"Figs/statusAndProgress/FADC/fadc_{dset}_kde_signal_vs_background_$#.pdf" % suffix) plotDset("fallTime") plotDset("riseTime") when false: let dfG = df.group_by("runNumber").summarize(f{float: "riseTime" << truncMean(col("riseTime").toSeq1D, 0.05)}, f{float: "fallTime" << truncMean(col("fallTime").toSeq1D, 0.05)}) ggplot(dfG, aes(runNumber, riseTime, color = fallTime)) + geom_point() + ggtitle("Comparison of FADC signal rise times in ⁵⁵Fe data for all runs in $#" % suffix) + ggsave("Figs/statusAndProgress/FADC/fadc_mean_riseTime_$#.pdf" % suffix) ggplot(dfG, aes(runNumber, fallTime, color = riseTime)) + geom_point() + ggtitle("Comparison of FADC signal fall times in ⁵⁵Fe data for all runsin $#" % suffix) + ggsave("Figs/statusAndProgress/FADC/fadc_mean_fallTime_$#.pdf" % suffix) template toEDF*(data: seq[float], isCumSum = false): untyped = ## Computes the EDF of binned data var dataCdf = data if not isCumSum: seqmath.cumsum(dataCdf) let integral = dataCdf[^1] let baseline = min(data) # 0.0 dataCdf.mapIt((it - baseline) / (integral - baseline)) import numericalnim / interpolate import arraymancer proc plotROC(dfB, dfC: DataFrame, suffix: string) = # 1. compute cumulative sum from each type of data that is binned in the same way # 2. plot cumsum, (1 - cumsum) when false: proc toInterp(df: DataFrame): InterpolatorType[float] = let data = df["riseTime", float].toSeq1D.sorted let edf = toEdf(data) ggplot(toDf(data, edf), aes("data", "edf")) + geom_line() + ggsave("/tmp/test_edf.pdf") result = newLinear1D(data, edf) let interpS = toInterp(dfC) let interpB = toInterp(dfB) proc doit(df: DataFrame) = let data = df["riseTime", float] let xs = linspace(data.min, data.max, 1000) let kde = kde(data) proc eff(data: seq[float], val: float, isBackground: bool): float = let cutIdx = data.lowerBound(val) result = cutIdx.float / data.len.float if isBackground: result = 1.0 - result let dataB = dfB["riseTime", float].toSeq1D.sorted let dataC = dfC["riseTime", float].toSeq1D.sorted var xs = newSeq[float]() var ysC = newSeq[float]() var ysB = newSeq[float]() var ts = newSeq[string]() for i in 0 ..< 200: # rise time xs.add i.float ysC.add dataC.eff(i.float, isBackground = false) ysB.add dataB.eff(i.float, isBackground = true) let df = toDf(xs, ysC, ysB) ggplot(df, aes("ysC", "ysB")) + geom_line() + ggtitle("ROC curve of FADC rise time cut (only upper), ⁵⁵Fe vs. background in $#" % suffix) + xlab("Signal efficiency [%]") + ylab("Background suppression [%]") + ggsave("Figs/statusAndProgress/FADC/fadc_rise_time_roc_curve.pdf", width = 800, height = 480) let dfG = df.gather(["ysC", "ysB"], "ts", "ys") ggplot(dfG, aes("xs", "ys", color = "ts")) + geom_line() + xlab("Rise time [clock cycles]") + ylab("Signal efficiency / background suppression [%]") + ggsave("Figs/statusAndProgress/FADC/fadc_rise_time_efficiencies.pdf", width = 800, height = 480) proc read(fname, typ: string, eLow, eHigh: float): DataFrame = var h5f = H5open(fname, "r") let fileInfo = h5f.getFileInfo() var peakPos = newSeq[float]() result = newDataFrame() for run in fileInfo.runs: if recoBase() & $run / "fadc" notin h5f: continue # skip runs that were without FADC var df = h5f.readRunDsets( run, #chipDsets = some((chip: 3, dsets: @["eventNumber"])), # XXX: causes problems?? Removes some FADC data # but not due to events! fadcDsets = @["eventNumber", "baseline", "riseStart", "riseTime", "fallStop", "fallTime", "minvals", "argMinval"] ) # in calibration case filter to if typ == "⁵⁵Fe": let xrayRefCuts = getXrayCleaningCuts() let cut = xrayRefCuts["Mn-Cr-12kV"] let grp = h5f[(recoBase() & $run / "chip_3").grp_str] let passIdx = cutOnProperties( h5f, grp, crSilver, # try cutting to silver (toDset(igRmsTransverse), cut.minRms, cut.maxRms), (toDset(igEccentricity), 0.0, cut.maxEccentricity), (toDset(igLength), 0.0, cut.maxLength), (toDset(igHits), cut.minPix, Inf), (toDset(igEnergyFromCharge), eLow, eHigh) ) let dfChip = h5f.readRunDsets(run, chipDsets = some((chip: 3, dsets: @["eventNumber"]))) let allEvNums = dfChip["eventNumber", int] let evNums = passIdx.mapIt(allEvNums[it]).toSet df = df.filter(f{int: `eventNumber` in evNums}) df["runNumber"] = run result.add df result["Type"] = typ echo result proc main(back, calib: string, year: int, energyLow = 0.0, energyHigh = Inf, riseTimeHigh = Inf ) = let is2017 = year == 2017 let is2018 = year == 2018 if not is2017 and not is2018: raise newException(IOError, "The input file is neither clearly a 2017 nor 2018 calibration file!") let yearToRun = if is2017: 2 else: 3 let suffix = "Run-$#" % $yearToRun var df = newDataFrame() let dfC = read(calib, "⁵⁵Fe", energyLow, energyHigh) let dfB = read(back, "Background", energyLow, energyHigh) plotROC(dfB, dfC, suffix) df.add dfC df.add dfB plotFallTimeRiseTime(df, suffix, riseTimeHigh) when isMainModule: import cligen dispatch main
UPDATE: See the subsection below for updated plots.
When looking at these fall and rise time plots:
we can clearly see there is something like a "background" or an offset that is very flat under both the signal and background data (in run 2 and 3).
Let's see what this might be using plotData looking at event
displays of clusters that pass the following requirements:
- X-ray cleaning cuts
- fall time < 400 (from there we clearly don't see anything that should be real in calibration data)
- energies around the escape peak (not strictly needed)
NOTE: This should not have been run with --chips 3!
plotData --h5file ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --runType rtCalibration \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --eventDisplay -1 \ --cuts '("rmsTransverse", 0.1, 1.1)' \ --cuts '("eccentricity", 0.0, 1.3)' \ --cuts '("energyFromCharge", 2.5, 3.5)' \ --cuts '("fadc/fallTime", 0.0, 400.0)' \ --region crSilver \ --applyAllCuts \ --septemboard
the --septemboard flag activates plotting of the full septemboard
layout with FADC data on the side.
See these events here:
By looking at them intently, we can easily recognize what the issue
is:
See for example fig.
looking at the
fallStop (not the fall time). It is at 820. What else
is at 820? The 0 register we set from the first few entries of the raw
data files!
Comparing this with other events in the file proves that this is indeed the reason. So time to fix the calculation of the rise and fall time by making it a bit more
import nimhdf5, ggplotnim import std / [strutils, os, sequtils] import ingrid / [tos_helpers, fadc_helpers, ingrid_types, fadc_analysis] proc stripPrefix(s, p: string): string = result = s result.removePrefix(p) proc plotIdx(df: DataFrame, fadcData: Tensor[float], idx: int) = let xmin = df["argMinval", int][idx] let xminY = df["minvals", float][idx] let xminlineX = @[xmin, xmin] # one point for x of min, max let fData = fadcData[idx, _].squeeze let xminlineY = linspace(fData.min, fData.max, 2) let riseStart = df["riseStart", int][idx] let fallStop = df["fallStop", int][idx] let riseStartX = @[riseStart, riseStart] let fallStopX = @[fallStop, fallStop] let baseline = df["baseline", float][idx] let baselineY = @[baseline, baseline] let df = toDf({ "x" : toSeq(0 ..< 2560), "baseline" : baseline, "data" : fData, "xminX" : xminlineX, "xminY" : xminlineY, "riseStart" : riseStartX, "fallStop" : fallStopX }) # Comparison has to be done by hand unfortunately let path = "/t/fadc_spectrum_baseline.pdf" ggplot(df, aes("x", "data")) + geom_line() + geom_point(color = color(0.1, 0.1, 0.1, 0.1)) + geom_line(aes = aes("x", "baseline"), color = "blue") + geom_line(data = df.head(2), aes = aes("xminX", "xminY"), color = "red") + geom_line(data = df.head(2), aes = aes("riseStart", "xminY"), color = "green") + geom_line(data = df.head(2), aes = aes("fallStop", "xminY"), color = "pink") + ggtitle("riseStart: " & $riseStart & ", fallStop: " & $fallStop) + ggsave(path) proc getFadcData(fadcRun: ProcessedFadcRun) = let ch0 = getCh0Indices() let fadc_ch0_indices = getCh0Indices() # we demand at least 4 dips, before we can consider an event as noisy n_dips = 4 # the percentile considered for the calculation of the minimum min_percentile = 0.95 numFiles = fadcRun.eventNumber.len var fData = ReconstructedFadcRun( fadc_data: newTensorUninit[float]([numFiles, 2560]), eventNumber: fadcRun.eventNumber, noisy: newSeq[int](numFiles), minVals: newSeq[float](numFiles) ) let pedestal = getPedestalRun(fadcRun) for i in 0 ..< fadcRun.eventNumber.len: let slice = fadcRun.rawFadcData[i, _].squeeze let data = slice.fadcFileToFadcData( pedestal, fadcRun.trigRecs[i], fadcRun.settings.postTrig, fadcRun.settings.bitMode14, fadc_ch0_indices ).data fData.fadc_data[i, _] = data.unsqueeze(axis = 0) fData.noisy[i] = data.isFadcFileNoisy(n_dips) fData.minVals[i] = data.calcMinOfPulse(min_percentile) let recoFadc = calcRiseAndFallTime( fData.fadcData, false ) let df = toDf({ "baseline" : recoFadc.baseline, "argMinval" : recoFadc.xMin.mapIt(it.float), "riseStart" : recoFadc.riseStart.mapIt(it.float), "fallStop" : recoFadc.fallStop.mapIt(it.float), "riseTime" : recoFadc.riseTime.mapIt(it.float), "fallTime" : recoFadc.fallTime.mapIt(it.float), "minvals" : fData.minvals }) for idx in 0 ..< df.len: plotIdx(df, fData.fadc_data, idx) sleep(1000) proc main(fname: string, runNumber: int) = var h5f = H5open(fname, "r") let fileInfo = h5f.getFileInfo() for run in fileInfo.runs: if run == runNumber: let fadcRun = h5f.readFadcFromH5(run) fadcRun.getFadcData() when isMainModule: import cligen dispatch main
Based on this we've now implemented the following changes:
- instead of median + 0.1 · max: truncated mean of 30-th to 95-th percentile
- instead of times to exact baseline, go to baseline - 2.5%
- do not compute threshold based on individual value, but on a moving average of window size 5
- Also: use all registers and do not set first two registers to 0!
These should fix the "offsets" seen in the rise/fall time histograms/kdes.
The actual spectra that come out of the code hasn't really changed in case it works (slightly more accurate baseline + rise/fall time not to baseline, but slightly below; but those are details), but the broken cases are now fixed.
An example event after the fixes is:
- EXPLANATION FOR FLAT BACKGROUND IN RISE / FALL TIME: The "dead" register causes our fall / rise time calculation to break! This leads to a 'background' of homogeneous rise / fall times -> THIS NEEDS TO BE FIXED FIRST!!
8.2.1. Updated look at rise/fall time data (signal vs background) after FADC fixes [/]
NOTE: The plots shown here are still not the final ones. More FADC
algorithm changes where done after, refer to
improved_rise_fall_algorithm plots with a 10percent_top_offset
suffix and sections below, in particular
sec. 8.3.
The 10 percent top offset was deduced from this section: 8.2.2.1.6.
Let's recompile and rerun the
/tmp/fadc_rise_fall_signal_vs_background.nim code.
We reran the whole analysis chain by doing:
cd $TPA/Analysis/ingrid ./runAnalysisChain -i ~/CastData/data \ --outpath ~/CastData/data \ --years 2017 --years 2018 \ --calib --back \ --reco
which regenerated all the files:
- ./../../CastData/data/CalibrationRuns2017_Reco.h5
- ./../../CastData/data/CalibrationRuns2018_Reco.h5
- ./../../CastData/data/DataRuns2017_Reco.h5
- ./../../CastData/data/DataRuns2018_Reco.h5
(the old ones have a suffix *_old_fadc_rise_fall_times)
For completeness sake, let's reproduce the old and the new plots together, starting with the old:
cd /tmp/ mkdir OldPlots cd OldPlots /tmp/fadc_rise_fall_signal_vs_background -b ~/CastData/data/DataRuns2017_Reco_old_fadc_rise_fall_time.h5 \ -c ~/CastData/data/CalibrationRuns2017_Reco_old_fadc_rise_fall_time.h5 \ --year 2017 /tmp/fadc_rise_fall_signal_vs_background -b ~/CastData/data/DataRuns2018_Reco_old_fadc_rise_fall_time.h5 \ -c ~/CastData/data/CalibrationRuns2018_Reco_old_fadc_rise_fall_time.h5 \ --year 2018 pdfunite /tmp/OldPlots/Figs/statusAndProgress/FADC/*.pdf /tmp/old_fadc_plots_rise_fall_time_signal_background.pdf
And now the new ones:
cd /tmp/ mkdir NewPlots cd NewPlots /tmp/fadc_rise_fall_signal_vs_background -b ~/CastData/data/DataRuns2017_Reco.h5 \ -c ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --year 2017 /tmp/fadc_rise_fall_signal_vs_background -b ~/CastData/data/DataRuns2018_Reco.h5 \ -c ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --year 2018 pdfunite /tmp/NewPlots/Figs/statusAndProgress/FADC/*.pdf /tmp/new_fadc_plots_rise_fall_time_signal_background.pdf
Holy fuck are the differences big!
Copied over to:
(and the individual plots as well, the old ones have the
*_with_offset suffix and the other ones no suffix).
Most impressive is the difference in the rise time
Rise time:
vs.
and
vs.
Fall time:
vs.
and
vs.
Two questions that come up immediately:
[X]How does the Run-2 data split up by the different FADC settings? -> See sec. [BROKEN LINK: sec:fadc:rise_time_different_fadc_amp_settings] for more.[ ]What are the peaks in the background data where we have super short rise times? I assume those are just our noise events? Verify!
The code above also produces data for the percentiles of the rise / fall time for the calibration data, which is useful to decide on the cut values.
For 2017:
============================== fallTime ============================== Percentiles: 1-th: 448.0 5-th: 491.0 95-th: 603.0 99-th: 623.0 ============================== riseTime ============================== Percentiles: 1-th: 82.0 5-th: 87.0 95-th: 134.0 99-th: 223.0
For 2018:
============================== fallTime ============================== Percentiles: 1-th: 503.0 5-th: 541.0 95-th: 630.0 99-th: 651.0 ============================== riseTime ============================== Percentiles: 1-th: 63.0 5-th: 67.0 95-th: 125.0 99-th: 213.0
Comparing these with the plots shows that the calculation didn't do anything too dumb.
So from these let's eye ball values of:
- rise time: 65 - 200
- fall time: 470 - 640
- Investigate peaks in FADC fall time < 200
The plots:
show small peaks in the background data at values below 200, more pronounced in the Run-2 data. My theory would be that these are noise events, but let's find out:
NOTE: This should not have been run with
--chips 3!plotData --h5file ~/CastData/data/DataRuns2017_Reco.h5 \ --runType rtBackground \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --eventDisplay -1 \ --cuts '("fadc/fallTime", 0.0, 200.0)' \ --region crSilver \ --applyAllCuts \ --septemboard
some of these generated events are found here:

There are a mix of the following events presents in this type of data:
- Events that saturate the FADC completely and resulting in a very sharp run back to the baseline. This is somewhat expected and fine. (e.g. page 2)
- pure real noise events based on extremely noisy activity on the septemboard (e.g. page 1). This is pretty irrelevant, as these Septemboard events will never be interesting for anything.
- regular Septemboard events with low frequency noise on the FADC
(e.g. page 3). These are problematic and we must make sure not to
apply the FADC veto for these. Fortunately they seem to be detected
correctly by the
noisyflag usually. Sometimes they are a bit higher frequency too (e.g. page 7, 12, …). - regular Septemboard events with very low frequency noise on the FADC, which does not trigger our noisy detection. (e.g. page 19, 39, …). These are very problematic and we need to fix the noise detection for these.
Takeaways from this: The noisy event detection actually works really well already! There are very few events in there that should be considered noisy, but are not!
- DONE Bug in properties on plots
[0/1]
Crap (fixed, see below):
page 23 (and page 42) is an interesting event of a very high energy detection on the septemboard with a mostly noise like signal in the FADC. HOWEVER the energy from charge, number of hits etc. properties DO NOT match what we see on the center chip! Not sure what's going on, but I assume we're dealing with a different cluster from the same event number? (Note: page e.g. 37 looks similar but has a reasonable energy! so not all events are problematic).
[X]Investigate raw data by hand first. Event number 23943, index 29975. -> Takeaway 1: event indices in plotData titles don't make sense. They are larger than the event numbers?! Mixing of indices over all runs? Or what. -> Takeaway 2: The entries in the rows of the raw data that match the event number printed on the side does match the numbers printed on the plot! So it seems like the data seen does not match the numbers. -> Takeaway 3:
- Chips 0, 1, 4, 5, 6 have no data for event number 23943
Chips 2 (idx 6481)), 3 (idx 6719) have data for event number
However, Chip 2 also only has 150 hits at index 6481 (2.41 keV)
This means there is no data at this event number on the whole chip that can explain the data. Is
inner_joinat fault here? :/ Orgroup_by? Uhhhhhhhhhhhhhhhhhhhhhhhhhhhhhhh I think I just figured it out. Ouch. It's just that the data does not match the event. The event "index" in the title is the event number nowadays! For some reason it gets screwed up for the annotations! The issue is likely that we simply walk through our cluster prop data index by index instead of making sure we get the index for the correct event number.
-> FIXED : Fixed by filtering to the event number manually (makes sure we get correct event number instead of aligning indices, even if latter is more efficient). If there are more than 1 cluster on the event, the properties of the cluster with the lowest lnL value is printed and a
numClusterfield is added that tells how many clusters found on the event.[ ]VERIFY SEPTEMBOARD EVENTS USED ELSEWHERE ABOVE HAVE CORRECT MATCHES!
8.2.2. Behavior of rise and fall time against energy
import nimhdf5, ggplotnim import std / [strutils, os, sequtils, sets, strformat] import ingrid / [tos_helpers, ingrid_types] import ingrid / calibration / [calib_fitting, calib_plotting] import ingrid / calibration proc plotFallTimeRiseTime(df: DataFrame, suffix: string, isCdl: bool) = ## Given a full run of FADC data, create the ## Note: it may be sensible to compute a truncated mean instead proc plotDset(dset: string) = for (tup, subDf) in groups(group_by(df, "Type")): echo "============================== ", dset, " ==============================" echo "Type: ", tup echo "Percentiles:" echo "\t 1-th: ", subDf[dset, float].percentile(1) echo "\t 5-th: ", subDf[dset, float].percentile(5) echo "\t50-th: ", subDf[dset, float].percentile(50) echo "\t mean: ", subDf[dset, float].mean echo "\t80-th: ", subDf[dset, float].percentile(80) echo "\t95-th: ", subDf[dset, float].percentile(95) echo "\t99-th: ", subDf[dset, float].percentile(99) df.writeCsv("/tmp/fadc_data_$#.csv" % suffix) #let df = df.filter(f{`Type` == "Cu-Ni-15kV"}) ggplot(df, aes(dset, fill = "Type")) + geom_histogram(position = "identity", bins = 100, hdKind = hdOutline, alpha = 0.7) + ggtitle(&"Comparison of FADC signal {dset} in ⁵⁵Fe vs background data in $#" % suffix) + ggsave(&"Figs/statusAndProgress/FADC/fadc_{dset}_energy_dep_$#.pdf" % suffix) ggplot(df, aes(dset, fill = "Type")) + geom_density(normalize = true, alpha = 0.7, adjust = 2.0) + ggtitle(&"Comparison of FADC signal {dset} in ⁵⁵Fe vs background data in $#" % suffix) + ggsave(&"Figs/statusAndProgress/FADC/fadc_{dset}_kde_energy_dep_$#.pdf" % suffix) let df = df.filter(f{`riseTime` < 200}) ggplot(df, aes(dset, fill = "Type")) + geom_histogram(position = "identity", bins = 100, hdKind = hdOutline, alpha = 0.7) + ggtitle(&"Comparison of FADC signal {dset} in ⁵⁵Fe vs background data in $#" % suffix) + ggsave(&"Figs/statusAndProgress/FADC/fadc_{dset}_energy_dep_less_200_rise_$#.pdf" % suffix) ggplot(df, aes(dset, fill = "Type")) + geom_density(normalize = true, alpha = 0.7, adjust = 2.0) + ggtitle(&"Comparison of FADC signal {dset} in ⁵⁵Fe vs background data in $#" % suffix) + ggsave(&"Figs/statusAndProgress/FADC/fadc_{dset}_kde_energy_dep_less_200_rise_$#.pdf" % suffix) if isCdl: let xrayRef = getXrayRefTable() var labelOrder = initTable[Value, int]() for idx, el in xrayRef: labelOrder[%~ el] = idx ggplot(df, aes(dset, fill = "Type")) + ggridges("Type", overlap = 1.5, labelOrder = labelOrder) + geom_density(normalize = true, alpha = 0.7, adjust = 2.0, color = "black") + ggtitle(&"Comparison of FADC signal {dset} in ⁵⁵Fe vs background data in $#" % suffix) + ggsave(&"Figs/statusAndProgress/FADC/fadc_{dset}_ridgeline_kde_energy_dep_less_200_rise_$#.pdf" % suffix) ggplot(df, aes(dset, fill = "Settings")) + geom_density(normalize = true, alpha = 0.7, adjust = 2.0, color = "black") + ggtitle(dset & " of different FADC settings used") + ggsave(&"Figs/statusAndProgress/FADC/fadc_{dset}_kde_different_fadc_ampb_settings_$#.pdf" % suffix) ggplot(df, aes(dset, fill = factor("runNumber"))) + geom_density(normalize = true, alpha = 0.7, adjust = 2.0, color = "black") + ggtitle(dset & " of different runs") + ggsave(&"Figs/statusAndProgress/FADC/fadc_{dset}_kde_different_runs_$#.pdf" % suffix) plotDset("fallTime") plotDset("riseTime") proc read(fname, typ: string, eLow, eHigh: float, isCdl: bool): DataFrame = var h5f = H5open(fname, "r") let fileInfo = h5f.getFileInfo() var peakPos = newSeq[float]() result = newDataFrame() for run in fileInfo.runs: if recoBase() & $run / "fadc" notin h5f: continue # skip runs that were without FADC var df = h5f.readRunDsets( run, fadcDsets = @["eventNumber", "baseline", "riseStart", "riseTime", "fallStop", "fallTime", "minvals", "noisy", "argMinval"] ) let xrayRefCuts = getXrayCleaningCuts() let runGrp = h5f[(recoBase() & $run).grp_str] let tfKind = if not isCdl: tfMnCr12 else: runGrp.attrs["tfKind", string].parseEnum[:TargetFilterKind]() let cut = xrayRefCuts[$tfKind] let grp = h5f[(recoBase() & $run / "chip_3").grp_str] let passIdx = cutOnProperties( h5f, grp, crSilver, # try cutting to silver (toDset(igRmsTransverse), cut.minRms, cut.maxRms), (toDset(igEccentricity), 0.0, cut.maxEccentricity), (toDset(igLength), 0.0, cut.maxLength), (toDset(igHits), cut.minPix, Inf), (toDset(igEnergyFromCharge), eLow, eHigh) ) let dfChip = h5f.readRunDsets(run, chipDsets = some((chip: 3, dsets: @["eventNumber"]))) let allEvNums = dfChip["eventNumber", int] let evNums = passIdx.mapIt(allEvNums[it]).toSet # filter to allowed events & remove any noisy events df = df.filter(f{int: `eventNumber` in evNums and `noisy`.int < 1}) df["runNumber"] = run if isCdl: df["Type"] = $tfKind df["Settings"] = "Setting " & $(@[80, 101, 121].lowerBound(run)) result.add df if not isCdl: result["Type"] = typ echo result proc main(fname: string, year: int, energyLow = 0.0, energyHigh = Inf, isCdl = false) = if not isCdl: var df = newDataFrame() df.add read(fname, "escape", 2.5, 3.5, isCdl = false) df.add read(fname, "photo", 5.5, 6.5, isCdl = false) let is2017 = year == 2017 let is2018 = year == 2018 if not is2017 and not is2018: raise newException(IOError, "The input file is neither clearly a 2017 nor 2018 calibration file!") let yearToRun = if is2017: 2 else: 3 let suffix = "run$#" % $yearToRun plotFallTimeRiseTime(df, suffix, isCdl) else: let df = read(fname, "", 0.0, Inf, isCdl = true) plotFallTimeRiseTime(df, "CDL", isCdl) when isMainModule: import cligen dispatch main
ntangle ~/org/Doc/StatusAndProgress.org && nim c -d:danger /t/fadc_rise_fall_energy_dep.nim ./fadc_rise_fall_energy_dep -f ~/CastData/data/CalibrationRuns2017_Reco.h5 --year 2017
Output for 2017:
============================== fallTime ==============================
Type: @[("Type", (kind: VString, str: "escape"))]
Percentiles:
1-th: 406.49
5-th: 476.0
50-th: 563.0
mean: 559.6049042145594
95-th: 624.0
99-th: 660.0
============================== fallTime ==============================
Type: @[("Type", (kind: VString, str: "photo"))]
Percentiles:
1-th: 462.0
5-th: 498.0
50-th: 567.0
mean: 561.2259466025087
95-th: 601.0
99-th: 616.0
============================== riseTime ==============================
Type: @[("Type", (kind: VString, str: "escape"))]
Percentiles:
1-th: 78.0
5-th: 84.0
50-th: 103.0
mean: 114.0039846743295
95-th: 177.0
99-th: 340.5100000000002
============================== riseTime ==============================
Type: @[("Type", (kind: VString, str: "photo"))]
Percentiles:
1-th: 83.0
5-th: 89.0
50-th: 104.0
mean: 107.4761626684731
95-th: 130.0
99-th: 196.0
Output for 2018:
./fadc_rise_fall_energy_dep -f ~/CastData/data/CalibrationRuns2018_Reco.h5 --year 2018
============================== fallTime ==============================
Type: @[("Type", (kind: VString, str: "escape"))]
Percentiles:
1-th: 456.0
5-th: 512.0
50-th: 585.0
mean: 582.0466121605112
95-th: 640.0
99-th: 677.6000000000004
============================== fallTime ==============================
Type: @[("Type", (kind: VString, str: "photo"))]
Percentiles:
1-th: 515.0
5-th: 548.0
50-th: 594.0
mean: 592.7100718941074
95-th: 629.0
99-th: 647.0
============================== riseTime ==============================
Type: @[("Type", (kind: VString, str: "escape"))]
Percentiles:
1-th: 60.0
5-th: 66.0
50-th: 86.0
mean: 96.70284747674091
95-th: 160.0
99-th: 309.2000000000007
============================== riseTime ==============================
Type: @[("Type", (kind: VString, str: "photo"))]
Percentiles:
1-th: 63.0
5-th: 68.0
50-th: 84.0
mean: 88.30370221057582
95-th: 118.0
99-th: 182.0
These values provide the reference to the estimation we will perform next.
- Looking at the CDL data rise / fall times
Time to look at the rise and fall time of the CDL data. We've added a filter for the events to not be noisy events (sec. 8.2.2.1.1).
./fadc_rise_fall_energy_dep -f ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --year 2019 \ --isCdl
(note that the year is irrelevant here)
============================fallTime============================Type: @[("Type", (kind: VString, str: "Ag-Ag-6kV"))] Percentiles: 1-th: 548.0 5-th: 571.0 50-th: 612.0 mean: 610.8296337402886 80-th: 631.0 95-th: 647.0 99-th: 660.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Al-Al-4kV"))] Percentiles: 1-th: 506.53 5-th: 538.0 50-th: 602.0 mean: 598.7740798747063 80-th: 629.0 95-th: 654.0 99-th: 672.0============================fallTime============================Type: @[("Type", (kind: VString, str: "C-EPIC-0.6kV"))] Percentiles: 1-th: 304.0 5-th: 357.0 50-th: 519.0 mean: 510.6200390370852 80-th: 582.0 95-th: 630.0 99-th: 663.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-0.9kV"))] Percentiles: 1-th: 365.35 5-th: 445.7 50-th: 556.0 mean: 549.2081310679612 80-th: 601.0 95-th: 637.0 99-th: 670.0599999999999============================fallTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-2kV"))] Percentiles: 1-th: 433.62 5-th: 487.0 50-th: 581.0 mean: 575.539179861957 80-th: 614.0 95-th: 651.0 99-th: 671.3800000000001============================fallTime============================Type: @[("Type", (kind: VString, str: "Cu-Ni-15kV"))] Percentiles: 1-th: 539.0 5-th: 575.0 50-th: 606.0 mean: 604.7243749086124 80-th: 618.0 95-th: 629.0 99-th: 640.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Mn-Cr-12kV"))] Percentiles: 1-th: 540.0 5-th: 568.0 50-th: 604.0 mean: 602.9526100904054 80-th: 620.0 95-th: 634.0 99-th: 646.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Ti-Ti-9kV"))] Percentiles: 1-th: 551.0 5-th: 575.0 50-th: 611.0 mean: 610.1495433789954 80-th: 627.0 95-th: 640.0 99-th: 655.0============================riseTime============================Type: @[("Type", (kind: VString, str: "Ag-Ag-6kV"))] Percentiles: 1-th: 61.0 5-th: 66.0 50-th: 84.0 mean: 84.54994450610432 80-th: 93.0 95-th: 105.0 99-th: 119.0============================riseTime============================Type: @[("Type", (kind: VString, str: "Al-Al-4kV"))] Percentiles: 1-th: 63.53 5-th: 70.0 50-th: 87.0 mean: 91.58535630383712 80-th: 103.0 95-th: 123.0 99-th: 146.0============================riseTime============================Type: @[("Type", (kind: VString, str: "C-EPIC-0.6kV"))] Percentiles: 1-th: 57.0 5-th: 63.0 50-th: 89.0 mean: 97.01626545217957 80-th: 113.0 95-th: 149.0 99-th: 184.6400000000001============================riseTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-0.9kV"))] Percentiles: 1-th: 59.0 5-th: 67.0 50-th: 89.0 mean: 96.92839805825243 80-th: 110.0 95-th: 138.0 99-th: 182.53============================riseTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-2kV"))] Percentiles: 1-th: 63.0 5-th: 71.0 50-th: 90.0 mean: 95.50669914738124 80-th: 109.0 95-th: 132.0 99-th: 166.3800000000001============================riseTime============================Type: @[("Type", (kind: VString, str: "Cu-Ni-15kV"))] Percentiles: 1-th: 61.0 5-th: 65.0 50-th: 82.0 mean: 84.01476824097091 80-th: 90.0 95-th: 99.0 99-th: 206.2399999999998============================riseTime============================Type: @[("Type", (kind: VString, str: "Mn-Cr-12kV"))] Percentiles: 1-th: 61.0 5-th: 65.0 50-th: 81.0 mean: 83.31525226013414 80-th: 89.0 95-th: 98.0 99-th: 185.4300000000003============================riseTime============================Type: @[("Type", (kind: VString, str: "Ti-Ti-9kV"))] Percentiles: 1-th: 63.0 5-th: 69.0 50-th: 85.0 mean: 87.13078930202218 80-th: 93.0 95-th: 105.0 99-th: 153.6899999999996we copy over the CSV file generated by the above command from
/tmp/fadc_data_CDL.csvto ./../resources/FADC_rise_fall_times_CDL_data.csv so that we can plot the positions separately in sec. 8.2.2.1.4.which produces the following plots:
- Look at Cu-EPIC-0.9kV events between rise 40-60
The runs for this target/filter kind are: 339, 340
Let's plot those events: NOTE: This should not have been run with
--chips 3!plotData --h5file ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --runType rtCalibration \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --cuts '("fadc/riseTime", 40, 60)' \ --applyAllCuts \ --runs 339 --runs 340 \ --eventDisplay \ --septemboard

So these events are essentially all just noise events! Which is a good point to add a
noisyfilter to the rise time plot!Considering how the rise times change with energy, it might after all be a good idea to have an energy dependent cut? Surprising because in principle we don't expect an energy dependence, but rather a dependence on absorption length! So AgAg should be less wide than TiTi!
- Look at C-EPIC-0.6kV rise time contribution in range: 110 - 130
Similar to the above case where we discovered the contribution of the noisy events in the data, let's now look at the contributions visible in the range 110 to 130 in the rise time in plot:
The runs for the C-EPIC 0.6kV dataset are: 342, 343
Generate the plots: NOTE: This should not have been run with
--chips 3!plotData --h5file ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --runType rtCalibration \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --cuts '("fadc/riseTime", 110, 130)' \ --applyAllCuts \ --runs 342 --runs 343 \ --eventDisplay \ --septemboard
which are found at:

Looking at them reveals two important aspects:
- there are quite a lot of double events where the signal is made significantly longer by a second X-ray, explaining the longer rise time in cases where the minimum shifts towards the right.
- The data was taken with an extremely high amplification and thus
there is significantly more noise on the baseline. In many cases
then what happens is that the signal is randomly a bit below the
baseline and the
riseStartappears a bit earlier, extending the distance to the minimum.
Combined this explains that the events visible there are mainly a kind of artifact, however not necessarily one we would be able to "deal with". Double hits in real data of course can be neglected, but the variations causing randomly longer rise times not.
However, it is important to realize that this case is not in any way practical for the CAST data, because we do not have an FADC trigger at those energies! Our trigger in the lowest of cases was at ~1.5 keV and later even closer to 2.2 keV. And we didn't change the gain (outside the specific cases where we adjusted due to noise).
As such we can ignore the contribution of that second "bump" and essentially only look at the "main peak"!
- Initial look at rise / fall times with weird
Next up we modified the code above to also work with the CDL data & split each run according to its target/filter kind. In
cutOnPropertieswe currently only use the X-ray cleaning cuts (which may not be ideal as we will see):./fadc_rise_fall_energy_dep -f ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 --year 2019 --isCdl
which generated:
with the following percentile outputs:
============================fallTime============================Type: @[("Type", (kind: VString, str: "Ag-Ag-6kV"))] Percentiles: 1-th: 378.05 5-th: 610.0 50-th: 656.0 mean: 649.087680355161 80-th: 674.0 95-th: 693.0 99-th: 707.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Al-Al-4kV"))] Percentiles: 1-th: 141.06 5-th: 510.9 50-th: 632.0 mean: 614.8747063429914 80-th: 663.0 95-th: 690.0 99-th: 714.0============================fallTime============================Type: @[("Type", (kind: VString, str: "C-EPIC-0.6kV"))] Percentiles: 1-th: 23.0 5-th: 26.3 50-th: 515.0 mean: 459.5428914217156 80-th: 595.0 95-th: 653.7 99-th: 687.3399999999999============================fallTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-0.9kV"))] Percentiles: 1-th: 22.0 5-th: 23.0 50-th: 541.0 mean: 431.9965601965602 80-th: 608.0 95-th: 658.0 99-th: 692.6600000000001============================fallTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-2kV"))] Percentiles: 1-th: 23.79 5-th: 361.0 50-th: 608.0 mean: 583.2463709677419 80-th: 650.0 95-th: 684.0 99-th: 711.21============================fallTime============================Type: @[("Type", (kind: VString, str: "Cu-Ni-15kV"))] Percentiles: 1-th: 367.28 5-th: 626.0 50-th: 656.0 mean: 649.8090364088317 80-th: 667.0 95-th: 679.0 99-th: 691.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Mn-Cr-12kV"))] Percentiles: 1-th: 520.5699999999999 5-th: 614.0 50-th: 652.0 mean: 646.8802857976086 80-th: 667.0 95-th: 682.0 99-th: 694.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Ti-Ti-9kV"))] Percentiles: 1-th: 438.62 5-th: 615.0 50-th: 654.0 mean: 649.1258969341161 80-th: 669.0 95-th: 685.0 99-th: 700.0============================riseTime============================Type: @[("Type", (kind: VString, str: "Ag-Ag-6kV"))] Percentiles: 1-th: 67.0 5-th: 77.0 50-th: 110.0 mean: 126.059748427673 80-th: 151.0 95-th: 234.0 99-th: 326.0============================riseTime============================Type: @[("Type", (kind: VString, str: "Al-Al-4kV"))] Percentiles: 1-th: 65.53 5-th: 77.0 50-th: 102.5 mean: 111.3120595144871 80-th: 130.0 95-th: 179.0 99-th: 244.2299999999982============================riseTime============================Type: @[("Type", (kind: VString, str: "C-EPIC-0.6kV"))] Percentiles: 1-th: 12.66 5-th: 62.3 50-th: 92.0 mean: 100.3239352129574 80-th: 121.0 95-th: 157.7 99-th: 204.6799999999998============================riseTime============================Type: @[("Type", (kind: VString, str: "C-EPIC-0.6kV"))] Percentiles: 1-th: 12.66 5-th: 62.3 50-th: 92.0 mean: 100.3239352129574 80-th: 121.0 95-th: 157.7 99-th: 204.6799999999998============================riseTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-0.9kV"))] Percentiles: 1-th: 43.34 5-th: 69.7 50-th: 92.0 mean: 102.62457002457 80-th: 115.0 95-th: 159.0 99-th: 234.6400000000003============================riseTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-2kV"))] Percentiles: 1-th: 52.79 5-th: 74.0 50-th: 104.0 mean: 109.2959677419355 80-th: 131.0 95-th: 175.0 99-th: 224.4200000000001============================riseTime============================Type: @[("Type", (kind: VString, str: "Cu-Ni-15kV"))] Percentiles: 1-th: 68.0 5-th: 79.0 50-th: 146.0 mean: 216.1468050884632 80-th: 374.0 95-th: 516.1000000000004 99-th: 600.0============================riseTime============================Type: @[("Type", (kind: VString, str: "Mn-Cr-12kV"))] Percentiles: 1-th: 67.0 5-th: 77.0 50-th: 109.0 mean: 125.1246719160105 80-th: 147.0 95-th: 230.0 99-th: 337.8600000000006============================riseTime============================Type: @[("Type", (kind: VString, str: "Ti-Ti-9kV"))] Percentiles: 1-th: 70.0 5-th: 81.0 50-th: 114.0 mean: 143.5371819960861 80-th: 167.0 95-th: 324.4499999999998 99-th: 549.3799999999992where we can mainly see that the 95-th percentile of the data is actually quite high in many cases (e.g. MnCr12kV is still "somewhat fine" at 230 for 95, but CuNi15kV is 516 and TiTi9 is 324!). Looking at the distributions of the rise times we see an obvious problem, namely rise times on the order of 350-500 in the CuNi15kV dataset! Question is what is that? Others also have quite a long tail. -> These were just an artifact of our old crappy way to compute rise times, fall times and baselines!
Let's plot the event displays of those CuNi events that are in that latter blob.I couldn't runplotDataas CDL data wasn't ran with modernreconstructionfor FADC yet. After rerunning that, these disappeared! The runs for CuNi15 are: 319, 320, 345 (rmsTransverse of CuNi dataset is interesting. Essentially just a linear increase up to1mm! [[file:~/org/Figs/statusAndProgress/FADC/old_rise_fall_algorithm/CDL_riseTime_fallTime/onlyCleaningCuts/rmsTransverse_run319 320 345_chip3_0.03_binSize_binRange-0.0_6.0_region_crSilver_rmsTransverse_0.1_1.0_eccentricity_0.0_1.3_toaLength_-0.0_20.0_applyAll_true.pdf]] run below with ~--ingridand onlyrmsTransverse+eccentricitycut):Important note: As of right now the CDL data still suffers from the FADC 0, 1 register = 0 bug! This will partially explain some "background" in the rise/fall times. UPDATE: Uhh, I reran the
--only_fadcoption ofreconstructionon the CDL H5 file and having done that the weird behavior of the additional peak at > 350 is completely gone. What did we fix in there again?- rise / fall time not to baseline, but to offset below
- based on moving average instead of single value
- different way to calculate baseline based on truncated mean
- Rise time and fall time plots of percentile values
With the file ./../resources/FADC_rise_fall_times_CDL_data.csv we can generate plots of the percentiles of each target/filter kind to have an idea where a cutoff for that kind of energy and absorption length might be:
import ggplotnim, xrayAttenuation import arraymancer except readCsv import std / strutils import ingrid / tos_helpers proc absLength(E: keV): float = let ar = Argon.init() let ρ_Ar = density(1050.mbar.to(Pascal), 293.K, ar.molarMass) result = absorptionLength(E, numberDensity(ρ_Ar, ar.molarMass), ar.f2eval(E).float).float let df = readCsv("/home/basti/org/resources/FADC_rise_fall_times_CDL_data.csv") var dfP = newDataFrame() let dset = "riseTime" let lineEnergies = getXrayFluorescenceLines() let invTab = getInverseXrayRefTable() for (tup, subDf) in groups(group_by(df, "Type")): let data = subDf[dset, float] var percs = newSeq[float]() var percName = newSeq[string]() proc percentiles(percs: var seq[float], percName: var seq[string], name: string, val: int) = percName.add name percs.add data.percentile(val) percs.percentiles(percName, "1-th", 1) percs.percentiles(percName, "5-th", 5) percs.percentiles(percName, "50-th", 50) percName.add "mean" percs.add data.mean percName.add "MPV" let kdeData = kde(data) let xs = linspace(min(data), max(data), 1000) percs.add(xs[kdeData.argmax(0)[0]]) percs.percentiles(percName, "80-th", 80) percs.percentiles(percName, "95-th", 95) percs.percentiles(percName, "99-th", 99) let typ = tup[0][1].toStr let E = lineEnergies[invTab[typ]].keV let absLength = absLength(E) dfP.add toDf({"Value" : percs, "Percentile" : percName, "Type" : typ, "Energy" : E.float, "λ" : absLength}) ggplot(dfP, aes("Type", "Value", color = "Percentile")) + geom_point() + ggsave("/tmp/fadc_percentiles_by_tfkind.pdf") proc filterPlot(to: string) = let dfF = dfP.filter(f{`Percentile` == to}) let title = if to == "mean": to else: to & " percentile" ggplot(dfF, aes("λ", "Value", color = "Type")) + geom_point() + ggtitle("$# of FADC rise time vs absorption length λ" % title) + ggsave("/tmp/fadc_$#_vs_absLength_by_tfkind.pdf" % to) filterPlot("95-th") filterPlot("80-th") filterPlot("mean") filterPlot("MPV")
- CDL rise / fall times after FADC algorithm updates
Let's apply that to the CDL data, plot some events with baseline, rise / fall lines and then look at distributions.
reconstruction -i ~/CastData/data/DataRuns2018_Reco.h5 --only_fadc
and plot some events:
plotData --h5file ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --runType rtCalibration \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --eventDisplay --septemboard
WAIT The below only implies something about our calculation of the minimum value of the FADC data (i.e. the
minvalsdataset) as we use that to draw the lines to! -> Fixed this in the plotting. However, another issue appeared: The lines for start and stop were exactly the same! ->findThresholdValuenow returns the start and stop parameters. -> looks much more reasonable now.New ridge line plots here we come:
./fadc_rise_fall_energy_dep -f ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --year 2019 \ --isCdl
============================fallTime============================Type: @[("Type", (kind: VString, str: "Ag-Ag-6kV"))] Percentiles: 1-th: 529.0 5-th: 553.0 50-th: 595.0 mean: 592.8311135775065 80-th: 612.0 95-th: 629.0 99-th: 643.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Al-Al-4kV"))] Percentiles: 1-th: 493.53 5-th: 522.0 50-th: 585.0 mean: 583.2071260767424 80-th: 612.0 95-th: 638.3499999999999 99-th: 658.4699999999998============================fallTime============================Type: @[("Type", (kind: VString, str: "C-EPIC-0.6kV"))] Percentiles: 1-th: 296.36 5-th: 349.8 50-th: 509.0 mean: 500.733246584255 80-th: 573.0 95-th: 620.0 99-th: 653.6400000000001============================fallTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-0.9kV"))] Percentiles: 1-th: 354.94 5-th: 433.0 50-th: 544.0 mean: 537.0983009708738 80-th: 587.0 95-th: 627.0 99-th: 658.53============================fallTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-2kV"))] Percentiles: 1-th: 424.0 5-th: 476.1 50-th: 568.0 mean: 561.8043036946813 80-th: 601.0 95-th: 633.0 99-th: 656.3800000000001============================fallTime============================Type: @[("Type", (kind: VString, str: "Cu-Ni-15kV"))] Percentiles: 1-th: 524.0 5-th: 555.0 50-th: 588.0 mean: 586.4952478432519 80-th: 600.0 95-th: 611.0 99-th: 622.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Mn-Cr-12kV"))] Percentiles: 1-th: 524.0 5-th: 551.0 50-th: 586.0 mean: 585.2143482064741 80-th: 602.0 95-th: 617.0 99-th: 628.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Ti-Ti-9kV"))] Percentiles: 1-th: 532.0 5-th: 556.0 50-th: 594.0 mean: 592.3918786692759 80-th: 608.0 95-th: 623.0 99-th: 639.0============================riseTime============================Type: @[("Type", (kind: VString, str: "Ag-Ag-6kV"))] Percentiles: 1-th: 50.0 5-th: 54.0 50-th: 70.0 mean: 70.35627081021087 80-th: 78.0 95-th: 88.0 99-th: 103.0============================riseTime============================Type: @[("Type", (kind: VString, str: "Al-Al-4kV"))] Percentiles: 1-th: 53.0 5-th: 59.0 50-th: 73.0 mean: 77.530931871574 80-th: 87.0 95-th: 105.0 99-th: 128.4699999999998============================riseTime============================Type: @[("Type", (kind: VString, str: "C-EPIC-0.6kV"))] Percentiles: 1-th: 48.0 5-th: 54.0 50-th: 78.0 mean: 86.26350032530904 80-th: 102.8 95-th: 134.4000000000001 99-th: 175.6400000000001============================riseTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-0.9kV"))] Percentiles: 1-th: 51.0 5-th: 57.0 50-th: 78.0 mean: 84.87135922330097 80-th: 99.0 95-th: 127.0 99-th: 170.0599999999999============================riseTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-2kV"))] Percentiles: 1-th: 52.0 5-th: 58.0 50-th: 77.0 mean: 82.13560698335364 80-th: 96.0 95-th: 120.0 99-th: 152.3800000000001============================riseTime============================Type: @[("Type", (kind: VString, str: "Cu-Ni-15kV"))] Percentiles: 1-th: 50.0 5-th: 53.0 50-th: 68.0 mean: 70.41541160988449 80-th: 75.0 95-th: 83.0 99-th: 186.1999999999989============================riseTime============================Type: @[("Type", (kind: VString, str: "Mn-Cr-12kV"))] Percentiles: 1-th: 50.0 5-th: 53.0 50-th: 67.0 mean: 69.50597841936424 80-th: 74.0 95-th: 81.0 99-th: 171.4300000000003============================riseTime============================Type: @[("Type", (kind: VString, str: "Ti-Ti-9kV"))] Percentiles: 1-th: 52.0 5-th: 57.0 50-th: 71.0 mean: 73.02185257664709 80-th: 77.0 95-th: 89.0 99-th: 139.0699999999988and for Run-2:
./fadc_rise_fall_energy_dep -f ~/CastData/data/Calibration2017_Runs.h5 \ --year 2017============================fallTime============================Type: @[("Type", (kind: VString, str: "escape"))] Percentiles: 1-th: 390.0 5-th: 461.0 50-th: 548.0 mean: 543.7853639846743 80-th: 577.0 95-th: 607.0 99-th: 644.0============================fallTime============================Type: @[("Type", (kind: VString, str: "photo"))] Percentiles: 1-th: 449.0 5-th: 483.0 50-th: 550.0 mean: 544.8517320314872 80-th: 568.0 95-th: 584.0 99-th: 599.0============================riseTime============================Type: @[("Type", (kind: VString, str: "escape"))] Percentiles: 1-th: 66.0 5-th: 71.0 50-th: 88.0 mean: 99.69295019157089 80-th: 105.0 95-th: 161.0 99-th: 328.5100000000002============================riseTime============================Type: @[("Type", (kind: VString, str: "photo"))] Percentiles: 1-th: 71.0 5-th: 75.0 50-th: 89.0 mean: 92.85532923781271 80-th: 98.0 95-th: 114.0 99-th: 181.0and Run-3:
./fadc_rise_fall_energy_dep -f ~/CastData/data/Calibration2018_Runs.h5 \ --year 2018============================fallTime============================Type: @[("Type", (kind: VString, str: "escape"))] Percentiles: 1-th: 443.0 5-th: 498.0 50-th: 571.0 mean: 567.8352288748943 80-th: 599.0 95-th: 625.0 99-th: 664.0============================fallTime============================Type: @[("Type", (kind: VString, str: "photo"))] Percentiles: 1-th: 501.0 5-th: 533.0 50-th: 580.0 mean: 578.2391351089849 80-th: 597.0 95-th: 615.0 99-th: 632.0============================riseTime============================Type: @[("Type", (kind: VString, str: "escape"))] Percentiles: 1-th: 50.0 5-th: 55.0 50-th: 73.0 mean: 84.06936742175016 80-th: 91.0 95-th: 145.0 99-th: 298.0============================riseTime============================Type: @[("Type", (kind: VString, str: "photo"))] Percentiles: 1-th: 53.0 5-th: 57.0 50-th: 72.0 mean: 75.88621764721692 80-th: 81.0 95-th: 105.0 99-th: 168.8699999999953which yield the following plots of interest (all others are found in the path of these):
Comparing them directly with the equivalent plots in ./../Figs/statusAndProgress/FADC/old_rise_fall_algorithm/ shows that the biggest change is simply that the rise times have become a bit smaller, as one might expect.
Upon closer inspection in particular in the CDL data however, it seems like some of the spectra become a tad narrower, losing a part of the additional hump.
In the signal / background case it's hard to say. There is certainly a change, but unclear if that is an improvement in separation.
- Investigation of
riseTimetails in calibration data
Let's look at what events look like in the tail of this plot:
What kind of events are, say, above 140?
plotData --h5file ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --runType rtCalibration \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --cuts '("fadc/riseTime", 140.0, Inf)' \ --region crSilver \ --cuts '("rmsTransverse", 0.0, 1.4)' \ --applyAllCuts \ --eventDisplay --septemboard
Looking at these events:
 it is very easily visible that the root cause of the increased rise
time is simply slightly larger than normal noise on the baseline,
resulting in a drop 'before' the real rise and extending the signal.
This is precisely what the "offset" is intended to combat, but in
these cases it doesn't work correctly!
it is very easily visible that the root cause of the increased rise
time is simply slightly larger than normal noise on the baseline,
resulting in a drop 'before' the real rise and extending the signal.
This is precisely what the "offset" is intended to combat, but in
these cases it doesn't work correctly!
Let's tweak it a bit and see again. We'll rerun the
reconstructionwith an offset of 10% down, just to see what happens.After reconstructing the FADC data, we plot the same event number of the first event (maybe more?) of the first plot in the above PDF: run 239, event 1007
plotData --h5file ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --runType rtCalibration \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --runs 239 \ --events 1007 \ --eventDisplay --septemboard
-> fixed the event, now rise time of 60!
- run 239, event 1181:
-> also fixed.
- run 239, event 1068:
-> same.
Let's look at the distribution now:
./fadc_rise_fall_energy_dep -f ~/CastData/data/Calibration2018_Runs.h5 \ --year 2018============================fallTime============================Type: @[("Type", (kind: VString, str: "escape"))] Percentiles: 1-th: 333.38 5-th: 386.0 50-th: 468.0 mean: 463.4154525801297 80-th: 492.0 95-th: 517.0 99-th: 547.0============================fallTime============================Type: @[("Type", (kind: VString, str: "photo"))] Percentiles: 1-th: 372.0 5-th: 420.0 50-th: 469.0 mean: 466.0160856828016 80-th: 487.0 95-th: 503.0 99-th: 519.0============================riseTime============================Type: @[("Type", (kind: VString, str: "escape"))] Percentiles: 1-th: 42.0 5-th: 45.0 50-th: 56.0 mean: 61.34223141272676 80-th: 62.0 95-th: 76.0 99-th: 240.0============================riseTime============================Type: @[("Type", (kind: VString, str: "photo"))] Percentiles: 1-th: 44.0 5-th: 46.0 50-th: 55.0 mean: 56.65509178380412 80-th: 59.0 95-th: 64.0 99-th: 114.8699999999953yields:
We've essentially removed any tail still present in the data!
But does that mean we removed information, i.e. the background case now looks also more similar?
./fadc_rise_fall_signal_vs_background \ -c ~/CastData/data/CalibrationRuns2018_Reco.h5 \ -b ~/CastData/data/DataRuns2018_Reco.h5 \ --year 2018
which yields:
-> Holy crap! I didn't think we could leave the background data this "untouched", but narrow the calibration data as much! It also makes much nicer the fact that the escape and photo peak data have become even more similar! So one cut might after all be almost enough (barring FADC settings etc).
Let's also look at the CDL data again:
- reconstruct it again with new settings
plot it:
./fadc_rise_fall_energy_dep -f ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --year 2019 \ --isCdl
============================fallTime============================[110/1676]Type: @[("Type", (kind: VString, str: "Ag-Ag-6kV"))] Percentiles: 1-th: 359.05 5-th: 410.0 50-th: 471.0 mean: 466.7824639289678 80-th: 493.0 95-th: 510.0 99-th: 524.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Al-Al-4kV"))] Percentiles: 1-th: 352.0 5-th: 392.0 50-th: 470.0 mean: 465.9608457321848 80-th: 500.0 95-th: 527.0 99-th: 548.4699999999998============================fallTime============================Type: @[("Type", (kind: VString, str: "C-EPIC-0.6kV"))] Percentiles: 1-th: 204.36 5-th: 254.8 50-th: 375.0 mean: 376.6766428106702 80-th: 445.0 95-th: 501.0 99-th: 540.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-0.9kV"))] Percentiles: 1-th: 262.47 5-th: 304.0 50-th: 419.0 mean: 413.0831310679612 80-th: 472.5999999999999 95-th: 508.0 99-th: 548.53============================fallTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-2kV"))] Percentiles: 1-th: 300.62 5-th: 334.1 50-th: 448.0 mean: 440.717011774259 80-th: 489.0 95-th: 524.0 99-th: 548.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Cu-Ni-15kV"))] Percentiles: 1-th: 344.0 5-th: 395.0 50-th: 462.0 mean: 456.8689866939611 80-th: 481.0 95-th: 496.0 99-th: 509.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Mn-Cr-12kV"))] Percentiles: 1-th: 354.0 5-th: 407.0 50-th: 464.0 mean: 459.7802566345873 80-th: 483.0 95-th: 499.0 99-th: 513.0============================fallTime============================Type: @[("Type", (kind: VString, str: "Ti-Ti-9kV"))] Percentiles: 1-th: 367.31 5-th: 417.0 50-th: 472.0 mean: 468.1409001956947 80-th: 492.0 95-th: 508.0 99-th: 524.6899999999996============================riseTime============================[15/1676]Type: @[("Type", (kind: VString, str: "Ag-Ag-6kV"))] Percentiles: 1-th: 41.0 5-th: 44.0 50-th: 54.0 mean: 53.89345172031076 80-th: 59.0 95-th: 63.0 99-th: 67.0============================riseTime============================Type: @[("Type", (kind: VString, str: "Al-Al-4kV"))] Percentiles: 1-th: 43.0 5-th: 48.0 50-th: 57.0 mean: 58.23923257635082 80-th: 63.0 95-th: 71.0 99-th: 80.0============================riseTime============================Type: @[("Type", (kind: VString, str: "C-EPIC-0.6kV"))] Percentiles: 1-th: 38.0 5-th: 47.0 50-th: 67.0 mean: 68.42680546519193 80-th: 79.0 95-th: 98.0 99-th: 118.0============================riseTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-0.9kV"))] Percentiles: 1-th: 41.47 5-th: 48.0 50-th: 64.0 mean: 65.94174757281553 80-th: 74.0 95-th: 92.65000000000009 99-th: 116.0599999999999============================riseTime============================Type: @[("Type", (kind: VString, str: "Cu-EPIC-2kV"))] Percentiles: 1-th: 44.0 5-th: 49.0 50-th: 61.0 mean: 62.69549330085262 80-th: 71.0 95-th: 82.0 99-th: 96.0============================riseTime============================Type: @[("Type", (kind: VString, str: "Cu-Ni-15kV"))] Percentiles: 1-th: 41.0 5-th: 43.0 50-th: 53.0 mean: 54.76941073256324 80-th: 57.0 95-th: 62.0 99-th: 144.6199999999999============================riseTime============================Type: @[("Type", (kind: VString, str: "Mn-Cr-12kV"))] Percentiles: 1-th: 41.0 5-th: 43.0 50-th: 53.0 mean: 54.94998541848936 80-th: 57.0 95-th: 62.0 99-th: 152.7200000000012============================riseTime============================Type: @[("Type", (kind: VString, str: "Ti-Ti-9kV"))] Percentiles: 1-th: 42.0 5-th: 45.0 50-th: 55.0 mean: 56.16699282452707 80-th: 59.0 95-th: 63.0 99-th: 105.7599999999984yielding:
which also gives a lot more 'definition'. Keep in mind that the main important lines are those from Aluminum. These are essentially all more or less the same width with the aluminum in particular maybe a bit wider.
This is pretty good news generally. What I think is going on in detail here is that we see there is an additional "bump" in AgAg6kV, MnCr12kV and a bigger one in CuNi15kV. What do these have in common? They have a longer absorption length and therefore shorter average diffusion! This might actually be the thing we were trying to identify! As there is a larger and larger fraction of these it becomes a significant contribution and not just a 'tail' to lower rise times!
Question: What events are still in the tail of the calibration rise time data? i.e. above rise time of 100 ns? Let's check:
plotData --h5file ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --runType rtCalibration \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --cuts '("fadc/riseTime", 100.0, Inf)' \ --region crSilver \ --cuts '("rmsTransverse", 0.0, 1.4)' \ --applyAllCuts \ --eventDisplay --septemboard
yielding events like this:
 where we can see that it is almost entirely double hit events. As a
small fraction further it is events with a crazy amount of
noise. But the double hits make up the biggest fraction.
where we can see that it is almost entirely double hit events. As a
small fraction further it is events with a crazy amount of
noise. But the double hits make up the biggest fraction.
Does that mean we can filter the data better for our calculation of the percentiles? Ideally we only use single X-rays. Outside of counting the number of clusters on an event, what can we do?
Ah, many of these events are not actually split up and remain a single cluster, which means their eccentricity is very large. But in the plots that produce the rise time KDE we already have a cut on the eccentricity. So I suppose we first need to look at the events that are eccentricity filtered that way as well.
UPDATE: OUCH! The filter of the events in the FADC scripts that read data do not apply the cuts to the eccentricity at all, but only to the transverse RMS dataset by accident!!!!!!! -> Note: immediate impact seems to be essentially nil. There is a small change, but it's really very small.
plotData --h5file ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --runType rtCalibration \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --cuts '("fadc/riseTime", 100.0, Inf)' \ --region crSilver \ --cuts '("rmsTransverse", 0.0, 1.2)' \ --cuts '("eccentricity", 0.0, 1.4)' \ --applyAllCuts \ --eventDisplay --septemboard
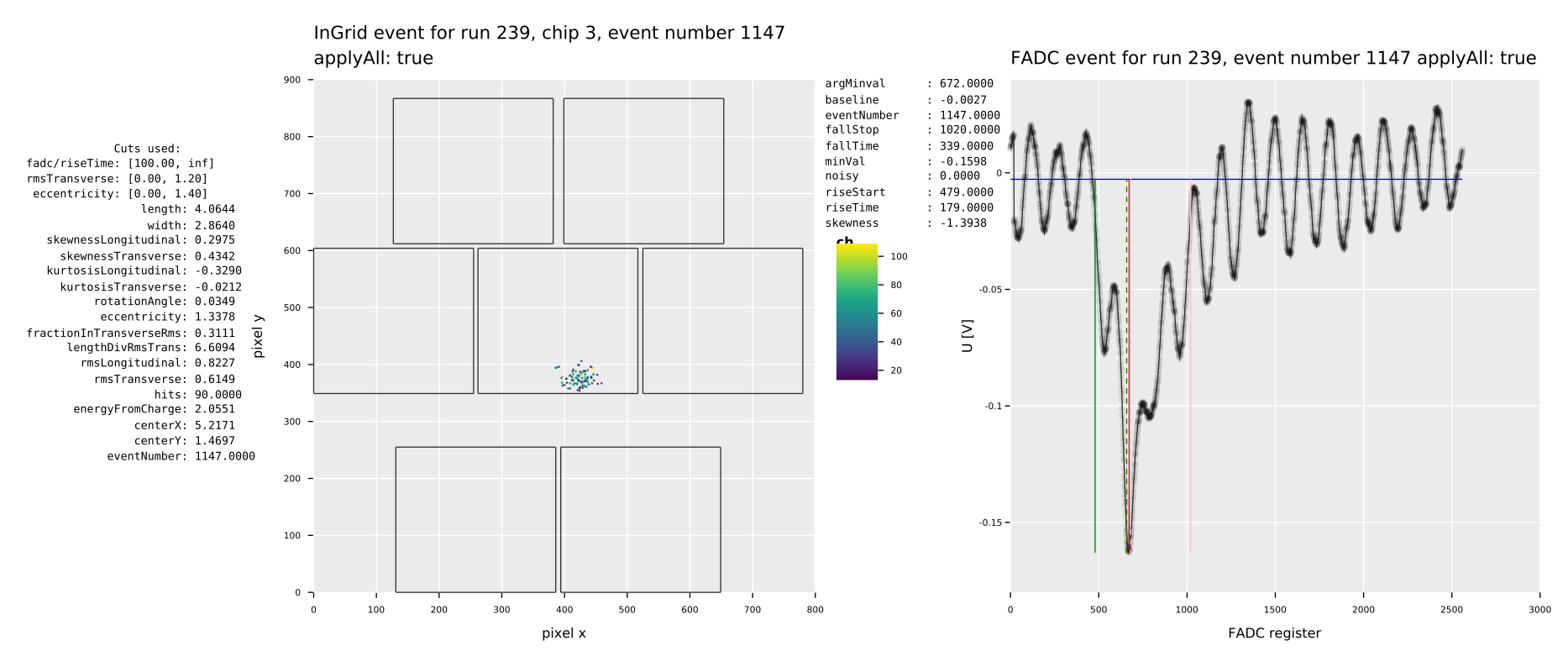 where we can see that what is left are events of one of the two cases:
where we can see that what is left are events of one of the two cases:
- clearly seperated clusters that are reconstructed as separate clusters
- clusters that are clearly double hits based on the FADC signal, but look like a perfect single cluster in the InGrid data
The latter is an interesting "problem". Theoretically a peak finding algorithm for the FADC data (similar to what used for noise detection) could identify those. But at the same time I feel that we have justification enough to simply cut away any events with a rise time larger X and compute the cut value only based on that. From the gaseous detector physics we know how this behaves. And our data describes our expectation well enough now. So a part of me says we should just take the maximum value and apply some multiplier to its rise time to get a hard cut for the data. Only all data below that will then be used to determine the desired percentile efficiency cut.
8.2.3. Difference in FADC rise times for different FADC amplifier settings
One of the big questions looking at the rise time as a means to improve the FADC veto is the effect of the different FADC amplifier settings used in 2017.
The code above /tmp/fadc_rise_fall_energy_dep.nim produces a plot
splitting up the different FADC settings if fed with the Run-2 data.
The result is fig. 65. The difference is very stark, implying we definitely need to pick the cut values on an, at least, per setting level.
However, I would have assumed that the distribution of setting 3 (the last one) would match the distribution for run 3, fig. 66. But the peak is at even lower values than even setting 1 (namely below 60!). What. Maybe we didn't rerun the 10 percent offset on calibration data yet? Nope, I checked, all up to date. Maybe the Run-3 data is not? Also up to date.
This brings up the questions whether the effect is not actually a "per setting", but a "per run" effect?
No, that is also not the case. Compare:
The Run-3 data clearly has all runs more or less sharing the same rise times (still though, different cuts may be useful?). And in the Run-2 data we see again more or less 3 distinct distributions.
This begs the question whether we actually ran with an even different setting in Run-3 than at the end of Run-2. This is certainly possible? In the end this is not worth trying to understand in detail. The reason will likely be that. All we care about then is to define cuts that are distinct for each run period & settings. So 4 different cuts in total, 3 for Run-2 and 1 for Run-3.
One weird aspect is the fall time of Run-2
namely the case for setting 2. That setting really seemed to shorten
the fall time significantly.
8.2.4. Estimating expected rise times
Generally speaking we should be able to estimate the rise time of the FADC signals from the gaseous detector physics.
The maximum diffusion possible for an X-ray photon should lead to a maximum time that an FADC signal should be long. This time then needs to be folded with the integration time. The result should be an expected FADC signal.
(Note that different energies have different penetration depths on average. The lower the energy the shorter the length in gas, resulting in on average more diffusion)
Going by
and
from
plotData --h5file ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --runType rtCalibration \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --ingrid \ --cuts '("rmsTransverse", 0.1, 1.1)' \ --applyAllCuts \ --region crSilver
we can conclude that typically the length is a bit less than 6 mm and the transverse RMS about 1 mm (which should be what we get from the transverse diffusion coefficient!) So let's go with that number.
Drift velocity is 2 cm·μs⁻¹ implies a drift time for the full X-ray of
import unchained let v = 2.cm•μs⁻¹ let s = 6.mm echo s / v
which are 300 ns. Naively that would equate to 300 clock cycles of the FADC. But our rise times are typically only less than 150, certainly less than 300 clock cycles. How come?
Inversely a time of 150 clock cycles corresponds to about 1.5 μs and so about half the size, namely 3 mm.
The length is related to the transverse diffusion. So what does the longitudinal diffusion look like in comparison? Surely not a factor of 2 off?
Refs:
- Talk that mentions relation of transverse & longitudinal diffusion: Diffusion coefficient: D = 1/3 v λ with longitudinal: 1/3 D and transverse: 2/3 D https://www.physi.uni-heidelberg.de/~sma/teaching/ParticleDetectors2/sma_GasDetectors_1.pdf
- Sauli ./../Papers/Gaseous Radiation Detectors Fundamentals and Applications (Sauli F.) (z-lib.org).pdf on page 92 mentions relation of σL to σT σT = σL / √( 1 + ω²τ²) where ω = EB/m (but our B = 0!?) and τ mean collision time. Naively I would interpret this formula to say σT = σL without a B field though.
Paper about gas properties for LHC related detectors. Contains (not directly comparable) plots of Argon mixtures longitudinal and transverse data: page 18 (fig 9): Ar longitudinal diffusion: Top right plot contains Ar Isobutane, but max 90/10. Best we have though: At 500 V/cm (drift field in detector) all mixtures are about 200 μm/cm. page 22 (fig 13): Ar transverse diffusion. Top right plot, closest listed is again Ar/Iso 90/10. That one at 500V/cm is 350 μm/cm. https://arxiv.org/pdf/1110.6761.pdf
However, the scaling between different mixtures is very large in transverse, but not in longitudinal. Assuming longitudinal is the same in 97.7/2.3 at 200 μm/cm, but transverse keeps jumping, it'll be easily more than twice as high.
- Our old paper (Krieger 2017) https://arxiv.org/pdf/1709.07631.pdf He reports a number of 474 and 500 μm/√cm (square root centimeter??)
[X]I think just compute with PyBoltz. -> Done.
UNRELATED HERE BUT GOOD INFO: https://arxiv.org/pdf/1110.6761.pdf contains good info on the Townsend coefficient and how it relates to the gas gain! page 11:
Townsend coefficient
The average distance an electron travels between ionizing collisions is called mean free path and its inverse is the number of ionizing collision per cm α (the first Townsend coefficient). This parameter determines the gas gain of the gas. If no is the number of primary electron without amplification in uniform electric field, and n is the number of electrons after distance x under avalanche condition. So n is given by n = noeαx and the gas gain G is given by G = n0/n = eαx. The first Townsend Coefficient depends on the nature of the gas, the electric field and pressure.
(also search for first Townsend coefficient in Sauli, as well as for "T/P") -> Also look at what Townsend coefficient we get for different temperatures using PyBoltz!
Where are we at now?
[ ]Use our computed values for the longitudinal / transverse diffusion to make a decision about the FADC rise time cut.[ ]Determine the Townsend coefficient from PyBoltz so that we can compute equivalent numbers to the temperature variation in the amplification region. Can we understand why gain changes the way it does?
Let's start with 1 by computing the values to our best knowledge.
- Pressure in the detector: \(\SI{1050}{mbar} = \SI{787.6}{torr}\)
- Gas: Argon/Isobutane: 97.7 / 2.3 %
- Electric field in drift region: \(\SI{500}{V.cm⁻¹}\)
- Temperature: \(\sim{30}{\degree\celsius}\) the temperature is by far the biggest issue to properly estimate of course. This value is on the higher end for sure, but takes into account that the detector will perform some kind of heating that also affects the gas in the drift region. But because of that we will simply simulate energies in a range.
- PyBoltz setup
To run the above on this machine we need to do:
cd ~/src/python/PyBoltz/ source ~/opt/python3/bin/activate source setup.sh python examples/argon_isobutane_cast.py
where the
setup.shfile was modified from the shipped version to:#!/usr/bin/env zsh # setup the enviorment export PYTHONPATH=$PYTHONPATH:$PWD export PATH=$PATH:$PWD echo $PYTHONPATH # build the code python3 setup_build.py clean python3 setup_build.py build_ext --inplace
- Diffusion coefficient and drift velocity results for CAST conditions
UPDATE: I wrote a version using NimBoltz, ./../../CastData/ExternCode/TimepixAnalysis/Tools/septemboardCastGasNimboltz/septemboardGasCastNimBoltz.nim to not depend on brittle Python anymore.
The code we use is ./../../CastData/ExternCode/TimepixAnalysis/Tools/septemboardCastGasNimboltz/septemboardGasCastPyboltz.nim which calls
PyBoltzfrom Nim and usescligen'sprocpoolto multiprocess this. It calculates the gas properties at the above parameters for a range of different temperatures, as this is the main difference we have experienced over the full data taking range.Running the code:
cd $TPA/Tools/septemboardCastGasPyboltz ./septemboardCastGasPyboltz --runPyBoltz
(to re-generate the output data by actually calling
PyBoltz. Note that this requiresPyBoltzavailable for the Python installation at highest priority in your PATH). orcd $TPA/Tools/septemboardCastGasPyboltz ./septemboardCastGasPyboltz --csvInput $TPA/resources/ar_iso_97_7_2_3_septemboard_cast_different_temps.csv
and it yields the following Org table as an output:
E [V•cm⁻¹] T [K] v [mm•μs⁻¹] Δv [mm•μs⁻¹] σTσL [UnitLess] ΔσTσL [UnitLess] σT [μm•√cm] σL [μm•√cm] ΔσT [μm•√cm] ΔσL [μm•√cm] 500 289.2 23.12 0.005422 2.405 0.04274 630.8 262.3 9.013 2.772 500 291.2 23.08 0.004498 2.44 0.05723 633.5 259.7 6.898 5.395 500 293.2 23.02 0.003118 2.599 0.06341 644.4 247.9 9.784 4.734 500 295.2 22.97 0.006927 2.43 0.06669 645.9 265.8 11.54 5.541 500 297.2 22.91 0.004938 2.541 0.05592 651.2 256.3 9.719 4.147 500 299.2 22.87 0.006585 2.422 0.05647 644.2 266 8.712 5.05 500 301.2 22.83 0.005237 2.362 0.06177 634.9 268.8 8.775 5.966 500 303.2 22.77 0.004026 2.539 0.07082 666.6 262.5 11.95 5.611 500 305.2 22.72 0.006522 2.492 0.07468 657.6 263.9 11.2 6.507 500 307.2 22.68 0.006308 2.492 0.05062 636.6 255.4 7.968 4.085 500 309.2 22.64 0.006007 2.472 0.06764 664.6 268.8 12.21 5.45 500 311.2 22.6 0.00569 2.463 0.05762 657.9 267.1 9.425 4.94 500 313.2 22.55 0.006531 2.397 0.0419 662.1 276.2 9.911 2.492 500 315.2 22.51 0.003245 2.404 0.04582 654.7 272.4 6.913 4.323 500 317.2 22.46 0.005834 2.593 0.07637 682 263 12.92 5.929 500 319.2 22.42 0.006516 2.594 0.06435 681.8 262.8 9.411 5.417 500 321.2 22.38 0.003359 2.448 0.05538 670.2 273.7 8.075 5.239 500 323.2 22.34 0.004044 2.525 0.08031 677.5 268.3 11.4 7.244 500 325.2 22.3 0.005307 2.543 0.06627 677.6 266.5 12.87 4.755 500 327.2 22.26 0.007001 2.465 0.05675 682.3 276.8 8.391 5.387 500 329.2 22.22 0.002777 2.485 0.07594 679.1 273.3 12.39 6.701 500 331.2 22.19 0.004252 2.456 0.06553 667.3 271.7 10 5.995 500 333.2 22.15 0.004976 2.563 0.06788 687.5 268.2 12.78 5.059 500 335.2 22.11 0.004721 2.522 0.06608 702 278.4 12.24 5.446 500 337.2 22.07 0.00542 2.467 0.09028 676.4 274.1 11.17 8.952 500 339.2 22.03 0.003971 2.527 0.04836 678.8 268.6 12.36 1.577 500 341.2 21.99 0.005645 2.575 0.07031 697 270.7 9.502 6.403 500 343.2 21.96 0.005118 2.535 0.06872 696.6 274.8 10.09 6.297 and these plots:
showing the drift velocity, transverse & longitudinal diffusion coefficients and the ratio of the two coefficients against the temperature.
The data file generated (essentially the above table) is available here:
- ./../../CastData/ExternCode/TimepixAnalysis/resources/ar_iso_97_7_2_3_septemboard_cast_different_temps.csv
- ./../resources/ar_iso_97_7_2_3_septemboard_cast_different_temps.csv
- ./../../phd/resources/ar_iso_97_7_2_3_septemboard_cast_different_temps.csv
(and by extension on Github).
- Computing an expected rise time from gas properties
Now that we know the properties of our gas, we can compute the expected rise times.
What we need are the following things:
- drift velocity
- transverse diffusion
- detector height
- (optional as check) length of the X-ray clusters
- longitudinal diffusion
The basic idea is just:
- based on height of detector compute:
- maximum transverse diffusion (which we can cross check!)
- maximum longitudinal diffusion a) based on gas property numbers b) based on known length data by scaling σT over σL
- maximum longitudinal length corresponds to a maximum time possibly seen in the drift through the grid
- this max time corresponds to an upper limit on rise times for real X-rays!
Let's do this by reading the CSV file of the gas and see where we're headed. When required we will pick a temperature of 26°C to be on the warmer side, somewhat taking into account the fact that the septemboard should itself heat the gas somewhat (it might actually be more in reality!).
[ ]MOVE THIS TO THESIS WHEN DONE!
NOTE: The code below is a bit tricky, as the handling of units in measuremancer is still problematic & the fact that unchained does neither support centigrade nor square root units!
import datamancer, unchained, measuremancer # first some known constants const FadcClock = 1.GHz DetectorHeight = 3.cm let MaxEmpiricalLength = 6.mm ± 0.5.mm # more or less! let df = readCsv("/home/basti/phd/resources/ar_iso_97_7_2_3_septemboard_cast_different_temps.csv") # compute the mean value (by accumulating & dividing to propagate errors correctly) var σT_σL: Measurement[float] for i in 0 ..< df.len: σT_σL += df["σT_σL [UnitLess]", i, float] ± df["ΔσT_σL [UnitLess]", i, float] σT_σL = σT_σL / (df.len.float) # Note: the temperature is centigrade and not kelvin as the header implies, oops. let df26 = df.filter(f{float -> bool: abs(idx("T [K]") - (26.0 + 273.15)) < 1e-4}) let v = df26["v [mm•μs⁻¹]", float][0].mm•μs⁻¹ ± df26["Δv [mm•μs⁻¹]", float][0].mm•μs⁻¹ let σT = df26["σT [μm•√cm]", float][0] ± df26["ΔσT [μm•√cm]", float][0] let σL = df26["σL [μm•√cm]", float][0] ± df26["ΔσL [μm•√cm]", float][0] # 1. compute the maximum transverse and longitudinal diffusion we expect # deal with the ugly sqrt units of the regular coefficient let maxDiffTransverse = (σT * sqrt(DetectorHeight.float) / 1000.0 * 1.0.mm) # manual conversion from μm to mm let maxDiffLongitudinal = (σL * sqrt(DetectorHeight.float) / 1000.0 * 1.0.mm) echo "Maximum transverse diffusion = ", maxDiffTransverse echo "Maximum longitudinal diffusion = ", maxDiffLongitudinal # however, the diffusion gives us only the `1 σ` of the diffusion. For that # reason it matches pretty much exactly with the transverse RMS data we have from # our detector! # First of all the length of the cluster will be twice the sigma (sigma is one sided!) # And then not only a single sigma, but more like ~3. let maxClusterSize = 3 * (2 * maxDiffTransverse) let maxClusterHeight = 3 * (2 * maxDiffLongitudinal) echo "Expected maximum (transverse) length of a cluster = ", maxClusterSize echo "Expected maximum longitudinal length of a cluster = ", maxClusterHeight # this does actually match our data of peaking at ~6 mm reasonably well. # From this now let's compute the expected longitudinal length using # the known length data and the known fraction: let maxEmpiricalHeight = MaxEmpiricalLength / σT_σL echo "Empirical limit on the cluster height = ", maxEmpiricalHeight # and finally convert these into times from a clock frequency ## XXX: Converson from micro secnd to nano second is broken in Measuremancer! ## -> it's not broken, but `to` is simply not meant for Unchained conversions yet. ## I also think something related to unit conversions in the errors is broken! ## -> Math is problematic with different units as of now.. Our forced type conversions ## in measuremancer remove information! ## -> maybe use `to` correctly everywhere? Well, for now does not matter. let maxTime = (maxClusterHeight / v) # * (1.0 / FadcClock).to(Meter) echo "Max rise time = ", maxTime # and from the empirical conversion: let maxEmpiricalTime = (maxEmpiricalHeight / v) # * (1.0 / FadcClock).to(Meter) echo "Max empirical rise time = ", maxEmpiricalTime
Investigate the errors on the maximum rise time!
[X]Done: Issue is that measuremancer screws up errors because of forced type conversion for errors.
From the above we can see that we expect a maximum rise time of something like 121 ns (clock cycles) from theory and if we use our empirical results about 105 ns.
These numbers match quite well with our median / mean and percentile values in the above section!
[ ]COMPUTE OUR GAS TEMPERATURE FROM RISE TIME &rmsTransversePROPERTY -> What temperature matches best to our measured transverse RMS? And our rise time? Is the gas property impact of variation in temperature even big enough or are other impacts (e.g. field distortions etc) more likely?
- Estimating typical diffusion distances
The typical distance that an X-ray of a known energy drifts in the first place depends on the typical absorption length in the material. If we look at the transverse RMS of our data, e.g.
we see a peak at about 1 mm. However, what does this RMS correspond to? It corresponds to those X-rays of the typical drift distance. And that is the typical distance of a 5.9 keV photon. So let's compute the typical absorption distance of such a photon to get a correction:
import xrayAttenuation, unchained, datamancer let ar = Argon.init() let ρ_Ar = density(1050.mbar.to(Pascal), 293.K, ar.molarMass) let E = 5.9.keV let dist = absorptionLength(E, numberDensity(ρ_Ar, ar.molarMass), ar.f2eval(E)) echo "Dist = ", dist, " in cm ", dist.to(cm)
2 cm??? -> Yes, this is correct and lead to the discussion in sec. 3.3.2! It all makes sense if one properly simulates it (the absorption is an 'exponential decay' after all!)
The big takeaway from looking at the correct distribution of cluster sizes given the absorption length is really that there are still a significant fraction of X-rays that diffuse to essentially the "cutoff" value. At very large values of λ > 2 this does lead to a general trend to smaller clusters than calculated based on the full 3 cm drift, but only by 10-20% at most.
- Preliminary results after first run of simple code snippet
The text below was written from the initial results we got from the very first snippet we ran for a single temperature in Python. The Nim code snippet printed here is already a second version that gets close to what we ended up running finally. The results below the snippet are from the very first Python run for a single data point!
The setup to have the PyBoltz library available (e.g. via a virtualenv) is of course also needed here.
The following is the script to run this code. It needs the
PyBoltzlibrary installed of course. (available in our virtualenv).[X]Better rewrite the below as a Nim script, then increase number of collisions (does that increase accuracy?) and useprocpoolto run 32 of these simulations (for different temps for example) at the same time. Also makes it much easier to deal with the data… -> Done.
import ggplotnim, unchained, measuremancer, nimpy import std / [strformat, json, times] defUnit(V•cm⁻¹) defUnit(cm•μs⁻¹) type MagRes = object E: V•cm⁻¹ T: K v: Measurement[cm•μs⁻¹] σT: Measurement[float] # μm²•cm⁻¹] # we currently do not support √unit :( σL: Measurement[float] # μm²•cm⁻¹] proc toMagRes(res: PyObject, temp: Kelvin): MagRes = result = MagRes(T: temp) let v = res["Drift_vel"].val[2].to(float) let Δv = res["Drift_vel"].err[2].to(float) result.v = v.cm•μs⁻¹ ± Δv.cm•μs⁻¹ # now get diffusion coefficients for a single centimeter (well √cm) let σ_T1 = res["DT1"].val.to(float) let Δσ_T1 = res["DT1"].err.to(float) result.σT = (σ_T1 ± Δσ_T1) let σ_L1 = res["DL1"].val.to(float) let Δσ_L1 = res["DL1"].err.to(float) result.σL = (σ_L1 ± Δσ_L1) proc `$`(m: MagRes): string = result.add &"T = {m.T}" result.add &"σ_T1 = {m.σT} μm·cm⁻⁰·⁵" result.add &"σ_L1 = {m.σL} μm·cm⁻⁰·⁵" proc toDf(ms: seq[MagRes]): DataFrame = let len = ms.len result = newDataFrame() for m in ms: var df = newDataFrame() for field, data in fieldPairs(m): when typeof(data) is Measurement: let uof = unitOf(data.value) let unit = &" [{uof}]" df[field & unit] = data.value.float df["Δ" & field & unit] = data.error.float else: let uof = unitOf(data) let unit = &" [{uof}]" df[field & unit] = data.float result.add df let pb = pyImport("PyBoltz.PyBoltzRun") # Set up helper object let PBRun = pb.PyBoltzRun() # Configure settings for our simulation var Settings = %* { "Gases" : ["ARGON","ISOBUTANE"], "Fractions" : [97.7,2.3], "Max_collisions" : 4e7, "EField_Vcm" : 500, "Max_electron_energy" : 0, "Temperature_C" : 30, "Pressure_Torr" : 787.6, "BField_Tesla" : 0, "BField_angle" : 0, "Angular_dist_model" : 1, "Enable_penning" : 0, "Enable_thermal_motion" : 1, "ConsoleOutputFlag" : 1} let t0 = epochTime() var res = newSeq[MagRes]() let temps = arange(14.0, 36.0, 2.0) for temp in temps: Settings["Temperature_C"] = % temp # commence the run! res.add(PBRun.Run(Settings).toMagRes((temp + 273.15).K)) let t1 = epochTime() echo "time taken = ", t1-t0 echo res[^1] let df = res.toDf() echo df.toOrgTable()
The output of the above is
Input Decor_Colls not set, using default 0 Input Decor_LookBacks not set, using default 0 Input Decor_Step not set, using default 0 Input NumSamples not set, using default 10 Trying 5.6569 Ev for final electron energy - Num analyzed collisions: 3900000 Calculated the final energy = 5.6568542494923815 Velocity Position Time Energy DIFXX DIFYY DIFZZ 22.7 0.3 11464558.7 1.1 3854.8 20838.4 0.0 22.7 0.5 22961894.0 1.1 8647.8 12018.0 0.0 22.7 0.8 34532576.4 1.1 7714.7 12014.1 202.4 22.7 1.0 46113478.7 1.1 6105.2 11956.1 641.4 22.7 1.3 57442308.9 1.1 5840.9 9703.4 739.7 22.8 1.6 68857082.2 1.1 7759.2 8817.9 608.6 22.8 1.8 80311917.6 1.1 7648.9 8248.2 574.8 22.8 2.1 91754361.3 1.1 7184.4 7322.1 611.2 22.8 2.3 103265642.2 1.1 7569.3 6787.9 656.5 22.8 2.6 114853263.8 1.1 7298.9 6968.7 764.8 time taken 103.45310592651367 σ_T = 7133.782820393255 ± 1641.0801103506417 σ_L = 764.8143711898326 ± 160.83754918535615 σ_T1 = 791.8143345965309 ± 91.07674206533429 σ_L1 = 259.263534784396 ± 27.261384253733745
What we gleam from this is that the diffusion coefficients we care about (namely the
*1versions) are:\[ σ_T = 791.8 ± 91.1 μm·√cm \]
and
\[ σ_L = 259.26 ± 27.3 μm·√cm \]
which turns out to be a ratio of:
\[ \frac{σ_T}{σ_L} = 3.05 \]
So surprisingly the transverse diffusion is a full factor 3 larger than the longitudinal diffusion!
In addition we can read off the drift velocity of \(\SI{2.28}{cm·μs⁻¹}\).
The main output being:
T v σT σL 14.0 23.12221717549076 ± 0.04451635054497993 720.4546229571265 ± 92.25686062895952 255.27791834496637 ± 25.183524735291876 16.0 23.103731285486774 ± 0.03595271262006956 616.587833132368 ± 53.89931070654909 222.14061731499962 ± 17.837640243065074 18.0 23.076301420096588 ± 0.036605366202092225 645.537278659896 ± 64.64445968202027 275.71926338282447 ± 37.91257063146355 20.0 22.997513931669804 ± 0.025816774253200406 640.31721992396 ± 68.9113486411086 236.43873330018673 ± 36.02017572086169 22.0 22.932268231504192 ± 0.0328347862828518 615.8682550046013 ± 74.24682912210032 242.31515490459608 ± 28.05523660699701 24.0 22.871239000070037 ± 0.04255711577757762 742.2002296818248 ± 72.94318786860077 263.0814747275606 ± 34.624811170582795 26.0 22.833848724962852 ± 0.03087355168336172 626.8271546734144 ± 67.1554564961464 260.00659390651487 ± 32.456334414972844 28.0 22.79969666113236 ± 0.04420068652428081 614.782404723097 ± 51.838235017654526 246.12174320906414 ± 29.60789215566301 30.0 22.72279250815483 ± 0.03699016950129097 698.6046486427862 ± 79.6459139815396 260.90895307103534 ± 27.98241664934684 32.0 22.72745917196911 ± 0.03166545537801199 681.6978915408016 ± 76.97738468648261 260.3776539762865 ± 31.627440708563316 34.0 22.60977721661218 ± 0.03555585123344388 621.0265075081438 ± 73.80599488874776 279.7425000247473 ± 29.957402479193366 The preliminary result for sure is though that this does indeed give a reasonably good explanation for why the rise time for X-rays is only of the order of 100 instead of 300 (as ~expected from drift velocity).
8.2.5. About the fall time
The fall time is dominated by the RC characteristics of the FADC readout chain.
The problem here is that we lack information. The FADC readout happens via a \(\SI{10}{nF}\) capacitor. However, we don't really know anything about the resistance and or whatever the real numbers are.
From the schematic from Deisting's MSc thesis we can gleam a resistor of \(\SI{12}{\mega\ohm}\) and a capacitor of \(\SI{470}{pF}\). These together still give an RC time of:
import unchained let R = 12.MΩ let C = 470.pF #10.nF echo "τ = ", R*C
about 5.6 ms! Waaayyy too long. So these are clearly not the relevant pieces of information. We'd likely need
8.3. Updating the FADC algorithms for rise & fall time as well as data structure
We've ended up performing the following changes in the commits from .
updated the algorithm that computes the rise and fall time of the FADC data such that we don't start from the minimum register, but an offset away from the mean minimum value.
const PercentileMean = 0.995 # 0.5% = 2560 * 0.005 = 12.8 registers around the minimum for the minimum val const OffsetToBaseline = 0.025 # 2.5 % below baseline seems reasonable let meanMinVal = calcMinOfPulse(fadc, PercentileMean) let offset = abs(OffsetToBaseline * (meanMinVal - baseline)) # relative to the 'amplitude' # ... (riseStart, riseStop) = findThresholdValue(fadc, xMin, meanMinVal + offset, baseline - offset)
where
meanMinVal + offsetis the lower threshold we need to cross before we start counting the rise or fall times. ThexMinin that sense is only a reference. The real calculation of the threshold is based on the minimum of the pulse using a 0.5% signal width around the minimum.Further, we change the data types that store the FADC data in order to change the workflow in which the FADC data is reconstructed. Instead of doing a weird mix of the reconstruction where we perform the conversion from the raw FADC data to the reconstructed, rotated, voltage based data together with the deduction of the pulse minimum and noisy-ness, this is now split.
The main motivation for this change is that previously the FADC reconstruction was tied to the noisy & minimum value calcs, which meant that these were only performed when reconstructing from a raw data file to a reco data file. This made changing the algorithms of the FADC parts very problematic, as there was no way to rerun only e.g. the noise detection. moved the calculation of whether a signal is noisy
In order to see what this looks like and in particular that events that saturate the FADC now have reasonable rise and fall times (from a well defined point not randomly in the total middle of the "minimum plateau), let's apply this to some data and plot some events.
plotData --h5file ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --runType rtCalibration \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --cuts '("energyFromCharge", 2.0, 2.5)' \ --eventDisplay --septemboard
which gives events like:
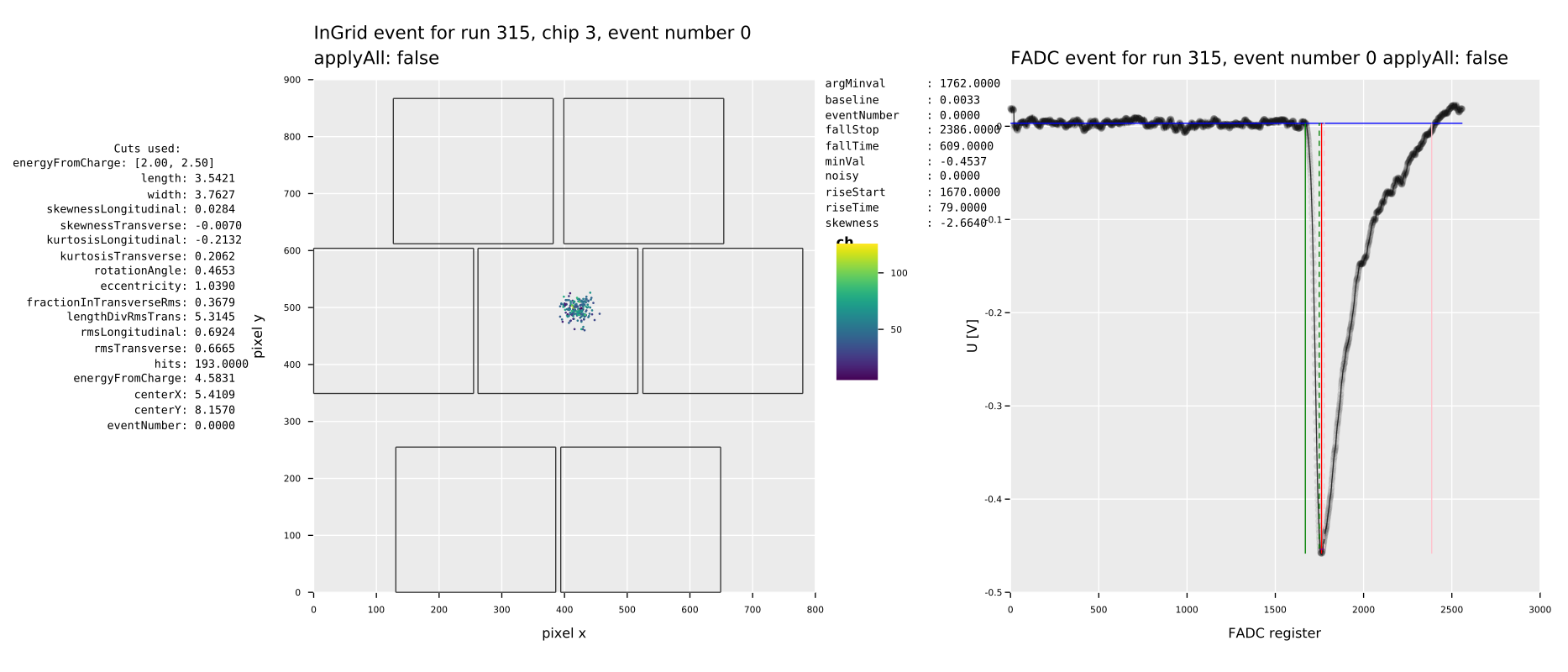
where the dashed lines represent the stop points for the rise and fall time (or start for the algorithm walking away from the minimum). The purple small line visible near the minimum is the actual minimum value that is used for other purposes.
Finally, let's look at one of the events from sec. 8.4.1, namely for example run 279, event 15997 (check the PDF in the linked section to see that event):
plotData --h5file ~/CastData/data/DataRuns2018_Reco.h5 \ --runType rtCalibration \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --eventDisplay \ --runs 279 \ --events 15997 \ --septemboard
which now looks like:
As far as I can tell & remember we now take care of all shortcomings that were known before.
8.4. FADC as veto
UPDATE:
The below lead to the discovery that the whole FADC veto had an
additional bug: namely we were not only checking if our index that we
need to check the rise/fall times for yield the same event number as
the number of the current cluster, but also check if the
fadcReadout flag is set. While that seems like a decent idea, it's
actually very bad, because the fadcReadout is a global dataset,
i.e. the indices of it do not match with the FADC dataset indices!
This all messed up the FADC veto. Fixed now and now the FADC veto acts
as one would expect, requiring some thoughts about how much efficiency
loss it is worth to us.
All files that were in /tmp/playground (/t/playground) referenced
here and in the meeting notes are backed up in
~/development_files/07_03_2023/playground
(to not make sure we lose something / for reference to recreate some
in development behavior etc.)
While investigating the effect of the FADC as a veto tool, I noticed that it seems like there is a large fraction of events that pass the logL cut that do not have FADC triggers. In the sense that this fraction seems to be much higher than expected (> 50%). This comes up by setting the FADC to trigger whenever there is an FADC trigger.
Let's look at the distribution of energies vs. fadc triggers.
import ggplotnim, nimhdf5 import ingrid / tos_helpers #let p1 = "/t/playground/lhood_2018_all_no_vetoes.h5"#"/home/basti/CastData/data/DataRuns2017_Reco.h5" let p1 = "/t/playground/lhood_2018_all_scinti_fadc.h5"#"/home/basti/CastData/data/DataRuns2017_Reco.h5" let h5f = H5open(p1, "r") let fileInfo = getFileInfo(h5f) var df = newDataFrame() for run in fileInfo.runs: let dfLoc = readRunDsets(h5f, run, chipDsets = some((chip: 3, dsets: @["energyFromCharge"])), commonDsets = @["fadcReadout"], basePath = "/likelihood/run_") df.add dfLoc ggplot(df, aes("fadcReadout")) + geom_bar() + ggsave("/t/events_with_fadcreadout.pdf") ggplot(df.filter(f{`energyFromCharge` < 15.0}), aes("energyFromCharge", fill = "fadcReadout")) + geom_histogram(bins = 50, alpha = 0.5, position = "identity", hdKind = hdOutline) + ggsave("/t/events_with_fadcreadout_energy.pdf")
Ok, something is fucked. As one would imagine of all the events
passing logL cut those above 2-2.5 keV in principle *ALL* have
~fadcReadout == 1! Therefore something MUST be broken in our FADC
veto!
-> Need comparison of our backgrounds after we fixed the issues!
With the above giving us an understanding about what was broken with the FADC veto and having fixed it, it was time to look at the background rates that could be achieved from that. The whole section 8.2 contains all the different ways we looked at the rise and fall times to deduce a reasonable cut value.
In the mean time we played around with different cut values (also using different values for Run-2 and Run-3).
Note that in order to generate the likelihood output files required
for the plots shown below, the likelihood.nim file had to be
modified before each run, as the FADC veto cuts are still hard coded!
This will be changed in the future.
All the plots were generated /tmp/playground at the time.
Run-2 for cut 105:
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out lhood_2017_all_scinti_fadc_105.h5 \ --region crGold --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --scintiveto --fadcveto \ --readOnly
Run-3 for cut 105:
likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out lhood_2018_all_scinti_fadc_105.h5 \ --region crGold --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --scintiveto --fadcveto \ --readOnly
After changing the value in likelihood.nim to 120:
Run-2 for cut 120:
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out lhood_2017_all_scinti_fadc_120.h5 \ --region crGold --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --scintiveto --fadcveto \ --readOnly
Run-3 for cut 120:
likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out lhood_2018_all_scinti_fadc_120.h5 \ --region crGold --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --scintiveto --fadcveto \ --readOnly
and now for some (maybe more appropriate?) numbers that are ~based on the 95-th percentiles for the Run-2 and Run-3 calibration data: Run-2 for cut 160:
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out lhood_2017_all_scinti_fadc_160.h5 \ --region crGold --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --scintiveto --fadcveto \ --readOnly
Run-3 for cut 120: Same as already computed above.
All these likelihood files with the same names are backed up in: ./../resources/fadc_rise_time_background_rate_studies/
From these we can generate different background rates (where we use the currently most up to date logL output files from the thesis resources):
Run-2 data with cut value of 160:
plotBackgroundRate ~/phd/resources/background/autoGen/likelihood_cdl2018_Run2_crGold.h5 \ lhood_2017_all_scinti_fadc_160.h5 \ --names "No vetoes" \ --names "fadc" \ --centerChip 3 \ --title "Background rate from CAST data (Run-2), incl. scinti and FADC veto (riseTime cut 160)" \ --showNumClusters --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_2017_crGold_scinti_fadc_160.pdf \ --outpath .
Dataset: No vetoes Integrated background rate in range: 0.0 .. 12.0: 2.2904e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.9087e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.0 .. 12.0: 1.9381e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.6150e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 2.5: 6.4023e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 3.2011e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.5 .. 2.5: 5.3849e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.6924e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 5.0: 1.1415e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.5367e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.5 .. 5.0: 9.1816e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.0403e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 2.5: 9.2560e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.7024e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.0 .. 2.5: 7.8416e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.1366e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 4.0 .. 8.0: 2.5063e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 6.2658e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 4.0 .. 8.0: 1.9108e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 4.7769e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 8.0: 1.6378e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 2.0472e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.0 .. 8.0: 1.3326e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.6657e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 2.0 .. 8.0: 7.7671e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.2945e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 2.0 .. 8.0: 6.0052e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.0009e-05 keV⁻¹·cm⁻²·s⁻¹
Run-3 data with cut value of 120:
plotBackgroundRate ~/phd/resources/background/autoGen/likelihood_cdl2018_Run3_crGold.h5 \ lhood_2018_all_scinti_fadc_120.h5 \ --names "No vetoes" \ --names "fadc" \ --centerChip 3 \ --title "Background rate from CAST data (Run-3), incl. scinti and FADC veto (riseTime cut 120)" \ --showNumClusters --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_2018_crGold_scinti_fadc_120.pdf \ --outpath .
Dataset: No vetoes Integrated background rate in range: 0.0 .. 12.0: 2.4289e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 2.0241e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.0 .. 12.0: 1.6004e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.3337e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 2.5: 5.6605e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.8303e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.5 .. 2.5: 4.6828e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.3414e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 5.0: 1.2196e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.7102e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.5 .. 5.0: 7.8733e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 1.7496e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 2.5: 8.1821e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.2728e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.0 .. 2.5: 6.8441e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 2.7376e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 4.0 .. 8.0: 2.8817e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 7.2043e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 4.0 .. 8.0: 1.7496e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 4.3741e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 8.0: 1.7033e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 2.1291e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.0 .. 8.0: 1.1424e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.4280e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 2.0 .. 8.0: 9.4171e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.5695e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 2.0 .. 8.0: 4.5799e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 7.6332e-06 keV⁻¹·cm⁻²·s⁻¹
Run-2 + Run-3 with cut value of 120:
plotBackgroundRate ~/phd/resources/background/autoGen/likelihood_cdl2018_Run2_crGold.h5 \ ~/phd/resources/background/autoGen/likelihood_cdl2018_Run3_crGold.h5 \ lhood_2017_all_scinti_fadc_120.h5 \ lhood_2018_all_scinti_fadc_120.h5 \ --names "No vetoes" --names "No vetoes" \ --names "fadc" --names "fadc" \ --centerChip 3 \ --title "Background rate from CAST data, incl. scinti and FADC veto (riseTime cut 120)" \ --showNumClusters --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_crGold_scinti_fadc_120.pdf \ --outpath .
Dataset: No vetoes Integrated background rate in range: 0.0 .. 12.0: 2.3355e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.9462e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.0 .. 12.0: 1.6005e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.3338e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 2.5: 6.1610e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 3.0805e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.5 .. 2.5: 4.7044e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.3522e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 5.0: 1.1669e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.5931e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.5 .. 5.0: 7.7849e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 1.7300e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 2.5: 8.9066e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.5626e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.0 .. 2.5: 7.0315e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 2.8126e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 4.0 .. 8.0: 2.6285e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 6.5711e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 4.0 .. 8.0: 1.5905e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 3.9762e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 8.0: 1.6591e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 2.0739e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.0 .. 8.0: 1.1468e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.4335e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 2.0 .. 8.0: 8.3039e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.3840e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 2.0 .. 8.0: 4.6375e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 7.7291e-06 keV⁻¹·cm⁻²·s⁻¹
Run-2 + Run-3 with cut value of 105:
plotBackgroundRate ~/phd/resources/background/autoGen/likelihood_cdl2018_Run2_crGold.h5 \ ~/phd/resources/background/autoGen/likelihood_cdl2018_Run3_crGold.h5 \ lhood_2017_all_scinti_fadc_105.h5 \ lhood_2018_all_scinti_fadc_105.h5 \ --names "No vetoes" --names "No vetoes" \ --names "fadc" --names "fadc" \ --centerChip 3 \ --title "Background rate from CAST data, incl. scinti and FADC veto (riseTime cut 105)" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_crGold_scinti_fadc_105.pdf \ --outpath .
Dataset: No vetoes Integrated background rate in range: 0.0 .. 12.0: 2.3355e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.9462e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.0 .. 12.0: 1.2690e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.0575e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 2.5: 6.1610e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 3.0805e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.5 .. 2.5: 4.5035e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.2518e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 5.0: 1.1669e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.5931e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.5 .. 5.0: 6.6297e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 1.4733e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 2.5: 8.9066e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.5626e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.0 .. 2.5: 6.8139e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 2.7256e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 4.0 .. 8.0: 2.6285e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 6.5711e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 4.0 .. 8.0: 9.5428e-06 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 2.3857e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 8.0: 1.6591e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 2.0739e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 0.0 .. 8.0: 9.7772e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.2221e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 2.0 .. 8.0: 8.3039e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.3840e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: fadc Integrated background rate in range: 2.0 .. 8.0: 3.0805e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 5.1341e-06 keV⁻¹·cm⁻²·s⁻¹
which now live here:
First of all these show that the FADC veto is indeed very worthwhile! We should make use of it somehow. These raise the following questions (from Klaus meeting notes on )
[ ]need to understand whether what we cut away for example in 3 keV region of background is real photons or not! -> Consider that MM detectors have Argon fluorescence in 1e-6 range! But they also have analogue readout of scintillator and thus higher veto efficiency![ ]Check the events which are removed in the background rate for riseTime cut of 105 that are below the FADC threshold! -> My current hypothesis for these events is that they are secondary clusters from events with main clusters above the FADC activation threshold! -> Indeed other event with large energy present. See below for more.
8.4.1. Investigate events removed in background rate at energies without FADC trigger [/]
Let's start with the events that are removed by the FADC, which have
energies lower than the threshold. How do we do that?
Easiest might be to do it in likelihood itself? Alternatively, take
the likelihood files without the FADC veto, but containing the FADC
data, then iterate those files and check for the FADC veto "after the
fact". This way we can check "which events do we remove?". And if
there's something that
- has lower energy than FADC trigger, but still has an
fadcReadout - lower rise time than cut
then create a plot?
Given that we need to access fadcReadout I'm not too certain how
plotData will fair, but we can give it a try.
Do
~/phd/resources/background/autoGen/likelihood_cdl2018_Run3_crGold.h5
already have the FADC in the output?
-> Nope.
So first generate some likelihood outputs using the same parameters,
but without FADC veto.
Run-2:
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out lhood_2017_all_no_vetoes_with_fadc_data.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --readOnly
Run-3:
likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out lhood_2018_all_no_vetoes_with_fadc_data.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --readOnly
The files live here:
- ./../resources/fadc_rise_time_background_rate_studies/lhood_2017_all_no_vetoes_with_fadc_data.h5
- ./../resources/fadc_rise_time_background_rate_studies/lhood_2018_all_no_vetoes_with_fadc_data.h5
First let's plot all events in this file that
- have
fadcReadout > 0-> No, becausefadcReadoutis a common dataset and those are not supported yet! Are a bit annoying to implement, but the same can be achieved by cutting on any FADC dataset (forces a read on FADC data) even if it's a dummy cut (i.e. from 0 to Inf for a positive variable). - have energy below 1 keV
(ok first attempt of cutting on fadcReadout did not work)
For Run-2 data:
plotData --h5file lhood_2017_all_no_vetoes_with_fadc_data.h5 \ --runType rtCalibration \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --cuts '("fadc/riseTime", 0.0, Inf)' \ --cuts '("energyFromCharge", 0.0, 1.0)' \ --applyAllCuts \ --eventDisplay --septemboard
Run-3:
plotData --h5file lhood_2018_all_no_vetoes_with_fadc_data.h5 \ --runType rtCalibration \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --cuts '("fadc/riseTime", 0.0, Inf)' \ --cuts '("energyFromCharge", 0.0, 1.0)' \ --applyAllCuts \ --eventDisplay --septemboard
the plots generated for these are:
where we have 5 events in the Run-3 data and 19 in the Run-2. NOTE: An important note about these event displays: As they are generated from the likelihood output files, they only contain those clusters that passed the lnL cut! That includes the center chip, but also the outer chips. Therefore the chips are as empty as they are!
If we closely look at the background rate for both of these (more easily seen in Run-3 data)
we can clearly count the number of counts less in the first two bins in green than in purple. In the Run-3 data we easily count 5 difference (check the height of a single count entry, then extrapolate). A bit harder to see, but still possible to get a rough idea this is the case in the Run-2 data.
Looking at these events, the majority (well, all but 7 in Run-2) are extremely high energy events that saturate the FADC. However the cluster visible in on the septemboard is very low energy of course. The explanation is that because the likelihood output only stores the clusters which pass lnL cuts, we simply don't see the other cluster that caused the FADC trigger. In many cases the events even look as if they might just be additional noise induced by the extremely ionizing event!
However, the more curious events are those that are not an FADC saturating signal in the FADC (those 7 events in Run-2). For these it would really be good to see the full events. They might be the same (another cluster present) or something different.
[X]IMPLEMENT the--eventsfunctionality inplotDatato plot individual event numbers. Idea is there, just needs to be finished! Then take the event numbers from the PDFs above of the event displays and generate the full event display of all the chip data to verify that indeed our hypothesis (if we can still call it that) is true. -> Done. -> Note: plotting multiple events at the same time is still broken!
All events in Run-3 are events with a huge spark in the top right chip towards the left edge.
[X]backup commands[X]backup plots
plotData --h5file ~/CastData/data/DataRuns2018_Reco.h5 --runType rtCalibration --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml --eventDisplay --runs 244 --events 12733 --septemboard plotData --h5file ~/CastData/data/DataRuns2018_Reco.h5 --runType rtCalibration --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml --eventDisplay --runs 274 --events 21139 --septemboard plotData --h5file ~/CastData/data/DataRuns2018_Reco.h5 --runType rtCalibration --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml --eventDisplay --runs 276 --events 26502 --septemboard plotData --h5file ~/CastData/data/DataRuns2018_Reco.h5 --runType rtCalibration --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml --eventDisplay --runs 279 --events 15997 --septemboard plotData --h5file ~/CastData/data/DataRuns2018_Reco.h5 --runType rtCalibration --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml --eventDisplay --runs 283 --events 15599 --septemboard
and so on for the ~/CastData/data/DataRuns2017_Reco.h5 file using
the run and event numbers mentioned here:
Let's look at a Run-2 event with "normal" structure:
run 76 event 83145 -> much bigger event present run 77 event 7427 -> same, super long track present on center chip
run 95, event number 102420 -> same run 95, event number 36017 -> different! almost looks like a normal event, at least the center chip part. However, there is still at track pointing at it. Why did it trigger? Maybe we "got lucky"? run 95, event number 39595 -> multiple clusters on center chip present run 97, event number 32219 -> another curious case like 95, 36017! run 97, event number 79922 -> classical track with long gaps between
And let's look at one FADC saturated event of Run-2 as well: 112, event number 12418 -> same as Run-3 data. Spark on top right towards left edge. In most cases these are induced by heavy activity on other chips (or that activity is noise and just happens to look quite natural?)
All these plots are found here:
The big question remains:
[ ]What do we do with this knowledge? Apply some additional filter on the noisy region in the top chip? For the events that look "reasonable" the FADC rise time there might precisely be the stuff that we want it for. The events are vetoed because they are too long. Given that there are tracks on the outside, this means they are definitely not X-rays, so fine to veto!
8.4.2. Updated background rate with FADC veto after algorithm update
After updating the FADC rise time / fall time algorithm, in particular , let's look at the background rate we get again.
likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out lhood_2018_crGold_scinti_fadc_new_algo.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --scintiveto --fadcveto \ --readOnly
./../resources/fadc_veto_new_rise_time_algo/lhood_2018_crGold_scinti_fadc_new_algo.h5
And compare it:
plotBackgroundRate \ ~/phd/resources/background/autoGen/likelihood_cdl2018_Run3_crGold.h5 \ lhood_2018_crGold_scinti_fadc_new_algo.h5 \ --names "No vetoes" \ --names "fadc" \ --centerChip 3 \ --title "Background rate from CAST data (Run-3), incl. scinti and FADC veto (riseTime cut 70)" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_2018_crGold_scinti_fadc_new_algo.pdf \ --outpath .
yields:
which looks surprisingly (and disappointingly?) similar to this plot:
and as a matter of fact it's worse.
But well, this was for cut values of:
const cutRiseLow = 40'u16 const cutRiseHigh = 70'u16 const cutFallLow = 350'u16 const cutFallHigh = 530'u16
which very well might be "less strict" now than the 120 were in the older plot!
For that reason, as the comparison without any more data as a basis is problematic let's create a ROC curve of the signal vs background data to have a better basis for what's going on and where we should cut.
In addition we should finally work on splitting up the FADC settings in Run-2.
- ROC curve
The script
/tmp/fadc_rise_fall_signal_vs_background.nimfrom section 8.2 also computes a ROC curve for the rise time. What this means is really the efficiency, if the rise time veto were performed purely on the upper end, disregarding both the lower cut as well as the cuts on the fall time (or skewness). So it should not be intended as a direct reference for the efficiency / suppression mapping found in the FADC data. However, it should only be better.The reason for not having a "realistic" ROC curve is that it is non trivial to define the existing FADC veto such that one can compute efficiencies over some range (as we have multiple parameters to vary instead of a single one!).
Figure 67: ROC curve based on the FADC rise time by cutting on the upper end only. The "dip" close to 0 on the x axis (signal efficiency) is due to a "bump" in the background rise time data at very low values. This is cut away in the FADC veto using an additional low rise time cut value!
8.4.3. Adaptive FADC veto cuts based on percentile of ⁵⁵Fe calibration data
As discussed in the meeting notes (e.g. 26.60) and to an extent in sections above, the different FADC amplifier settings used imply that a fixed set of cuts on the rise and fall times is not suitable (sec. 8.2.3).
Instead the idea is to perform the percentile calculations as done in
for example /tmp/fadc_rise_fall_signal_vs_background.nim (and the
energy dep. snippet) and use it as a basis for a set of cuts with a
desired percentile. This allows us to directly quantify the induced
dead time / signal efficiency of the cuts.
This has now been implemented in likelihood.nim. The desired
percentile is adjusted using a command line argument
--vetoPercentile and defaults to the 99-th (and 1st on lower
end). Note the two ended-ness and therefore the signal efficiency is
twice the percentile!
In addition to the percentile used, there is an additional parameter,
the --fadcScaleCutoff, which defines a scale factor for a hard cut
towards the upper end of the data before the percentile is
determined. Because we know from
sec. 8.2.2.1.6 (end of that section)
that the major contribution to the very long tail even left after the
10% top offset was implemented is double X-ray hits (with clusters too
close to separate), we want to remove the impact of those events on
the calculation of the percentile. We have no perfect way of
determining those events and so a hard cut at a value where we are
reasonably certain no real X-rays will be found above is chosen. The
scale factor is 1.45 by default, which corresponds to the factor
multiplied to the peak position of the rise/fall time distribution.
For now the code also generates a plot for the rise and fall time
each, for each FADC setting in /tmp/, which indicate the percentile
cuts and the hard cut off.
So now let's apply likelihood to both Run-2 and Run-3 datasets with
different veto percentiles and see both the plots of the distributions
with the cuts as well as the background rates we can achieve.
Example Run-2:
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out lhood_2017_crGold_scinti_fadc_adaptive_fadc_cuts_99perc.h5 \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --scintiveto --fadcveto \ --readOnly \ --vetoPercentile 0.99
Example Run-3:
likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out lhood_2018_crGold_scinti_fadc_adaptive_fadc_cuts_99perc.h5 \ --calibFile ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --scintiveto --fadcveto \ --readOnly \ --vetoPercentile 0.99
(Note the replacement of the years in the background data file, the output file name and the calibration file! If you run with the wrong calibration file you will be greeted by an assertion error though).
In addition we run the same for the veto percentiles 90 and 80 as well. Note that the 80 percentile case implies removing 40% of the data!
The FADC distribution plots are found here:
and the data files here:
- ./../resources/fadc_veto_percentiles/lhood_2017_crGold_scinti_fadc_adaptive_fadc_cuts_80perc.h5
- ./../resources/fadc_veto_percentiles/lhood_2017_crGold_scinti_fadc_adaptive_fadc_cuts_90perc.h5
- ./../resources/fadc_veto_percentiles/lhood_2017_crGold_scinti_fadc_adaptive_fadc_cuts_99perc.h5
- ./../resources/fadc_veto_percentiles/lhood_2018_crGold_scinti_fadc_adaptive_fadc_cuts_80perc.h5
- ./../resources/fadc_veto_percentiles/lhood_2018_crGold_scinti_fadc_adaptive_fadc_cuts_90perc.h5
- ./../resources/fadc_veto_percentiles/lhood_2018_crGold_scinti_fadc_adaptive_fadc_cuts_99perc.h5
As is pretty obvious the hard cut off in each of these plots (black vertical line) is very conservative. There shouldn't be anything to worry about using the scale factor 1.45 as done here.
Let's plot the background rate comparing to the no veto case: Run-2:
plotBackgroundRate \ ~/phd/resources/background/autoGen/likelihood_cdl2018_Run2_crGold.h5 \ lhood_2017_crGold_scinti_fadc_adaptive_fadc_cuts_99perc.h5 \ lhood_2017_crGold_scinti_fadc_adaptive_fadc_cuts_90perc.h5 \ lhood_2017_crGold_scinti_fadc_adaptive_fadc_cuts_80perc.h5 \ --names "No vetoes" \ --names "fadc_99perc" \ --names "fadc_90perc" \ --names "fadc_80perc" \ --centerChip 3 \ --title "Background rate from CAST data (Run-2), incl. scinti and FADC veto" \ --showNumClusters --showTotalTime --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_2017_crGold_scinti_fadc_adaptive_cuts.pdf \ --outpath .
Run-3:
plotBackgroundRate \ ~/phd/resources/background/autoGen/likelihood_cdl2018_Run3_crGold.h5 \ lhood_2018_crGold_scinti_fadc_adaptive_fadc_cuts_99perc.h5 \ lhood_2018_crGold_scinti_fadc_adaptive_fadc_cuts_90perc.h5 \ lhood_2018_crGold_scinti_fadc_adaptive_fadc_cuts_80perc.h5 \ --names "No vetoes" \ --names "fadc_99perc" \ --names "fadc_90perc" \ --names "fadc_80perc" \ --centerChip 3 \ --title "Background rate from CAST data (Run-3), incl. scinti and FADC veto" \ --showNumClusters --showTotalTime --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_2017_crGold_scinti_fadc_adaptive_cuts.pdf \ --outpath .
which are here:
The major aspect sticking out is that going from a percentile of 90 to 80 does not actually improve the background rate much more (only in the 3 keV peak, which makes sense if these are real photons). In addition it is quite interesting to see that in the 80% case we manage to remove essentially all background in the range after the 3 keV and before the 8 keV peak! For the latter:
Dataset: fadc_80perc
Integrated background rate in range: 4.0 .. 8.0: 5.4593e-06 cm⁻² s⁻¹
Integrated background rate/keV in range: 4.0 .. 8.0: 1.3648e-06 keV⁻¹·cm⁻²·s⁻¹
i.e. in the very low 1e-6 range there!
8.4.4. TODO Investigate how cutting rise time percentile impacts fall time data [/]
[ ]When we perform a percentile cut on the rise time, e.g. 5% this will
remove some of the tail of the fall time data at the same time. So saying that 5% on rise and 5% on fall time implies only 0.95·0.95 = 0.9025 remaining data is simply wrong. I assume that cutting on one is almost perfectly correlated with the other. But we should investigate this.
[ ]At the same time there is a question about how much the fall time veto even helps at all! How much more does it even remove? Does it remove anything the rise time doesn't already remove?
8.5. Noisy events
From section 8.2.1.1
Ideas for better noise detection:
- some events are completely in the negative, no positive at all. Very unlikely to be non noise, unless an event was extremely long, which physically doesn't make much sense.
- real noise events, independent of their frequency have extreme peaks not only to negative from baseline, but also to positive! This likely means we can look at the data away from the baseline. If it has significant outliers towards both sides it's likely noise. We could look at a histogram of these types of events compared to others?
I think one problem is using mean & standard deviation. If the fluctuations are generally very large in the data, then of course the σ is also very large. Need to think this over.
- Skewness of the FADC data is usually < -2! -> We've now added the skewness as an FADC dataset in TPA.
Back to this: let's try to implement the improvements to the noise filtering.
The skewnees is a very useful property. Let's look at all skewness values of all FADC data:
import nimhdf5, ggplotnim import std / [strutils, os, sequtils, stats] import ingrid / [tos_helpers, fadc_helpers, ingrid_types, fadc_analysis] proc getFadcSkewness(h5f: H5File, run: int): DataFrame = let fadcRun = readRecoFadcRun(h5f, run) let recoFadc = readRecoFadc(h5f, run) let num = fadcRun.eventNumber.len var skews = newSeqOfCap[float](num) for idx in 0 ..< fadcRun.eventNumber.len: skews.add fadcRun.fadcData[idx, _].squeeze.toSeq1D.skewness() result = toDf({skews, "riseTime" : recoFadc.riseTime.asType(float), "noisy" : recoFadc.noisy}) echo result proc main(fname: string) = let tmpFile = "/tmp/" & fname.extractFilename.replace(".h5", ".csv") var df = newDataFrame() if not fileExists tmpFile: var h5f = H5open(fname, "r") let fileInfo = h5f.getFileInfo() var dfs = newSeq[DataFrame]() for run in fileInfo.runs: echo "Run = ", run let fadcGroup = fadcRecoPath(run) if fadcGroup in h5f: # there were some runs at end of data taking without any FADC (298, 299) dfs.add h5f.getFadcSkewness(run) df = assignStack(dfs) df.writeCsv(tmpFile) else: df = readCsv(tmpFile) echo df ggplot(df, aes("skews")) + geom_density() + ggsave("/tmp/fadc_skewness_kde.pdf") ggplot(df, aes("skews", "riseTime", color = "noisy")) + #geom_point(size = 1.0, alpha = 0.2) + geom_point(size = 0.5, alpha = 0.75) + themeLatex(fWidth = 0.9, width = 600, baseTheme = singlePlot) + ggsave("/tmp/fadc_risetime_skewness.pdf", dataAsBitmap = true) when isMainModule: import cligen dispatch main
./fadc_data_skewness -f ~/CastData/data/DataRuns2017_Reco.h5
Having looked at these plots
 I wanted to understand where the blotch of non noise events near rise
time 180-210 and skewness -0.7 to 0.3 come from. These plots are here:
I wanted to understand where the blotch of non noise events near rise
time 180-210 and skewness -0.7 to 0.3 come from. These plots are here:
 from
NOTE: This should not have been run with
from
NOTE: This should not have been run with --chips 3!
plotData --h5file ~/CastData/data/DataRuns2017_Reco.h5 \ --runType rtBackground \ --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --eventDisplay \ --cuts '("fadc/riseTime", 150.0, 210.0)' --cuts '("fadc/skewness", -0.7, 0.3)' \ --applyAllCuts --septemboard
It seems there are both a mix of events in there that are noisy, as well as some that are not noisy, but likely just multi clusters within a short time (i.e. having ionization gaps) thus leading to long events. However, at skewness values above -0.4 there does not seem to be anything of 'value'. So in the FADC veto we'll include a cut on the skewness. Pretty clearly from the skewness vs risetime plot we can see that there is a hard edge closer to -1, so we won't be removing much of value.
Notes:
- below fall time of 20 there are essentially only events that
contain either noise or high energy events inducing noise on the
septemboard. All FADC events are noisy and our current algo
identifies it as such.

NOTE: This should not have been run with --chips 3!
plotData --h5file ~/CastData/data/DataRuns2017_Reco.h5 --runType rtBackground \ --chips 3 --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --eventDisplay --cuts '("fadc/fallTime", 0.0, 20.0)' --applyAllCuts --septemboard
- below rise time of 20 there are a mix of noise events similar to
the fall time case, but also quite some that are real, sensible
looking signals, where the FADC signal just drops immediately to
the minimum amplitude of the signal.
These are not identified as noise (which is correct). Just look
through the following:
 and you'll see plenty of them.
and you'll see plenty of them.
NOTE: This should not have been run with --chips 3!
plotData --h5file ~/CastData/data/DataRuns2017_Reco.h5 --runType rtBackground \ --chips 3 --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --eventDisplay --cuts '("fadc/riseTime", 0.0, 20.0)' --applyAllCuts --septemboard
[X]The only final part is what is the data at rise times < 20 or so and skewnesses -2 to -0.5 or so that are not noisy? The same kind of super steep rising events? (I mean partially must be, because we looked at those before, just without skewness info! But just verify). from
from
NOTE: This should not have been run with --chips 3!
plotData --h5file ~/CastData/data/DataRuns2017_Reco.h5 \ --runType rtBackground --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --eventDisplay \ --cuts '("fadc/riseTime", 0.0, 20.0)' --cuts '("fadc/skewness", -2.2, -0.6)' \ --applyAllCuts --septemboard
So, YEP. It's exactly those kind of events.
9. Clustering algorithms
The default clustering algorithm is, among other places, discussed in 13.
It's just a simple radius based clustering algorithm.
For the meeting with Klaus on I implemented the DBSCAN clustering algorithm into TPA.
It's very successful in particular for the septem veto producing more "realistic" clustering.
Reference: https://scikit-learn.org/stable/auto_examples/cluster/plot_cluster_comparison.html#sphx-glr-auto-examples-cluster-plot-cluster-comparison-py compare the DBSCAN column, in particular the "track" example (row 4).
9.1. Discussion of the clustering of ~/org/Mails/KlausUpdates/klaus_update_03_08_21/
The first two files are a continuation of the septem events of all clusters that pass the logL cuts in the gold region.
Instead of using the old clustering algorithm we now use the DBSCAN clustering algorithm. The two files correspond to two different settings.
The minimum number of samples in a cluster is 5 in both cases. The ε parameter (something like the search radius) is 50 and 65 respectively. The latter gives the better results (but the final number needs to be determined, as that is still just a number from my ass).
In addition the ε = 65 case contains lines that go through the cluster centers along the slope corresponding to the rotation angle of the clusters. These lines however are currently not used for veto purposes, but will be in the future.
Looking at the clustering in the ε = 65 case we learn:
- some center clusters that are still separate are still not passing now. Why? Is this because DBSCAN drops some pixels changing the geometry or because of the energy computation?
- many clusters where something is found on the outside are now correctly identified as being connected to something.
- few clusters are not connected to the outside cluster. Might be caught with a slight ε modification?
- some clusters (of those still passing / not passing due to bug, see above?) can be removed if we use the lines drawn as an additional veto (e.g. line going through 1 σ of passing cluster).
With this the veto stuff is essentially done now.
- Scintillator vetoes are implemented and will be used as a straight cut if < 80 clock cycles or something like this
- septem veto has just been discussed
- FADC: FADC will be dropped as a veto, as it doesn't provide enough information, is badly calibrated, was often noisy and won't be able to provide a lot of things to help.
If one computes the background rate based on the DBSCAN clustering septem veto, we get the background rate shown in the beginning. The improvement in the low energy area is huge (but makes sense from looking at the clusters!).
9.2. DBSCAN for Septemevent veto clustering
The septem veto works such that for all events that have a passing cluster on the central chip, we read the full data from all chip into a "Septemevent", i.e. a event with 768x768 pixel.
These septem events are processed from the beginning again, including clustering. For each cluster anew the logL (and everything required for it) is calculated and checked if any cluster passes the cut.
The effect of this clustering can be seen in the comparison of the following two files:
~/org/Mails/KlausUpdates/septem_events_clustered_2017.pdfUtilizes the 'normal' clustering algorithm with a search radius of 50 pixels~/org/Mails/KlausUpdates/klaus_update_03_08_21/septemEvents_2017_logL_dbscan_eps_65_w_lines.pdfUtilizes DBSCAN with an ε = 65.
The main take away is that DBSCAN performs better for clusters that are humanly separateable.
Compare the following examples. Fig. 68 shows the default clustering algorithm with a default search radius of 50 pixels. There are 4 clusters found in total. Arguably the most bottom right cluster should connect to the one on the right middle chip. Increasing the search radius to 65 pixels, seen in fig. 69, results in a correct unification of these two clusters. However, now the two parallel tracks are also part of the same cluster. Ideally, they remain separate.
The DBSCAN algorithm with an ε = 65 is seen in fig. 70. It produces exactly the 'intuitively expected' result. The parallel tracks are still separate, but the track pointing to the cloud are part of the same cluster.
This showcases the better behavior of DBSCAN. Where a human sees separate clusters due to density variations, the DBSCAN algorithm approaches that intuitive behavior instead of a 'dumb' distance computation.
9.3. TODO Investigate why default cluster PDF has 194 pages and DBSCAN only 186!!
9.4. TODO Investigate why cluster centers sometimes exactly in middle
9.5. TODO Investigate why sometimes clusters fail to pass that should [0/2]
Might be the energy calculation or actually the clustering being a problem. Investigate.
9.5.1. TODO check if clustering the reason (dropped pixels)
9.5.2. TODO check if energy calculation still the reason
9.6. TODO Change logic to only 'pass' septem event if new cluster in gold region passes
It's not good enough to check if any cluster passes, because we might get completely different clusters that end up passing on the outside chip. That's not desired. It must be a cluster that is actually where we are looking!
9.7. TODO Once clustering done reconstruct all data incl. CDL
This reconstruction should be done as part of the reproducibility steps. We write the code (well it mostly exists!) to reconstruct everything including the CDL reference data with the new clustering algorithm.
10. Ground up investigation of Marlin / TPA differences
UPDATE: While working on the evaluation of systematics, in particular of the software efficiency, another severe bug was found, which caused a wrong mapping of the logL cut values to the energy bins. This lead to some areas having artificially low / high software efficiencies, instead of the desired percentage. This seems to largely explain the remaining difference between the Marlin and TPA results, in particular at energies below 1 keV. See the background rate in 20.11 and discussion of the bug in sec. 14.7.
As mentioned in the section 7, a bug was discovered which caused the calculation of the total charge to be wrong. Since the charge calibration function is a non-linear approximation (see implementation here: https://github.com/Vindaar/TimepixAnalysis/blob/master/Analysis/ingrid/calibration.nim#L812-L832 ), essentially the inverse of
ToT[clock cycles] = a*TestPuseHeight [mV] + b - ( c / (TestPuseHeight [mV] -t) )
care has to be taken how the total charge of a cluster is calculated. In ToT mode the Timepix reports the ToT values for each pixel. So in order to calculate the total charge of a cluster, the easy and correct way is to calculate the charge values of each active pixel in a cluster and then sum those individual charges to a total charge. However, in the past the code summed the ToT values first and calculated the charge from that huge ToT value, which is way outside of the range of values for which the calibration function can be assumed to yield valid results. In pseudo code the wrong and correct way:
# assuming ToT is a sequence of all ToT values in a cluster and # calibrateCharge is the charge calibration function let wrong = ToT.sum.calibrateCharge # wrong, but previously done let correct = ToT.map(x => x.calibrateCharge).sum
This bug has since been fixed. However, the calculation of all previously shown results with this fix in place is somewhat more complicated. On the one hand simply due to having to recalculate all data and recreate all plots (\(\mathcal{O}(\SI{1}{\day})\) of work), but mainly due to the fact that such a bug could even go unnoticed in the first place.
Due to this and the still prevalent issue of the TimepixAnalysis code not recovering the same background rates as the MarlinTPC code even for the 2014/15 datasets, a "ground up investigation" was started.
As often the case when programming in research environments writing of unit tests is often neglected due to time constraints. The only real way to solve the above mentioned issues to actually start writing many unit tests.
Most work is spent on tests, which extract elements from the MarlinTPC data results and compare each single cluster property with the same data reconstructed with the TimepixAnalysis code.
This work is still ongoing.
Relevant PR: https://github.com/Vindaar/TimepixAnalysis/pull/40 Still WIP. Original PR was "only" a refactor of the raw data manipulation code, but for practical reason writing a large number of tests while refactoring delayed this process.
See the 13 section below for further information on this.
11. Ray tracer simulation of axions
Johanna worked on the implementation of a ray tracer to simulate the expected flux of axions from the Sun (using Arshia's code as a basis, which performs the calculation of the axion production in the Sun based on all axion electron production processes) and resulting flux of X-rays after conversion in the CAST magnet. It takes into account the properties of the LLNL X-ray telescope.
@Johanna: a couple of plots would be handy:
- solar axion image
- image of LLNL shells from simulation
The flux that is sampled from the differential flux and thus used as an input for the axions to be ray traced, is shown in fig. 71.
11.1. Performance enhancements of existing ray tracing code
Profiling the code using perf pointed at 3 main performance
bottlenecks:
- most of the time spent in
findPosXRT - also a lot spent in
getRandomPointFromSolarModel - as well as
getRandomEnergyFromSolarModel
11.1.1. Change implementation of findPosXRT
The old implementation for findPosXRT looks as follows:
proc findPosXRT*(pointXRT: Vec3, pointCB: Vec3, r1, r2, angle, lMirror, distMirr, uncer, sMin, sMax: float): Vec3 = ## this is to find the position the ray hits the mirror shell of r1. it is after ## transforming the ray into a coordinate system, that has the middle of the ## beginning of the mirror cones as its origin var point = pointCB s: float term: float sMinHigh = (sMin * 100000.0).int sMaxHigh = (sMax * 100000.0).int pointMirror = vec3(0.0) let direc = pointXRT - pointCB for i in sMinHigh.int .. sMaxHigh.int: s = i.float / 100000.0 term = sqrt((point[0] + s * direc[0]) * (point[0] + s * direc[0]) + (point[ 1] + s * direc[1]) * (point[1] + s * direc[1])) - ((r2 - r1) * (point[ 2] + s * direc[2] - distMirr) / (cos(angle) * lMirror)) if abs(r1 - term) < 2.0 * uncer: pointMirror = point + s * direc ## sometimes there are 3 different s for which this works, in that case the one with the highest value is taken result = pointMirror
where sMin is 1.0 and sMax is 1.1.
This means the implementation literally tries to find the optimal s
by walking over a for loop in 10_000 steps! No wonder it's a
bottleneck.
Our replacement does this with a binary search. We won't invest more time into this, because it will be replaced once we rewrite the ray tracing aspect.
proc findPosXRT*(pointXRT: Vec3, pointCB: Vec3, r1, r2, angle, lMirror, distMirr, uncer, sMin, sMax: float): Vec3 = ## this is to find the position the ray hits the mirror shell of r1. it is after ## transforming the ray into a coordinate system, that has the middle of the ## beginning of the mirror cones as its origin var point = pointCB s: float term: float sMinHigh = sMin sMaxHigh = sMax pointMirror = vec3(0.0) let direc = pointXRT - pointCB template calcVal(s: float): untyped = let res = sqrt((point[0] + s * direc[0]) * (point[0] + s * direc[0]) + (point[ 1] + s * direc[1]) * (point[1] + s * direc[1])) - ((r2 - r1) * (point[ 2] + s * direc[2] - distMirr) / (cos(angle) * lMirror)) res var mid = (sMaxHigh + sMinHigh) / 2.0 while abs(r1 - term) > 2.0 * uncer: if abs(sMinHigh - sMaxHigh) < 1e-8: break term = calcVal(mid) if abs(r1 - calcVal((sMinHigh + mid) / 2.0)) < abs(r1 - calcVal((sMaxHigh + mid) / 2.0)): # use lower half sMaxHigh = mid mid = (sMinHigh + mid) / 2.0 else: # use upper half sMinHigh = mid mid = (sMaxHigh + mid) / 2.0 pointMirror = point + mid * direc ## sometimes there are 3 different s for which this works, in that case the one with the highest value is taken result = pointMirror
NOTE: This new implementation still has some small bugs I think. But no matter for now.
11.1.2. Change implementation of getRandomPointFromSolarModel
The old implementation of the random sampling from the solar model is (already cleaned up a bit):
## proc gets `emRatesVecSums` which is: `var emRatesVecSums = emRates.mapIt(it.sum)` let angle1 = 360 * rand(1.0) angle2 = 180 * rand(1.0) ## WARN: loop over emRatesVecSums randEmRate = rand(emRatesVecSums.sum) ## Idea seems to be: ## - sample a random emission rate between 0 <= emRate <= totalSolarEmission ## - search for the radius that this emission rate belongs to in the sense ## that the radius up to that point combines emRate flux block ClearThisUp: var prevSum = 0.0 ## WARN: another loop over full vec! for iRad in 0 ..< emRateVecSums.len - 1: # TODO: why len - 1? Misunderstanding of ..< ? if iRad != 0 and randEmRate > prevSum and randEmRate <= emRateVecSums[iRad] + prevSum: # what are these exact values? related to solare model radial binning I presume. Should be clearer r = (0.0015 + (iRad).float * 0.0005) * radius elif iRad == 0 and randEmRate >= 0.0 and randEmRate <= emRateVecSums[iRad]: r = (0.0015 + (iRad).float * 0.0005) * radius prevSum += emRateVecSums[iRad] let x = cos(degToRad(angle1)) * sin(degToRad(angle2)) * r let y = sin(degToRad(angle1)) * sin(degToRad(angle2)) * r let z = cos(degToRad(angle2)) * r result = vec3(x, y, z) + center
which means we loop over the emRatesVecSums twice in total! First
for sums and then in the for loop explicitly.
Inserting some simple sampling + plotting code:
## sample from random point and plot var rs = newSeq[float]() var ts = newSeq[float]() var ps = newSeq[float]() for i in 0 ..< 100_000: let pos = getRandomPointFromSolarModel(vec3(0.0, 0.0, 0.0), radiusSun, emratesCumSum) # convert back to spherical coordinates let r = pos.length() rs.add r ts.add arccos(pos[2] / r) ps.add arctan(pos[1] / pos[0]) let df = toDf(rs, ts, ps) ggplot(df, aes("rs")) + geom_histogram(bins = 300) + ggsave("/tmp/rs.pdf") ggplot(df, aes("ts")) + geom_histogram() + ggsave("/tmp/ts.pdf") ggplot(df, aes("ps")) + geom_histogram() + ggsave("/tmp/ps.pdf") if true: quit()
The old implementation yields the following sampling for the radii, fig. 72.
The new implementation simply does the same idea using inverse
transform sampling. Instead of the emRatesVecSums we compute a
normalized CDF:
var emRatesCumSum = emRates.mapIt(it.sum).cumSum() # normalize to one let emRateSum = emRatesCumSum[^1] emRatesCumSum.applyIt(it / emRateSum)
and replace the above implementation simply by:
let angle1 = 360 * rand(1.0) angle2 = 180 * rand(1.0) ## random number from 0 to 1 corresponding to possible solar radii. randEmRate = rand(1.0) rIdx = emRateCumSum.lowerBound(randEmRate) r = (0.0015 + (rIdx).float * 0.0005) * radius let x = cos(degToRad(angle1)) * sin(degToRad(angle2)) * r let y = sin(degToRad(angle1)) * sin(degToRad(angle2)) * r let z = cos(degToRad(angle2)) * r result = vec3(x, y, z) + center
Which does the same except simpler and faster.
This yields the same result, fig. 73.
Note: The "spiking" behavior is due to the selection of a specific
radius using rIdx. Since only discrete radii are available, this
leads to an inherent binning artifact. Choosing more bins shows that
there are many "empty" bins and few with entries (i.e. the ones
available according to the radius computation).
A better approach might be to fuzz out the radii in between a single
radius. The problem with that is that then we need to recompute the
correct index for the getEnergyFromSolarModel proc.
Instead we should have a proc, which returns an index corresponding to a specific radius. Using this index we can access the correct CDFs for the energy and emission rate. Then the radius can be fuzzed as well as the energy.
This also leads to the same kind of behavior in the energy and emission distributions.
11.1.3. Change implementation of getRandomEnergyFromSolarModel
This procedure has the same kind of problem as the above. It performs internal for loops over data, which are completely unnecessary. Again we will do sampling using inverse CDF.
The old implementation (already modified from the original):
proc getRandomEnergyFromSolarModel(vectorInSun, center: Vec3, radius: float64, energies: seq[float], emissionRates: seq[seq[float]], computeType: string): float = ## This function gives a random energy for an event at a given radius, biased ## by the emissionrates at that radius. This only works if the energies to ## choose from are evenly distributed ## var rad = (vectorInSun - center).length r = rad / radius iRad: int indexRad = (r - 0.0015) / 0.0005 if indexRad - 0.5 > floor(indexRad): iRad = int(ceil(indexRad)) else: iRad = int(floor(indexRad)) ## WARN: another loop over full emrate seq let ffRate = toSeq(0 ..< energies.len).mapIt(emissionRates[iRad][it] * energies[it] * energies[it]) ## WARN: another loop over full ffrate seq let ffSumAll = ffRate.sum let sampleEmRate = rand(1.0) * ffSumAll ## WARN: another loop over full ffrate seq let ffCumSum = cumSum(ffRate) ## WARN: another ~half loop over full cumsum seq let idx = ffCumSum.lowerBound(sampleEmRate) ## TODO: this is horrible. Returning a physically different function from the same ## proc is extremely confusing! Especially given that we even calculate both at the ## same time! case computeType of "energy": let energy = energies[idx] * 0.001 result = energy of "emissionRate": let emissionRate = emissionRates[iRad][idx] result = emissionRate
Similar sampling code to the above:
var es = newSeq[float]() var ems = newSeq[float]() for i in 0 ..< 100_000: let pos = getRandomPointFromSolarModel(centerSun, radiusSun, emratesCumSum) let energyAx = getRandomEnergyFromSolarModel( pos, centerSun, radiusSun, energies, emrates, "energy" ) let em = getRandomEnergyFromSolarModel( pos, centerSun, radiusSun, energies, emrates, "emissionRate" ) es.add energyAx ems.add em let df = toDf(es, ems) ggplot(df, aes("es")) + geom_histogram(bins = 500) + ggsave("/tmp/es.pdf") ggplot(df, aes("ems")) + geom_histogram(bins = 500) + ggsave("/tmp/ems.pdf") ggplot(df.filter(f{`ems` > 0.0}), aes("ems")) + geom_histogram(bins = 500) + scale_x_log10() + ggsave("/tmp/ems_log_x.pdf")
yields the following two sampling plots, fig. 74, 75.
Note to self: when computing a CDF, don't drop non-linear terms that one feels are not important…
New implementation:
proc getRandomEnergyFromSolarModel(vectorInSun, center: Vec3, radius: float64, energies: seq[float], emissionRates: seq[seq[float]], emRateCDFs: seq[seq[float]], computeType: string): float = ## This function gives a random energy for an event at a given radius, biased ## by the emissionrates at that radius. This only works if the energies to ## choose from are evenly distributed ## var rad = (vectorInSun - center).length r = rad / radius iRad: int indexRad = (r - 0.0015) / 0.0005 if indexRad - 0.5 > floor(indexRad): iRad = int(ceil(indexRad)) else: iRad = int(floor(indexRad)) let cdfEmRate = emRateCDFs[iRad] let idx = cdfEmRate.lowerBound(rand(1.0)) ## TODO: this is horrible. Returning a physically different function from the same ## proc is extremely confusing! Especially given that we even calculate both at the ## same time! case computeType of "energy": let energy = energies[idx] * 0.001 result = energy of "emissionRate": let emissionRate = emissionRates[iRad][idx] result = emissionRate
This does indeed produce the same sampling results, fig. 76, 77.
11.1.4. TODO again if sampling of emission rate the same! Does look different
11.1.5. Summary
With these improvements we get from about 1 Mio. axions in 1 min on 32 threads to ~ 50 Mio. in 1 min on 12 threads.
So a ~133 speedup.
This is about 4,166,666 rays per minute and thread!
10 Mio. on 12 threads: 17.039 s.
11.2. Axion-photon conversion
As mentioned in section 17.5.2, the photon conversion done in the current ray tracing code:
func conversionProb*(B, g_agamma, length: float): float {.inline.} = result = 0.025 * B * B * g_agamma * g_agamma * (1 / (1.44 * 1.2398)) * (1 / (1.44 * 1.2398)) * (length * 1e-3) * (length * 1e-3) #g_agamma= 1e-12 echo conversionProb(1.0, 1.0, 1e3)
is likely to be wrong.
We will replace it by code using unchained, something like:
import unchained, math defUnit(GeV⁻¹) func conversionProb(B: Tesla, L: MilliMeter, g_aγ: GeV⁻¹): UnitLess = result = pow( (g_aγ * B.toNaturalUnit * L.toNaturalUnit / 2.0), 2.0 ) echo conversionProb(1.T, 1000.mm, 1.GeV⁻¹) echo conversionProb(9.T, 9.26.m.to(mm), 1e-10.GeV⁻¹)
| 0.245023 UnitLess |
| 1.70182e-17 UnitLess |
See also Klaus derivation of the magnetic field conversion in
.
The flux after the full raytracing through the magnet (using \(N =
\num{1e7}\) MC samples) before changing this conversion probability is shown in
fig. 78. After changing the code to use
unchained we get the flux shown in
fig. 79 for the exact same parameters.
The flux (i.e. weighted histogram of weights + traced axions / photons) remaining after traversing through the magnet and being converted without any detector window or gas absorption, is shown in fig. 80. It is also simulated using \(N = \num{1e7}\) number of axions and is thus directly comparable to fig. 79 with the detector efficiency being the only difference.
This flux will be used in a test limit calculation of the 2013 CAST paper data (because of their very high quantum efficiency).
\clearpage
11.3. DONE Can we compute the effective area of the telescope using the code?
Try to reproduce fig. 259.
UPDATE: This has been implemented a couple of months ago, but the numbers don't match either the PhD thesis by A. Jakobsen nor the JCAP CAST paper about the LLNL telescope. See more here: Sec. [BROKEN LINK: sec:journal:2023_07_13] in ./../journal.html and the couple of days after that.
Generally, for more information about the telescope, see sec. 2.11.
11.4. Axion image at CAST conditions [0/1]
UPDATE
In the last week I 'finished' (there are still things to do, but) the
interactive raytracer. Doing that I uncovered a major random sampling
bug in the old raytracing code in the randomPointFromSolarModel
procedure. It used a naive sampling approach for a random vector in a
sphere, which gave too much weight to z values, but too little to
x, y and thus reproduced the correct "radii", but did not
reproduce the correct angular distributions!
[ ]A more updated plot will be added soon.
UPDATE :
I've since learned (see journal.org and email "CAST detectors behind
LLNL telescope") that the design is such that the focal spot is in
the gas volume center and not at the readout plane! This changes the
calculation!
-> Instead of 1470 + 12.2 it is 1500 - (15 - 12.2) = 1500 - 2.8 =
1497.2
UPDATE 2: -> A few days after the above mail I redid the calculation of the average absorption depth in ./SolarAxionConversionPoint/axion_conversion_point.html and sec. 3.3.1.2, which actually yields a median value of about 0.3 cm behind the window! -> See further below for an updated axion image using the new found knowledge.
The following aspects are of interest for the CAST image:
- rotation of the window when the detector was installed at CAST: 30°
- window material and shape
- distance at which to compute the image
- the axion model to consider: axion electron
The latter was computed in 3.3.1 for CAST conditions and ends up being about 1.22 cm inside of the detector.
The resulting axion image (without the detector window!) is shown in:
The data for this case is stored in:
./../resources/axion_image_no_window_1470mm_plus_12_2mm.csv
For a version in the center of the detector (equivalent to 1.5 cm in the compared to the above 1.22 cm) with the window transmission included:
The data for this case is stored in:
./../resources/axion_image_30deg_1485mm.csv
UPDATE: The code was recently updated to correct the computation of the cone tracing algorithm in https://github.com/jovoy/AxionElectronLimit/pull/19
With this in place in particular for telescopes other than the LLNL it seems some significant changes took place.
The shape of the axion image changed slightly from before this PR to after. The shape before is the one shown in the figure above 97.
The new plot is shown in
When looking closely at the comparison it is visible that the new traced image is more focused in the center. There is less "bright yellow" around the center spot and it's not as wide in x direction. This should in theory mean that the limit should improve using this axion image. See the limit sanity section for an update on this, sec. 29.1.2.4.
11.4.1. Computing axion image based on numerical absorption depth
NOTE: We learned at some point from Jaime, that the actual focal point is \(\SI{1}{cm}\) behind the window, not \(\SI{1.5}{cm}\). The numbers here assume the 1.5!
From sec. 3.3.1.2 we know the real depth is closer to 0.3cm behind the window.
The correct number is 1487.93 mm behind the telescope. We need to have
this in our config.toml of raytracer:
distanceDetectorXRT = 1487.93 # mm
Let's generate the correct plot with the detector window included.
./raytracer --ignoreGasAbs --suffix "" --distanceSunEarth 0.9891144450781392.AU --title "Axion electron image for CAST GridPix setup behind LLNL telescope"
The resulting plot was moved here:
11.5. Development of the interactive raytracer, results and lessons learned
These are just some notes about the development of the interactive raytracer to simulate a LLNL like CAST telescope.
They were written after most of the hard problems were done and their aim is to document the bugs that were encountered.
In addition they contain preliminary results computed with this raytracer and certain ideas to play around with.
At some point after implementing the ImageSensor material I added an
option via F5 to save the current image buffer, count buffer and the
internal buffers of the SensorBuf objects. It stores these buffers
as a binary file (just dumping the raw buffers data to file).
To visualize those, I wrote plotBinary:
./../../CastData/ExternCode/RayTracing/plotBinary.nim.
The calls we ran over the last days are found in sec. [BROKEN LINK: sec:raytracer:interactive:plotBinaryCalls].
Before I started to work on the LLNL telescope, I had to implement lighting materials as well as the primitive shapes for cylinders and cones.
11.5.1. Initial attempts
After implementing the LLNL telescope initially, the first thing I tried is to place the camera into the focal point and see what happens. To be honest at that point I wasn't really sure what to expect.
The image I got (as a screenshot) is by running the raytracer from the
focal point (using --focalPoint) with a vfov of maybe 5 and the
background color as black:
 which shows (most of) the shells illuminated very well. (Note: The
different shells are illuminated differently, which was an initial
hint that something was a bit fishy with the telescope layout!) This
confused me for a bit until I realized this totally makes sense. A
camera like implemented in the raytracer can never see an image in
the focal spot. The camera essentially is infinitely small and follows
the rays under different incoming angles. So effectively, a regular
physical camera that could observe the projected image would receive
the entire image as seen above onto a single pixel. So to
reconstruct the image from the telescope, we need to invert the
principle. We need a sensor with a physical size that accumulates
information from all angles onto single pixels. Then different
pixels see different amounts of flux, which is the source of the
actual image!
which shows (most of) the shells illuminated very well. (Note: The
different shells are illuminated differently, which was an initial
hint that something was a bit fishy with the telescope layout!) This
confused me for a bit until I realized this totally makes sense. A
camera like implemented in the raytracer can never see an image in
the focal spot. The camera essentially is infinitely small and follows
the rays under different incoming angles. So effectively, a regular
physical camera that could observe the projected image would receive
the entire image as seen above onto a single pixel. So to
reconstruct the image from the telescope, we need to invert the
principle. We need a sensor with a physical size that accumulates
information from all angles onto single pixels. Then different
pixels see different amounts of flux, which is the source of the
actual image!
For this reason I started to think about ways to implement this. The
first idea was to have a separate raytracing mode, which works "in
reverse". This birthed the TracingType in the code with ttCamera
and ttLights.
Instead of shooting rays from the camera, in ttLights mode we emit
lights from a source towards an invisible target. This was initially
done in the rayColorAndPos procedure. The next step was to implement
a new material ImageSensor, which acts as a physical sensor in the
3D space. The idea is to have an object that the raytracing algorithm
than intersect with. Then once we found an intersection with it, we
record the position on the object using the \((u, v)\) coordinates. In
the ttLights mode we then went ahead sampled rays from the light
source (disk at the end of the magnet) towards the other end of the
magnet. Those that hit the image sensor were then recorded and their
\((u, v)\) coordinates mapped to the bufT tensor and correspondingly
displayed.
Doing this then finally produced an actual image, in

which is "Cris' candle" haha.
I made the following screenshots of the entire setup here:
which I sent to the GasDet discord CAST/IAXO channel.
Given that the ttLights implementation had the following
limitations:
- doesn't with multithreading, because the target position of each ray
is unknown at the time of sampling and therefore each thread cannot
safely write to a subset of the output buffer (unless one were to
use a
Lock) - requires a slightly hacky
rayColorAndPosimplementation that differs from the regularrayColorprocedure
I decided to start work on making the ImageSensor a more sane
object.
The idea was to make it have an internal buffer to record when it was hit by rays.
For that purpose I implemented a helper object SensorBuf, which is
just a very dumb buffer store. It allocates memory and keeps it as a
ptr UncheckedArray so that it can be copied around easily and only
a single buffer is actually allocated. Independent on which thread
reads and writes to it, it all ends up in one buffer. The =destroy
hook takes care of only freeing up the memory when the memory owner is
destroyed (which can be passed over by a =sink).
Initially this (obviously) caused segfaults. Memory access is now
guarded with a Lock, which seems to work fine. The code runs both
under ARC and ORC with no issues.
The ImageSensor then was implemented so that the hit count stored in
the buffer is increased each time the scatter procedure is hit. Note
that the internal buffer is effectively a 2D array. If one maps the
ImageSensor to a non rectangular object, the resulting distribution
may not accurately reflect the hit positions due to distortions!
The emit procedure simply returns the current value of the buffer at
that position. I then included the Viridis colormap into the project
so that I can base the emitted color on the Viridis color map. In
order to know where we are on the colormap, there is a currentMax
field which stores the currently largest value stored in the buffer.
The initial attempt at looking at the parallel disk seen in the screenshot above on the ImageSensor can be seen in the following image:

As we can see this works very well! The 'resolution' of the
ImageSensor is decided at construction.
Note that at this point the ImageSensor was still sensitive to
every incoming ray, whether it came from the camera or from a light
source. See sec. 11.5.4 for
more on this.
11.5.2. Implementing the graphite spacers
In the screenshots of the previous sections the graphite spacer was not implemented yet. This was done afterwards and is now present.
11.5.3. First X-ray finger result
Next I wanted to look at the X-ray finger based on the DTU PhD thesis numbers (14.2 m and 3mm radius). This yielded the following image:

We can see two things:
- it generally looks reasonable, the graphite spacer is visible, resulting in the typical two lobed structure.
- there is a very distinct kind of high focus area and regions with lower flux.
Shortly after we implemented writing the buffers and wrote
plotBinary. The same figure as a plot:
I ran the old raytracer with a similar setup (14.2 m and 3 mm):
The config file contains:
[TestXraySource] useConfig = true # sets whether to read these values here. Can be overriden here or useng flag `--testXray` active = true # whether the source is active (i.e. Sun or source?) sourceKind = "classical" # whether a "classical" source or the "sun" (Sun only for position *not* for energy) parallel = false energy = 3.0 # keV The energy of the X-ray source distance = 13960.64 #14200 #20000 # 9260.0 #106820.0 #926000 #14200 #9260.0 #2000.0 # mm Distance of the X-ray source from the readout radius = 3.0 #21.5 #44.661 #8.29729 #46.609 #4.04043 #3.0 #4.04043 #21.5 # #21.5 # mm Radius of the X-ray source offAxisUp = 0.0 # mm offAxisLeft = 0.0 # mm activity = 0.125 # GBq The activity in `GBq` of the source lengthCol = 0.0 #0.021 # mm Length of a collimator in front of the source
where the distance number is simply due to the pipe lengths in front of the telescope that is currently not part of the interactive simulation.
./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_xray_finger_1390mm_3mm"
This yields:
Comparing this to our result above shows three main differences:
- it is much smoother
- it is wider
- the focus seems more towards the "narrow" part of the image.
My first thought was on whether this could be due to us not making use of reflecitvities. So rerun it without taking reflectivity into account:
./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_no_refl_1390mm_3mm" --ignoreReflection
which gives us:
We can see that the general shape is the same, but the flux becomes "wider" and moves slightly more towards the left, i.e. to the wide part of the image.
It's closer to the new raytracer, but not the same.
At this point we continued along with a few other things before coming back to this difference.
See sections 11.5.6 and 11.5.8 for further work in understanding the differences and fixing the X-ray finger result.
11.5.4. Making the ImageSensor insensitive to camera rays
As the ImageSensor was also sensitive to the Camera based rays, it
was problematic to go close to the sensor. At that point we would be
shooting orders of magnitudes more rays at it from the camera than
from light sources (we sample camera and light rays in 1 to 1
ratio). This overwhelmed the current max value rendering the image
'broken'.
As such we added a new enum RayType and extended the Ray type to
store such a field. The initRay now receives the origin type of the
ray. That way we know whether a ray comes from a light source
initially or from the camera. The hits are only added to the
ImageSensor when a ray comes from a light source. On each bounce the
ray type is copied.
This solves the problem nicely and allows to easily differentiate the two types of rays.
From this point on the ImageSensor is only sensitive to rays from
light sources.
11.5.5. Making light sources emit into specific directions
In order to sample rays from a light source, we need a general direction in which to sample. If we simply sampled in any direction (for a diffuse light) as it theoretically emits into, it would end up incredibly computationally inefficient. At least for the purpose of an image sensor behind an X-ray telescope we know that we only care about a very specific direction.
(This would of course be very different if we simply want to sample rays into a room and measure e.g. the amount of light received due to bounces from all possible directions or something like this!)
Initially the sampling of rays for the ImageSensor was handled in a
hardcoded fashion in the sampleRay procedure. In it first we sample
a point using samplePoint from a light source. We defined a
hardcoded target towards / inside the magnet which we sampled another
point from and then connected the two using the vector describing the
difference of the two points. Alternatively, if we wanted parallel
light we simply sampled towards the -z direction.
In order to make this nicer, we added another material, LightTarget,
which is added to the scene geometry just like any other object.
Then when sampling rays in sampleRay we construct targets from the
light source to any target. This is done in a random fashion. We just
uniformly sample light sources and targets (so we do not currently
take area / volume of a source / target into account!).
These LightTargets can either be visible for Camera rays or
invisible. This is changed by using the --visibleTarget argument.
11.5.6. Fix placement of the first mirrors (height)
From the initial X-ray finger result mentioned above, I thought about looking into the exact placement of the mirrors again.
This revealed that my initial placement code was wrong. The initial implementation just used
\[ y_i = R_{1,0} - R_{1,i} \]
where \(R_{1,i}\) is the radius 1 of layer \(i\). And in addition then moved the layer position by \(δ = \sin(α_i) l_M / 2\) before the rotation. But we did not move the position by \(δ\) in y after rotation. This effectively meant we did not correctly take into account the difference in height of the cone at the center. As a result the second layers were also slightly off, because they are based on the first ones.
Doing this massively improved the homogeneity of the X-ray finger image as well as the illumination of the shells as seen from the camera, see this screenshot:
 We can see that the illumination is pretty much perfect. Maybe there
is still a reason why the upper and lower mirrors do not reflect
everything perfectly though. Who knows. I don't think so though, I
think this is just the fact that the focal spot is also not a perfect
point. This seems to me like a limitation of constructing a telescope
from cones. Perfect coverage of all layers at the same time is likely
not possible?
-> NOTE: The likely reason for the still not perfect illumination is
the wrong focal length! At this point we are at 1500 mm, but it seems
like the correct focal length is 1530mm!
We can see that the illumination is pretty much perfect. Maybe there
is still a reason why the upper and lower mirrors do not reflect
everything perfectly though. Who knows. I don't think so though, I
think this is just the fact that the focal spot is also not a perfect
point. This seems to me like a limitation of constructing a telescope
from cones. Perfect coverage of all layers at the same time is likely
not possible?
-> NOTE: The likely reason for the still not perfect illumination is
the wrong focal length! At this point we are at 1500 mm, but it seems
like the correct focal length is 1530mm!
[ ]REVISE THIS ONCE FOCAL LENGTH SECTION DONE[ ]Potentially check with 'constructed' telescope from a bottom mandrel.
This change meant the resulting X-ray finger image was then:
We can see a much cleaner image with better flux coverage near the center and less structure visible. Still some visible though!
Next we fixed the calculation of the y displacement, due to some
cosine / sine stuff:
which is a tiny change.
We made another small change along the same lines for:
See the next section 11.5.7 for further improvements later.
- Note about mirror angles
Also another important point about the layers: Because we are constructing cones, we don't actually have to rotate the mirrors themselves. They get their correct rotation BY CONSTRUCTION. Because we construct cones precisely such that they have the desired angle using
\[ h = \tan(α) / R \]
which gives us the required height of the cone. A cone described by radius \(R\) and height \(h\) has the opening angle \(α\).
11.5.7. Improving mirror placement by using R1, R5 and angles
Up to this point the telescope system was computed using the numbers used in the old raytracer. This meant we used the R1, angle and xSep values. At the same time we did not use the tabulated R5 values.
As discussed in great length in the following document ./LLNL_def_REST_format/llnl_def_rest_format.html this is not a wise choice.
Instead the basic idea should be the following:
Mirrors centered at lMirror/2.
Entire telescope
lMirror xsep lMirror
| |----| | ( Magnet bore )
^-- center 1
^-- center 2
^-- z = 0
due to rotation around the *centers* of the mirrors, the real x separation increases.
`xSep = 4 mm` exactly, `lMirror = 225 mm` and the correct cones are computed based on the
given angles and the R1, R5 values. R4 is the relevant radius of the cone for the
second set of mirrors. Therefore R1 and R4 are actually relevant.
meaning the telescope in theory is exactly 2 · lMirror + xSep long, where xSep is exactly 4 mm and given along the x direction!
Then by rotating the mirrors the x separation changes. And the mirrors are rotated around their center.
Finally, we should use the R5 angles in order to compute R4 required for the second mirrors and make the changes accordingly to compute the height differences based on a fixed xSep and lMirror length of each "part" of the telescope.
This is done in the code now.
The result is
for the 14.2 m, 3 mm case and
This was a good improvement, but there is still some parts with more flux than expected. I don't fully remember what actually fixed it in the end, because I worked on other stuff for a while, rewriting a whole bunch of stuff, refactoring the scene construction etc etc. In the end now it looks very reasonable, see the next section plot!
11.5.8. Reconstructing the X-ray finger result from the old raytracer
At this point the X-ray finger (14.2m, 3mm) looks as follows:
./raytracer --width 1200 --maxDepth 10 --speed 10.0 --nJobs 32 --vfov 30 --llnl --focalPoint --sourceKind skXrayFinger ./plotBinary -f out/image_sensor_0_2023-08-21T19:44:22+02:00__dx_14.0_dy_14.0_dz_0.1_type_int_len_160000_width_400_height_400.dat --dtype int --outfile /tmp/image_sensor_14x14_llnl_xrayFinger_14.2m_21_08_23.pdf
which looks damn near identical to the result from the old raytracer
when excluding the reflectivity
in terms of the distribution. The size of the result is still too
large though!
We can reconstruct something that looks more similar (i.e. wider), if we wrongly use the radius R1 again for the second sets of mirrors. Meaning instead of constructing a cone that naturally has the correct radius, we construct one of a too large radius and move it to the correct position. In order to get the correct angle simply the total height of the cone changes, but it is possible to construct such a cone.
The following plot shows the result for an X-ray finger (same distance
etc) using R1 again for the second set:
Note that this needs to be compared with the result from the previous section. One can see that the main change is the result becomes much wider than in the correct case.
Rerunning this case with the current version of the code yields:
[[~/org/Figs/statusAndProgress/rayTracing/debugAxionImageDifference/imagesensor14x14llnlxrayFinger14.2msamer1forsecondcones210823.pdf]
Note that this is still different. The center position is further to the right / left in the two. For the new raytracer the position is more 'to the center' for the entire blob, the old one is centered more towards the side. The reason for this is likely the position of the focal point, see sec. 11.5.9.
11.5.9. Computing the position of the focal point
Initially the focal point was computed based on the spurious \(\SI{2.75}{°}\) found in the old raytracing code. However, the placement was always slightly wrong. In addition the old raytracer also contains a number, \(\SI{-83.0}{mm}\) for the apparent offset from the center (?) of the telescope to the focal spot in the direction that is "away" from the axis of the coldbore. This neither worked out for us for a position of the focal spot!
Thinking about the telescope layout more (also in terms of a theoretical full telescope, see sec. 11.5.12), I thought it should be pretty easy to compute the position of the focal spot by hand.
Essentially, we know the focal spot is precisely on the axis of the cones! And we know their radii exactly. That means at the z axis that defines each cone, the focal spot is found.
Nowadays we have this function in the code to compute the point based on the
smallest radius R1. We use that one, because the center of the first mirror
shell is aligned exactly with the bottom of the magnet bore. Our coordinate
system center is on the center of the magnet bore and at the end of the
telescope in z. This means the center of the first mirror shell is precisely
at z = -boreRadius. With the angle we can thus compute the center of the
focal point along the y axis (or x axis if we rotate the telescope the way it
is at CAST) by:
proc llnlFocalPoint(fullTelescope: bool): (Point, Point) = let lMirror = 225.0 let α0 = 0.579.degToRad let xSep = 4.0 ## xSep is exactly 4 mm let r1_0 = 63.006 let boreRadius = 43.0 / 2 let yOffset = if fullTelescope: 0.0 else: (r1_0 - (sin(α0) * lMirror) / 2.0) + boreRadius let zOffset = lMirror + xSep / 2.0 let focalDist = 1500.0 let lookFrom = point(0.0, -yOffset, - focalDist + zOffset) let lookAt = point(0.0, 0.0, 0.0) # Telescope entrance/exit center is at origin result = (lookFrom, lookAt)
This is likely the reason the new raytracer spots are at a different position compared to the old raytracer, which uses a somewhat random approximation of the position!
11.5.10. Missing intersections for rays from Sun
When testing for solar emission for the first time there were multiple
issues. In essence there was no image visible on the ImageSensor, because
no sampled rays actually got through the telescope.
While the exact details are still unclear to me, the reason was likely due to floating point uncertainty. The numbers for 1 AU, solar radius etc. in milli meter are obviously large O(1e14), but that should still be enough accuracy within a 64 bit floating point number (given the mantissa has precision up to 16 digits), at least assuming that the initial math is not sensitive to the last digits. But likely that is precisely the case due to the calculation of the hit intersection for the cones of the telescope. The calculation of the \(a, b, c\) coefficients squares the components, including \(z\) (which is the largest). When solving the square polynomial equation this likely causes drastic issues with the floating point accuracy.
Ideally we would handle this case similar to it is done in pbrt, namely by
using a dual floating point type that carries the floating point uncertainty
in all calculations.
But for the time being we work around it in a simple way.
Our ray sampling algorithm, as explained earlier, works by
- sample a point on the light source, \(p\)
- sample a point on the
LightTarget, \(p_t\) - create a vector \(d = p_t - p\)
- the final ray is defined by origin \(p\) and direction \(d\)
As we know the target we aim for, we simply propagate the initial ray to the target point \(p_t\) minus a unit length.
There is one serious downside to this: If the user defines a LightTarget
that might be blocked from the direction of the light source, we simply
skip through the potential target. So it is up to the user to make sure
that the light target can never be occluded from the light source, otherwise
faulty tracing can happen!
This solves the problem nicely.
11.5.11. Random sampling bug
Looking at the solar emission after fixing all the bugs above - based on homogeneous emission in the center 20% of the Sun - yielded an image as seen in:
As we can see it is significantly bigger than the axion image that we normally construct from the other raytracer.
Trying to understand this behavior led down another rabbit hole. I
implemented another kind of emissive material, SolarEmission. The idea is
to use the radial flux emission as described by the solar model, same as it
is done in the other raytracer. I copied over the data parsing, preparation
routines as well as the getRandomFromSolarModel procedure.
Then when sampling from a Sphere that has the SolarEmission material,
instead of relying on homoegenous sampling of random vectors, we sample based
on the above procedure.
Doing this lead to the following axion image in the focal point:
As we can see this much more resembles the axion image from the other raytracer, but it is a bit smaller. It turned out that I had a small bug due to keeping the size of the Sun at 20% instead of the full radius, making the emission another factor 0.2 smaller in radial distribution.
Fixing this then produced
which is pretty close to the axion image from the other raytracer, as seen here:
However, given that the sampled radii are mostly from a between the 10-20% of the sun makes the stark difference extremely confusing.
I then did some debug hacking of the code to produce plots about the sampled points in the Sun.
The sampled radii for the homogeneous emission looked like this:
which might look a bit confusing at first, but makes sense: the volume of a sphere grows by \(R³\). Thus the possible points to sample also grows the same way with the radius, which means we expect to sample points according to \(R³\). Perfect.
Sampling for the SolarEmission (i.e. the solar model CDF sampling via
getRandomFromSolarModel):
NOTE: There is a big difference here, because this plot includes the entire
real solar radius, whereas the one above only goes to 0.2 · Rsun!
(The spiky behavior is due to binning artifacts). This also looks exactly like expected! We can see the same \(R³\) behavior at low radii and then the decay as expected.
The two plots are in contradiction to the difference in the axion images. Which lead me to the conclusion that the difference must be coming from the individual components of the sampled vectors.
Plotting these then yielded
which is as expected the same distribution for each component!
But for the points from getRandomFromSolarModel we found instead:
which, uhhhhhhhhhhhhhh, is not what we want.
As it turns out the sampling code is naive. :(
Without comments the code looked like this:
proc getRandomPointFromSolarModel(radius: float, fluxRadiusCDF: seq[float], rnd: var Rand): Point = let φ = 360 * rnd.rand(1.0) θ = 180 * rnd.rand(1.0) randEmRate = rnd.rand(1.0) rIdx = fluxRadiusCDF.lowerBound(randEmRate) r = (0.0015 + (rIdx).float * 0.0005) * radius # in mm let x = cos(degToRad(φ)) * sin(degToRad(θ)) * r let y = sin(degToRad(φ)) * sin(degToRad(θ)) * r let z = cos(degToRad(θ)) * r result = point(x,y,z)
I remember thinking about whether this is valid random sampling from a sphere back when working on the performance improvements and refactorings of the old raytracer, but dumb me only looked at the combined sampled RADII and not at the individual components! The issue is - as discussed before - that the volume changes with the radius. Sampling uniformly for the angles does not correctly reflect that.
See for example also this ref. about sampling from a uniform sphere: https://corysimon.github.io/articles/uniformdistn-on-sphere/ it has some nice plots visualizing what happens here.
We fixed this by simply using rejection sampling (as done elsewhere in the code) to first get a random vector on a unit sphere, and then scaling this by the target radius.
This then results in the following \((x,y,z)\) distributions:
which now works as intended! :partyingface:
The resulting axion image for the SolarEmission is now
which is great news in terms of the comparison to the homogeneous emission case, but terrible news for the actual solar axion image going into the limit calculation, because the area sensitive to axions becomes bigger. :) Also one needs to keep in mind that this is the focal spot and not the point where the median conversions happen!
11.5.12. Simulating a 'full LLNL telescope'
[ ]Include images of the setup
Out of pure curiosity I wanted to see what the LLNL telescope would look like as a full telescope in front of a larger magnet. Given the simplicity in the change (keeping the image sensor at \(x = y = 0\) and changing the mirror size to \(\SI{360}{°}\)), here it is:
From the front (incl disk blocking center):
 from the side:
from the side:
 from slightly above:
from slightly above:
 towards the image sensor:
towards the image sensor:

Note: the first of these plots were done before the random sampling bug and others were fixed.
Note 2: the first screenshot is at \(\SI{1500}{mm}\) distance, which I believe is not the real focal length of the telescope as described in the DTU PhD thesis, see sec. 11.5.15.
Our initial attempt at producing a plot from parallel light at 12m away looked like this:
The reason for this was plainly that we did not block the inner part of the telescope! When constructing a full LLNL telescope with the few mirrors that we have radii for (without computing the correct radii for lower ones), the entire center of the telescope is empty of course! This means emission of a parallel source is dominated by the light that just arrives straight ahead (there's a question about the flux of the focused light and the center unfocused, but at this point here we can ignore that due to the way the sampling worked).
After putting in a disk to block the center region, we got the following
which is a very neat looking focus!
There is a very slight deviation visible from a perfect symmetry.
- because of the graphite spacer (we did not put in N spacers, but left the single spacer)
- there is minutely more flux at the bottom compared to the top. I assume this is because of us copying the exact numbers that were built for the LLNL telescope. Extending them to a full circle does likely not produce perfect alignment for all shells.
Also we can see deviations from a perfect point due to the fact that we have a cone approximation instead of a true Wolter I type telescope.
Producing the image for a 12m away X-ray finger source looked like this:
Fun! The fact it is so unfocused is likely due to the very short distance of only 12 m combined with our telescope now having a radius of \(\SI{105}{mm}\) radius, but the X-ray finger still only being \(\SI{3}{mm}\) in radius. In addition the center of the telescope is insensitive, resulting in all rays having to have very large angles. Thus they are not focused correctly.
For solar emission we got this:
Again we can see a very slight difference in amount of flux from top to bottom, same as in the parallel light case. The sampling in this case is homogeneous within the center 20% of the solar radius.
11.5.13. Simulating a 'double LLNL telescope'
One thing that occurred to me as a crackpot idea as to why the axion image
from Michael Pivovaroff from LLNL is symmetric around the center,
might be due to a misunderstanding.
What if he simulated two LLNL like telescopes, one for each magnet bore with a single focal point in between the bores?
Given the fact that this raytracer makes it trivial to test such an idea, I went ahead and implemented this idea (hey it's just a copy the beamline, rotate by 180° and move a bit away!).
Behold the mighty double LLNL setup:
From above:
 From the front:
From the front:
 Towards the readout
Towards the readout ImageSensor in between the two:

Emission from the sun using homogeneous emission for this case is given by:
and using the realistic SolarEmission we get:
As one would expect, the result in this case is indeed symmetric around the center, just like the LLNL result. However, we still don't produce those excessively long tails visible in that! Also the image has a somewhat narrower shape than the LLNL result (that one is more elliptical).
11.5.14. Simulation of the 'figure error' similar to NuSTAR of 1 arcmin
In the DTU PhD thesis by Jakobsen, the caption of the raytracing results from
Pivovaroff,
 it talks about the NuSTAR optic having a 'figure error' of 1 arc minute.
it talks about the NuSTAR optic having a 'figure error' of 1 arc minute.
If I understand this correctly (I'm not aware of a technical interpretation of the term 'figure error') I assume this means the following:
Instead of having perfect reflection, the reflected rays are scattered into a cone around the ideal reflection with a size of 1 arc minute.
Assuming we consider the reflected ray to have unit length, we produce such rays by sampling a vector from a unit sphere \(\vec{r}\) with radius \(r_{\text{fuzz}}\). If \(\vec{r}\) points orthogonal to the reflected ray, the required \(r_{\text{fuzz}}\) required for 1 arc minute is:
\[ \tan(1') = r_{\text{fuzz}} / 1 \]
where the 1 denominator is because of the unit length of the scattered ray. This implies \(r_{\text{fuzz}} = 0.0002908\).
Also see the fuzz section here: https://raytracing.github.io/books/RayTracingInOneWeekend.html#metal/fuzzyreflection
Using this approach results in an axion image from the real solar
emission (after fixing the sampling) of:
and for a figure error about twice as large:
And for an X-ray finger using fuzz value of r_fuzz = 0.0005 yields
In all cases we can see that as expected such a figure error smoothes out the result significantly.
11.5.15. Focal distance weirdness
So while writing the above I started to notice that the telescope does not appear to be fully illuminated. This started when I created the illumination screenshot for sec. 11.5.6 and then later when I created the screenshot for the full LLNL telescope, when I got really skeptical.
Consider the screenshot for the full telescope again, which is taken at 1500 mm distance from the telescope center, i.e. apparently in the focal point:

We can clearly see that not all layers are fully illuminated!
I started playing around, moving the camera further back (yay for interactivity!) showed a much better
At 1530 mm:
 at 1550 mm:
at 1550 mm:
 at 1570 mm:
at 1570 mm:

CLEARLY the illlumination of the shells is much better in all of these than in the initial one at 1500 mm. While they get slightly 'better' illumination in 1550 and 1570 mm than in 1530 mm, the bright red part of it becomes much less after 1530 mm.
So I looked at the parallel light for the full telescope at 1530 mm only to find:
WHAT you're telling me the full telescope produces an almost perfect focal point 30 mm behind the supposed actual focal point??
So I looked at a few homogeneous sun emission cases for the regular LLNL telescope at different distances:
1525 mm:
1530 mm:
1535 mm:
And then I thought, uhh, what the hell, what does the axion image look like in the old raytracer (NOTE: using the bugged sampling code still!) at different distances?
default 1500 mm, comets points left:
1550 mm, comet points right:
1525 mm, holy fuck, it is symmetric after all:
JFC, you're telling me the symmetric image was always about 25 to 30mm BEHIND where we were looking? I don't believe it.
A few more plots about the correct solar emission from the new
raytracer, at 1525 mm:
quite a bit further away:
These following two plots while at rayAt = 1.0 were done by modifying the angles α
by
\[
α' = α · 1.02
\]
because this is \(1530/1500 = 1.02\):
and without the graphite spacer to see if the visible horizontal line
is actually just the graphite spacer:
and the answer is no, because it is still visible!
The same but with a 1 arcminute figure error:
And now (*still the same settings as last three plots, i.e. angles
modified!) trying to reproduce the LLNL raytracing result by running
with the 1 arc minute figure error and also using the 3 arc minute
apparent size of the sun. First 15% of solar radius contributing:
and then a real 3 arc minute source, i.e. 0.094 % of the solar radius contributing:
which is actually not that different than the LLNL result! It's just a
bit more flat.
All of this is extremely disturbing and confusing. Let's calculate the theoretical focal distance using the Wolter equation.
According to the NuSTAR DTU thesis and its reference the "The Rainwater Memorial Calibration Facility (RaMCaF) for X-ray optics" https://downloads.hindawi.com/archive/2011/285079.pdf minimum radius of a NuSTAR optic is 54.4 mm.
In our telescope it is 63.006 mm if one uses R1. 53.821 for R5.
Taking the code from llnldefrestformat.org:
import unchained, math const lMirror = 225.mm const xSep = 4.mm const d_glass = 0.21.mm const R1_1 = 63.006.mm const α1 = 0.579.degToRad const α2 = 0.603.degToRad proc calcR3(r1, lMirror: mm, α: float): mm = let r2 = r1 - lMirror * sin(α) result = r2 - 0.5 * xSep * tan(α) # 1. compute R3 of layer 1 let R3_1 = calcR3(R1_1, lMirror, α1) echo R3_1 # 2. compute R1 of layer 0 (mandrel) # based on R3_i+1 = R1_i + d_glass # -> R1_i-1 = R3_i - d_glass let R1_0 = R3_1 - d_glass # 3. approximate α0 as α1 - (α2 - α1) let α0 = α1 - (α2 - α1) echo α0 # 4. compute R3_0 using α0 let R3_0 = calcR3(R1_0, lMirror, α0) echo R3_0 # 5. use Wolter equation to compute `f`! # -> tan(4α) = R3 / f let f = R3_0 / tan(4 * α0) echo "Approximate focal length: ", f echo "Using layer 1: ", R3_1 / tan(4 * α1)
| 60.7121 mm | |||
| 0.009686577348568527 | |||
| 58.3033 mm | |||
| Approximate | focal | length: | 1503.99 mm |
| Using | layer | 1: | 1501.15 mm |
Pretty close to 1500…
So at least the existing radii and angles seem to match our focal length of 1500mm. At least compared to our 'experimental' value of closer to 1525-1530mm.
I think what's going on is that our placement of the mirrors is slightly off. I don't understand why that leads to an apparent shift of the focal point, but that's what seems to be happening. -> NO: This was a mistake yesterday evening. I changed the angles of all shells from \(α ↦ 1.02 · α\). That was what changed the focal spot to the target 1500mm. I got that number from 1530 / 1500 = 1.02, knowing the focal spot is 1.02 too far behind.
This here:
contains analytical calculations about X-ray optics which follow the
conical approximation. They explicitly state (page 3):
 that the focal length of a conical approximation is always slightly
larger than the focal length of a real Wolter type 1 optic!
Unfortunately they don't specify by how much (at least I haven't seen
it yet).
that the focal length of a conical approximation is always slightly
larger than the focal length of a real Wolter type 1 optic!
Unfortunately they don't specify by how much (at least I haven't seen
it yet).
I figured it out!
See:
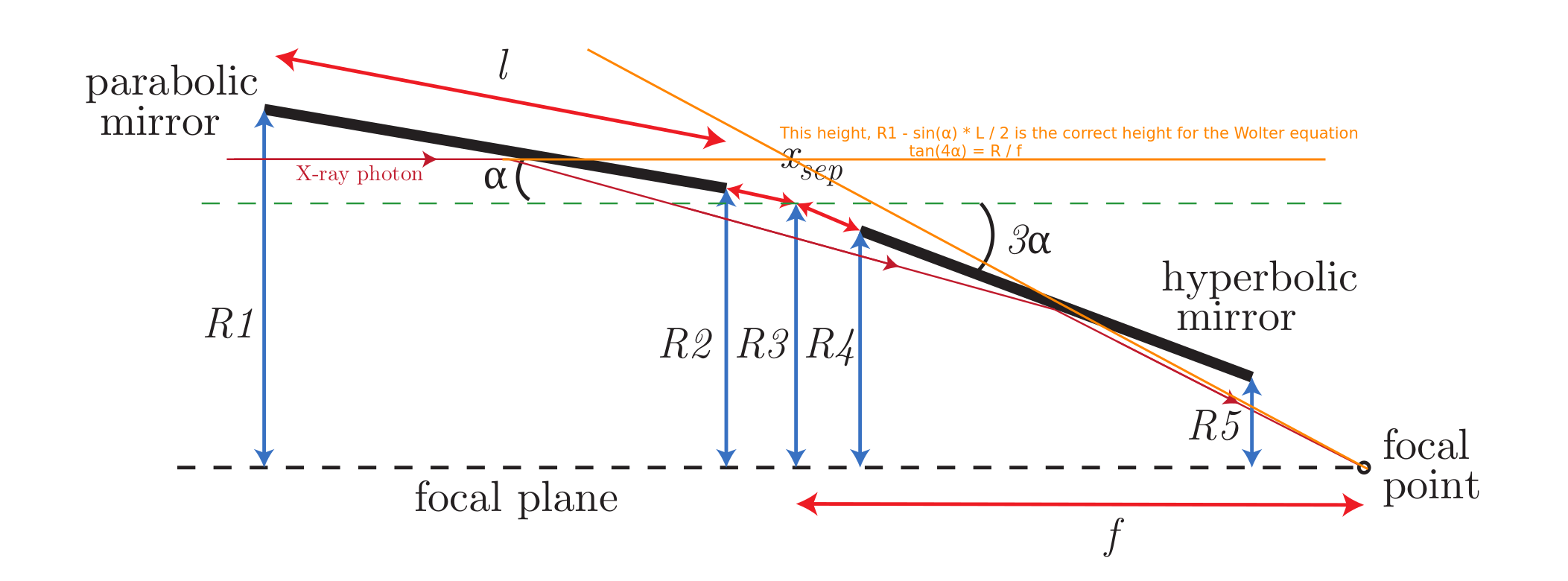
The point is that the naive way presented in the DTU PhD thesis mentions the following Wolter equation
\[ \tan(4 α) = \frac{R_3}{f} \]
where \(R_3\) is the radius at the middle of \(x_{\text{sep}}\).
Let's compute the focal length using the Wolter equation first based on \(R_3\) and then based on the height indicated in the annotated screenshot above, namely at \(R_1 - \sin(α) · l_M / 2\):
import unchained, sequtils, math let R1s = @[63.006, 65.606, 68.305, 71.105, 74.011, 77.027, 80.157, 83.405, 86.775, 90.272, 93.902, 97.668, 101.576, 105.632].mapIt(it.mm) let R5s = @[53.821, 56.043, 58.348, 60.741, 63.223, 65.800, 68.474, 71.249, 74.129, 77.117, 80.218, 83.436, 86.776, 90.241].mapIt(it.mm) let αs = @[0.579, 0.603, 0.628, 0.654, 0.680, 0.708, 0.737, 0.767, 0.798, 0.830, 0.863, 0.898, 0.933, 0.970].mapIt(it.Degree) const lMirror = 225.mm const xSep = 4.mm proc calcR3(r1, lMirror: mm, α: float): mm = let r2 = r1 - lMirror * sin(α) result = r2 - 0.5 * xSep * tan(α) for i in 0 ..< R1s.len: let r1 = R1s[i] let α = αs[i].to(Radian).float let r3 = calcR3(r1, lMirror, α) let r1minus = r1 - sin(α) * lMirror/2 echo "Focal length at i ", i, " f = ", r3 / tan(4 * α), " using r1mid f_m = ", r1minus / tan(4 * α)
| Focal | length | at | i | 0 | f | = | 1501.15 mm | using | r1mid | fm | = | 1529.75 mm |
| Focal | length | at | i | 1 | f | = | 1500.8 mm | using | r1mid | fm | = | 1529.41 mm |
| Focal | length | at | i | 2 | f | = | 1500.25 mm | using | r1mid | fm | = | 1528.85 mm |
| Focal | length | at | i | 3 | f | = | 1499.55 mm | using | r1mid | fm | = | 1528.16 mm |
| Focal | length | at | i | 4 | f | = | 1501.14 mm | using | r1mid | fm | = | 1529.74 mm |
| Focal | length | at | i | 5 | f | = | 1500.4 mm | using | r1mid | fm | = | 1529.01 mm |
| Focal | length | at | i | 6 | f | = | 1499.82 mm | using | r1mid | fm | = | 1528.41 mm |
| Focal | length | at | i | 7 | f | = | 1499.43 mm | using | r1mid | fm | = | 1528.03 mm |
| Focal | length | at | i | 8 | f | = | 1499.29 mm | using | r1mid | fm | = | 1527.89 mm |
| Focal | length | at | i | 9 | f | = | 1499.46 mm | using | r1mid | fm | = | 1528.06 mm |
| Focal | length | at | i | 10 | f | = | 1500.01 mm | using | r1mid | fm | = | 1528.6 mm |
| Focal | length | at | i | 11 | f | = | 1499.18 mm | using | r1mid | fm | = | 1527.77 mm |
| Focal | length | at | i | 12 | f | = | 1500.58 mm | using | r1mid | fm | = | 1529.16 mm |
| Focal | length | at | i | 13 | f | = | 1500.82 mm | using | r1mid | fm | = | 1529.4 mm |
We can see we get exactly the correct focal distance of about \SI{1530}{mm} instead of the wrong number of \(\SI{1500}{mm}\) (according to our raytracer).
How the hell does this make any sense?
11.5.16. Other misc changes
- the entire code now uses Malebolgia instead of weave
- instead of having many, many arguments to the render functions, we
now have a
RenderContext - the
RenderContexthas differentHittablesListfields for the entire world, for sources only, for targets only and the world without any elements that the light sampling should see (i.e. no sources or targets in there). - any ref object in
hittablesis marked asacyclicto make ORC happy. We have no intention of storing cyclic ref structures, so this is correct. - the different LLNL scenes are now using the same procedures to construct the entire scene, e.g. one proc for magnet bore taking a radius etc.
- the
TelescopeandMagnettypes were taken from the old raytracing code to be a bit more 'compatible' with it, if the desired arises (but likely this will simply replace the old code at this point due to being much clearer. The additional features should be easy enough to add) - not only individual
Hittables, but also wholeHittableListscan now be transformed. This is just done by applying the same transformation to each element. This allows to easily rotate the entire telescope by adding all elements first to a combinedHittableListfor example.
11.6. Fixing our implementation of a figure error extended
tl;dr: To reproduce the determination of the final parameters we use, run ./../../CastData/ExternCode/RayTracing/optimize_figure_error.nim using
./optimize_figure_error --hpd 206 --c80 397 --c90 568 --bufOutdir out_GN_DIRECT_L --shmFile /dev/shm/image_sensor_GN_DIRECT_L.dat
which calls the raytracer with a set of fuzzing parameters, computes
the EEF for the HPD, c80 and c90 radii comparing them to the given
arguments (these are the target values for the PANTER data of the Al
Kα line). It uses nlopt for the optimization using the GN_DIRECT_L
algorithm (a global, derivative free algorithm).
See here: https://nlopt.readthedocs.io/en/latest/NLopt_Algorithms/#direct-and-direct-l
While writing the appendix of the thesis about raytracing, I finally had some ideas about how to fix the figure error. The issue with our implementation of course was that we sampled into all possible directions for the fuzzing. But of course, light tends to scatter most dominantly still into the plane defined by incoming, normal and reflected vectors and to a much lesser extent into the orthogonal direction (defined by normal cross reflected for example).
So I've implemented that idea now.
\(i\): incoming vector \(n\): normal vector of the reflecting surface \(r\): reflected vector
Then we want the vector orthogonal to \(n\) and \(r\) in order to do the orthogonal fuzzing (of which we want little):
\[ n_{\perp} = n \times r. \]
More importantly, we want the vector orthogonal to both \(n_{\perp}\) and \(r\), along which we do most of the fuzzing. We can get that by another cross product
\[ p = n_{\perp} \times r. \]
Now we just have to find the correct fuzzing values along \(p\) and along \(n_{\perp}\).
I implemented two environment variables FUZZ_IN and FUZZ_ORTH to
adjust the parameter at runtime (well, program startup).
We will run this command with different values:
FUZZ_IN=7.0 FUZZ_ORTH=0.5 \ ./raytracer \ --width 1200 --speed 10.0 --nJobs 32 --vfov 10 --maxDepth 10 \ --llnl --focalPoint --sourceKind skXrayFinger --rayAt 1.013 --sensorKind sSum \ --energyMin 1.48 --energyMax 1.50 \ --usePerfectMirror=false \ --ignoreWindow --sourceDistance 130.297.m --sourceRadius 0.42.mm \ --telescopeRotation 90.0 --sourceOnOpticalAxis \ --ignoreMagnet --targetRadius 40.mm
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer2023-11-11T11:47:45+01:00typeuint32len1440000width1200height1200.dat [INFO] Writing file: out/counts2023-11-11T11:47:45+01:00typeintlen1440000width1200height1200.dat [INFO] Writing file: out/imagesensor02023-11-11T11:47:45+01:00_dx14.0dy14.0dz0.1typefloatlen1000000width1000height1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T11:47:45+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_7.0_fuzz_orth_0.5.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 10.0, Orth 0.7:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T11:44:51+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T11:44:51+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T11:44:51+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T11:44:51+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_10.0_fuzz_orth_0.7.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 7.0, Orth 0.3:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T11:51:12+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T11:51:12+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T11:51:12+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T11:51:12+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_7.0_fuzz_orth_0.3.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 7.0, Orth 0.1:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T11:56:54+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T11:56:54+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T11:56:54+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T11:56:54+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_7.0_fuzz_orth_0.1.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 5.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T11:59:37+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T11:59:37+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T11:59:37+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T11:59:37+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_5.0_fuzz_orth_0.5.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 5.0, Orth 0.75:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:04:32+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:04:32+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:04:32+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:04:32+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_5.0_fuzz_orth_0.75.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 8.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:07:25+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:07:25+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:07:25+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:07:25+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_8.0_fuzz_orth_0.5.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
From all the above, it seems like we struggle with the correct
shape. Look at the hpd_y plots and the EEF. If we get a value close
to 90%, we get a too large radius for 50%. The HPD y shape is too
gaussian. The real data is more "flat" near the top.
Can we modify the random sampling to not be gaussian?
Now we factor * factor of the fuzzIn sampled values.
In 8.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:20:07+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:20:07+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:20:07+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:20:07+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_8.0_fuzz_orth_0.5_squared.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 4.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:23:19+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:23:19+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:23:19+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:23:19+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_4.0_fuzz_orth_0.5_squared.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 2.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:26:02+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:26:02+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:26:02+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:26:02+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_2.0_fuzz_orth_0.5_squared.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 2.5, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:29:26+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:29:26+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:29:26+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:29:26+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_2.5_fuzz_orth_0.5_squared.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
I forgot to take care of keeping the sign! Fixed now. Rerunning
In 2.5, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:33:02+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:33:02+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:33:02+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:33:02+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_2.5_fuzz_orth_0.5_squared_sgn.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
Looks better, but got larger.
In 2.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:35:58+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:35:58+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:35:58+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:35:58+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_2.0_fuzz_orth_0.5_squared_sgn.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
Testing:
let n_orth = cross(reflected, rec.normal).normalize let r_orth = cross(reflected, n_orth).normalize #echo "FUZZ: ", fuzzIn, " vs ", fuzzOrth let factor = rnd.gauss(mu = 0.0, sigma = fuzzIn) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let sgn = sign(factor) let fzFz = m.fuzz * 2.0 scattered = initRay(rec.p, reflected + fzFz * (sgn * factor * factor * r_orth) + m.fuzz * (factor_orth * n_orth), r_in.typ)
now with
In 1.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:42:08+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:42:08+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:42:08+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:42:08+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_1.0_fuzz_orth_0.5_squared_sgn_fzFz2.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 1.5, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:43:58+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:43:58+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:43:58+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:43:58+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_1.5_fuzz_orth_0.5_squared_sgn_fzFz2.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
Time for cubic! In 1.5, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:49:01+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:49:01+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:49:01+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:49:01+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_1.5_fuzz_orth_0.5_cubic_sgn_fzFz2.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 1.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:51:39+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:51:39+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:51:39+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:51:39+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_1.0_fuzz_orth_0.5_cubic_sgn_fzFz2.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
Hmm, let's try sqrt (m.fuzz unchanged): In 7.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:55:18+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:55:18+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:55:18+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:55:18+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_7.0_fuzz_orth_0.5_sqrt.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 14.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T12:56:53+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T12:56:53+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T12:56:53+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T12:56:53+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_14.0_fuzz_orth_0.5_sqrt.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 24.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:00:02+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:00:02+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:00:02+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:00:02+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_24.0_fuzz_orth_0.5_sqrt.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 50.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:01:49+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:01:49+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:01:49+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:01:49+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_50.0_fuzz_orth_0.5_sqrt.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
Double gaussian, one narrow, one wide!
let factor = rnd.gauss(mu = 0.0, sigma = fuzzIn) let factorOuter = rnd.gauss(mu = 0.0, sigma = fuzzIn * 4) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let fc = if factor > fuzzIn: factorOuter else: factor scattered = initRay(rec.p, reflected + fzFz * (fc * r_orth) + m.fuzz * (factor_orth * n_orth), r_in.typ)
In 7.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:11:00+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:11:00+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:11:00+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:11:00+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_7.0_fuzz_orth_0.5_narrowWide.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 5.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:13:11+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:13:11+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:13:11+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:13:11+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_5.0_fuzz_orth_0.5_narrowWide.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
Double gaussian with wide *6!
let factor = rnd.gauss(mu = 0.0, sigma = fuzzIn) let factorOuter = rnd.gauss(mu = 0.0, sigma = fuzzIn * 6) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let fc = if factor > fuzzIn: factorOuter else: factor scattered = initRay(rec.p, reflected + fzFz * (fc * r_orth) + m.fuzz * (factor_orth * n_orth), r_in.typ)
In 5.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:15:26+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:15:26+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:15:26+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:15:26+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_5.0_fuzz_orth_0.5_narrowWide_times6.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 4.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:16:57+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:16:57+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:16:57+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:16:57+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_4.0_fuzz_orth_0.5_narrowWide_times6.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
We're getting somewhere…
let factorOuter = rnd.gauss(mu = 0.0, sigma = fuzzIn * 7) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let fc = if factor > fuzzIn * 0.9: factorOuter else: factor
In 4.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:22:32+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:22:32+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:22:32+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:22:32+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_4.0_fuzz_orth_0.5_narrowWide_0.9_times7.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 3.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:24:32+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:24:32+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:24:32+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:24:32+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_3.0_fuzz_orth_0.5_narrowWide_0.9_times7.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
…
let factorOuter = rnd.gauss(mu = 0.0, sigma = fuzzIn * 7) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let fc = if factor > fuzzIn * 1.1: factorOuter else: factor
In 3.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:26:59+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:26:59+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:26:59+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:26:59+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_3.0_fuzz_orth_0.5_narrowWide_1.1_times7.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
…
let factorOuter = rnd.gauss(mu = 0.0, sigma = fuzzIn * 8) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let fc = if factor > fuzzIn * 1.1: factorOuter else: factor
In 3.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:29:43+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:29:43+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:29:43+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:29:43+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_3.0_fuzz_orth_0.5_narrowWide_1.1_times8.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 4.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:31:02+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:31:02+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:31:02+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:31:02+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_4.0_fuzz_orth_0.5_narrowWide_1.1_times8.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
…
let factorOuter = rnd.gauss(mu = 0.0, sigma = fuzzIn * 8) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let fc = if factor > fuzzIn * 1.2: factorOuter else: factor
In 4.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:33:12+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:33:12+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:33:12+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:33:12+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_4.0_fuzz_orth_0.5_narrowWide_1.2_times8.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
let factor = rnd.gauss(mu = 0.0, sigma = fuzzIn * 0.8) let factorOuter = rnd.gauss(mu = 0.0, sigma = fuzzIn * 8) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let fc = if factor > fuzzIn * 1.1: factorOuter else: factor scattered = initRay(rec.p, reflected + fzFz * (fc * r_orth) + m.fuzz * (factor_orth * n_orth), r_in.typ)
In 4.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:35:15+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:35:15+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:35:15+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:35:15+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_4.0_fuzz_orth_0.5_narrowWide_0.8f_1.1_times8.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
let factor = rnd.gauss(mu = 0.0, sigma = fuzzIn * 0.6) let factorOuter = rnd.gauss(mu = 0.0, sigma = fuzzIn * 8) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let fc = if factor > fuzzIn * 0.8: factorOuter else: factor scattered = initRay(rec.p, reflected + fzFz * (fc * r_orth) + m.fuzz * (factor_orth * n_orth), r_in.typ)
In 4.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:38:42+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:38:42+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:38:42+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:38:42+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_4.0_fuzz_orth_0.5_narrowWide_0.6f_0.8_times8.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
Forgot to take 0.6 into account
let fzIn = fuzzIn * 0.6 let factor = rnd.gauss(mu = 0.0, sigma = fzIn) let factorOuter = rnd.gauss(mu = 0.0, sigma = fuzzIn * 8) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let fc = if factor > fzIn * 0.8: factorOuter else: factor scattered = initRay(rec.p, reflected + fzFz * (fc * r_orth) + m.fuzz * (factor_orth * n_orth), r_in.typ)
In 4.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:41:10+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:41:10+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:41:10+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:41:10+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_4.0_fuzz_orth_0.5_narrowWide_fixed_0.6f_0.8_times8.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
Update 1.0:
let fzIn = fuzzIn * 0.6 let factor = rnd.gauss(mu = 0.0, sigma = fzIn) let factorOuter = rnd.gauss(mu = 0.0, sigma = fuzzIn * 8) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let fc = if factor > fzIn * 1.0: factorOuter else: factor scattered = initRay(rec.p, reflected + m.fuzz * (fc * r_orth) + m.fuzz * (factor_orth * n_orth), r_in.typ)
In 4.0, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:44:03+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:44:03+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:44:03+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:44:03+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_4.0_fuzz_orth_0.5_narrowWide_fixed_0.6f_1.0_times8.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
In 3.5, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:46:02+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:46:02+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:46:02+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:46:02+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_3.5_fuzz_orth_0.5_narrowWide_fixed_0.6f_1.0_times8.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
let fzIn = fuzzIn * 0.6 let factor = rnd.gauss(mu = 0.0, sigma = fzIn) let factorOuter = rnd.gauss(mu = 0.0, sigma = fuzzIn * 8) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let fc = if factor > fzIn * 1.1: factorOuter else: factor scattered = initRay(rec.p, reflected + m.fuzz * (fc * r_orth) + m.fuzz * (factor_orth * n_orth), r_in.typ)
In 3.5, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:48:26+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:48:26+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:48:26+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:48:26+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_3.5_fuzz_orth_0.5_narrowWide_fixed_0.6f_1.1_times8.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
let fzIn = fuzzIn * 0.5 let factor = rnd.gauss(mu = 0.0, sigma = fzIn) let factorOuter = rnd.gauss(mu = 0.0, sigma = fuzzIn * 8) let factor_orth = rnd.gauss(mu = 0.0, sigma = fuzzOrth) #1.0 / sqrt(2.0)) let fc = if factor > fzIn * 1.0: factorOuter else: factor scattered = initRay(rec.p, reflected + m.fuzz * (fc * r_orth) + m.fuzz * (factor_orth * n_orth), r_in.typ)
In 3.5, Orth 0.5:
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer_2023-11-11T13:51:02+01:00_type_uint32_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/counts_2023-11-11T13:51:02+01:00_type_int_len_1440000_width_1200_height_1200.dat [INFO] Writing file: out/image_sensor_0_2023-11-11T13:51:02+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T13:51:02+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out ~/org/Figs/statusAndProgress/rayTracing/fixFigureError/al_kalpha_fuzz_in_3.5_fuzz_orth_0.5_narrowWide_fixed_0.5f_1.0_times8.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5 # same range as full plots in
I just noticed that I had two outer / orth factor variables in the entire snippets above, so my modifications to those didn't even do anything! Anyway, writing an optimizer now…
Leaving it running for a while using LN_COBYLA,
./optimize_figure_error --hpd 206 --c80 397 --c90 568
Current difference: 33.29940305450022 from : @[218.1545760531755, 402.3834526133487, 563.2028920473765] param: 6.109924329596698 param: 0.7021469719954326 param: 0.7672823036143021 param: 6.392485426989791 param: 0.9462750420513524 Params: @[6.109924329596698, 0.7021469719954326, 0.7672823036143021, 6.392485426989791, 0.9462750420513524]
(where the difference is from:
# penalize HPD 3x more than the other two result = (data.hpd - hpdR)^2 / sqrt(data.hpd) * 3.0 + (data.c80 - c80R)^2 / sqrt(data.c80) + (data.c90 - c90R)^2 / sqrt(data.c90) echo "Current difference: ", result, " from : ", resSpl
When running as:
./optimize_figure_error --hpd 200 --c80 391 --c90 545
we get:
Current difference: 51.06462856407439 from : @[215.293904603881, 391.688010954991, 539.2377947764865] param: 5.866112831835506 param: 0.6942075732787438 param: 0.7243055192027785 param: 6.173377076415993 param: 0.9050670563207799 Params: @[5.866112831835506, 0.6942075732787438, 0.7243055192027785, 6.173377076415993, 0.9050670563207799]
Let's try BOBYQA:
./optimize_figure_error --hpd 200 --c80 391 --c90 545
Current difference: 76.5355776638029 from : @[217.272373487044, 394.343054506765, 527.7921276534421] param: 4.589373255722458 param: 0.5492832831983449 param: 0.927345898543579 param: 4.781368702342282 param: 0.9265644878944641 Params: @[4.589373255722458, 0.5492832831983449, 0.927345898543579, 4.781368702342282, 0.9265644878944641]
So yeah, it seems like our double gaussian approach does not approach the shape very well.
Hmm, or maybe it is actually related to local minima. The LN_*
algorithms are local after all. Running LN_COBYLA again with
different starting parameters, generally looks better. So maybe we
should try a global algorithm.
Using:
# define starting parameters let params = @[ 3.5, # FUZZ_IN 0.5, # FUZZ_ORTH 0.5, # FUZZ_IN_SCALE 8.0, # FUZZ_OUTER_SCALE 1.0 ] # FUZZ_IN_RATIO]
with LN_COBYLA
Current difference: 40.80516707794559 from : @[216.6796117074927, 378.758920745324, 570.5178328618443] param: 3.694297126415748 param: 0.5098297588328489 param: 0.4894695519681594 param: 8.148799193763727 param: 0.5190522194705663 Params: @[3.694297126415748, 0.5098297588328489, 0.4894695519681594, 8.148799193763727, 0.5190522194705663]
with
./optimize_figure_error --hpd 206 --c80 397 --c90 568
I tried GN_DIRECT and GN_DIRECT_L. The latter attempts some local
optimizations as well, good for problems that do not suffer from a
large number of local minima.
LNCOBYLA:
./optimize_figure_error --hpd 206 --c80 397 --c90 568 --bufOutdir out_LN_COBYLA --shmFile /dev/shm/image_sensor_LN_COBYLA.dat
Current difference: 25.96944923605506 from : @[216.868040602679, 392.0808702690267, 566.7386829846979] param: 3.252194308182035 param: 0.2596708930642096 param: 1.000973731445702 param: 9.23060790188306 param: 1.083648328960083 Params: @[3.252194308182035, 0.2596708930642096, 1.000973731445702, 9.23060790188306, 1.083648328960083]
GNDIRECTL:
./optimize_figure_error --hpd 206 --c80 397 --c90 568 --bufOutdir out_GN_DIRECT_L --shmFile /dev/shm/image_sensor_GN_DIRECT_L.dat
Current difference: 25.77930908807127 from : @[216.7270630094659, 391.2699941926571, 566.6228749201783] param: 3.257544581618656 param: 0.2242798353909466 param: 0.9814814814814816 param: 9.22976680384088 param: 1.083333333333333 Params: @[3.257544581618656, 0.2242798353909466, 0.9814814814814816, 9.22976680384088, 1.083333333333333]
Let's look at the last GNDIRECTL case:
./plotBinary \ --dtype float \ -f out_GN_DIRECT_L/image_sensor_0_2023-11-11T21:05:39+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out /t/testme.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5
When compared with the EEFs found in the slides
(slide 17) the shape is actually quite similar now!
Let's run a regular raytrace with them:
FUZZ_IN=3.257544581618656 \ FUZZ_ORTH=0.2242798353909466 \ FUZZ_IN_SCALE=0.9814814814814816 \ FUZZ_OUTER_SCALE=9.22976680384088 \ FUZZ_IN_RATIO=1.083333333333333 \ ./raytracer \ --width 1200 --speed 10.0 --nJobs 32 --vfov 10 --maxDepth 10 \ --llnl --focalPoint --sourceKind skXrayFinger \ --rayAt 1.013 --sensorKind sSum --energyMin 1.48 --energyMax 1.50 \ --usePerfectMirror=false \ --ignoreWindow --sourceDistance 130.297.m --sourceRadius 0.42.mm \ --telescopeRotation 90.0 --sourceOnOpticalAxis \ --ignoreMagnet --targetRadius 40.mm
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer2023-11-11T21:10:58+01:00typeuint32len1440000width1200height1200.dat [INFO] Writing file: out/counts2023-11-11T21:10:58+01:00typeintlen1440000width1200height1200.dat [INFO] Writing file: out/imagesensor02023-11-11T21:10:58+01:00_dx14.0dy14.0dz0.1typefloatlen1000000width1000height1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T21:10:58+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out /t/al_kalpha_params_figure_error.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5
Looks pretty good I guess. A bit more narrow than I would have expected, but clearly shows the much longer bow tie than we otherwise got!
Axion image:
FUZZ_IN=3.257544581618656 \ FUZZ_ORTH=0.2242798353909466 \ FUZZ_IN_SCALE=0.9814814814814816 \ FUZZ_OUTER_SCALE=9.22976680384088 \ FUZZ_IN_RATIO=1.083333333333333 \ ./raytracer \ --width 1200 --speed 10.0 --nJobs 32 --vfov 10 --maxDepth 10 \ --llnl --focalPoint --sourceKind skSun \ --sensorKind sSum \ --usePerfectMirror=false \ --ignoreWindow \ --solarModelFile ~/CastData/ExternCode/AxionElectronLimit/resources/solar_model_dataframe_fluxKind_fkAxionElectronPhoton_0.989AU.csv
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer2023-11-11T21:16:32+01:00typeuint32len1440000width1200height1200.dat [INFO] Writing file: out/counts2023-11-11T21:16:32+01:00typeintlen1440000width1200height1200.dat [INFO] Writing file: out/imagesensor02023-11-11T21:16:32+01:00_dx14.0dy14.0dz0.1typefloatlen1000000width1000height1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T21:16:32+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out /t/axion_image_figure_errors.pdf \ --inPixels=false \ --title "Axion image, axion-electron" \ --xrange 7.0
NOTE: GNDIRECTL was extremely useful finding good general parameters in the entire range! I fed some of the good ones into LNCOBYLA to start!
So… :)
Continue !
Let's try to decrease the fuzzing size slightly and see if that decreases the size as expected.
We decrease the FUZZ_IN from 3.25… to 3.15…. Everything else unchanged:
FUZZ_IN=3.157544581618656 \ FUZZ_ORTH=0.2242798353909466 \ FUZZ_IN_SCALE=0.9814814814814816 \ FUZZ_OUTER_SCALE=9.22976680384088 \ FUZZ_IN_RATIO=1.083333333333333 \ ./raytracer \ --width 1200 --speed 10.0 --nJobs 32 --vfov 10 --maxDepth 10 \ --llnl --focalPoint --sourceKind skXrayFinger \ --rayAt 1.013 --sensorKind sSum --energyMin 1.48 --energyMax 1.50 \ --usePerfectMirror=false \ --ignoreWindow --sourceDistance 130.297.m --sourceRadius 0.42.mm \ --telescopeRotation 90.0 --sourceOnOpticalAxis \ --ignoreMagnet --targetRadius 40.mm
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer2023-11-12T11:18:10+01:00typeuint32len1440000width1200height1200.dat [INFO] Writing file: out/counts2023-11-12T11:18:10+01:00typeintlen1440000width1200height1200.dat [INFO] Writing file: out/imagesensor02023-11-12T11:18:10+01:00_dx14.0dy14.0dz0.1typefloatlen1000000width1000height1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-12T11:18:10+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out /t/al_kalpha_params_figure_error_bit_smaller.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5
So it mainly makes the upper end smaller (by 9 arc seconds), the middle a bit (3 arc seconds) and the inner most one by only 1 arc second.
Let's go back to 3.25… and adjust the inscale down a bit. 0.98… to 0.90…
FUZZ_IN=3.257544581618656 \ FUZZ_ORTH=0.2242798353909466 \ FUZZ_IN_SCALE=0.9014814814814816 \ FUZZ_OUTER_SCALE=9.22976680384088 \ FUZZ_IN_RATIO=1.083333333333333 \ ./raytracer \ --width 1200 --speed 10.0 --nJobs 32 --vfov 10 --maxDepth 10 \ --llnl --focalPoint --sourceKind skXrayFinger \ --rayAt 1.013 --sensorKind sSum --energyMin 1.48 --energyMax 1.50 \ --usePerfectMirror=false \ --ignoreWindow --sourceDistance 130.297.m --sourceRadius 0.42.mm \ --telescopeRotation 90.0 --sourceOnOpticalAxis \ --ignoreMagnet --targetRadius 40.mm
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer2023-11-12T11:22:56+01:00typeuint32len1440000width1200height1200.dat [INFO] Writing file: out/counts2023-11-12T11:22:56+01:00typeintlen1440000width1200height1200.dat [INFO] Writing file: out/imagesensor02023-11-12T11:22:56+01:00_dx14.0dy14.0dz0.1typefloatlen1000000width1000height1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-12T11:22:56+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out /t/al_kalpha_params_figure_error_smaller_in.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5
0.9 -> 0.8
FUZZ_IN=3.257544581618656 \ FUZZ_ORTH=0.2242798353909466 \ FUZZ_IN_SCALE=0.7014814814814816 \ FUZZ_OUTER_SCALE=9.22976680384088 \ FUZZ_IN_RATIO=1.083333333333333 \ ./raytracer \ --width 1200 --speed 10.0 --nJobs 32 --vfov 10 --maxDepth 10 \ --llnl --focalPoint --sourceKind skXrayFinger \ --rayAt 1.013 --sensorKind sSum --energyMin 1.48 --energyMax 1.50 \ --usePerfectMirror=false \ --ignoreWindow --sourceDistance 130.297.m --sourceRadius 0.42.mm \ --telescopeRotation 90.0 --sourceOnOpticalAxis \ --ignoreMagnet --targetRadius 40.mm
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer2023-11-12T11:24:39+01:00typeuint32len1440000width1200height1200.dat [INFO] Writing file: out/counts2023-11-12T11:24:39+01:00typeintlen1440000width1200height1200.dat [INFO] Writing file: out/imagesensor02023-11-12T11:24:39+01:00_dx14.0dy14.0dz0.1typefloatlen1000000width1000height1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-12T11:24:39+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out /t/al_kalpha_params_figure_error_smaller_in_even.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5
0.7: [INFO] Writing file: out/imagesensor02023-11-12T11:26:01+01:00_dx14.0dy14.0dz0.1typefloatlen1000000width1000height1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-12T11:26:01+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out /t/al_kalpha_params_figure_error_smaller_in_even_more.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5
Yeah, this does not have a great impact.
Fuzz in ratio increase: 1.08 -> 1.2
FUZZ_IN=3.257544581618656 \ FUZZ_ORTH=0.2242798353909466 \ FUZZ_IN_SCALE=0.9814814814814816 \ FUZZ_OUTER_SCALE=9.22976680384088 \ FUZZ_IN_RATIO=1.23333333333333 \ ./raytracer \ --width 1200 --speed 10.0 --nJobs 32 --vfov 10 --maxDepth 10 \ --llnl --focalPoint --sourceKind skXrayFinger \ --rayAt 1.013 --sensorKind sSum --energyMin 1.48 --energyMax 1.50 \ --usePerfectMirror=false \ --ignoreWindow --sourceDistance 130.297.m --sourceRadius 0.42.mm \ --telescopeRotation 90.0 --sourceOnOpticalAxis \ --ignoreMagnet --targetRadius 40.mm
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer2023-11-12T11:28:14+01:00typeuint32len1440000width1200height1200.dat [INFO] Writing file: out/counts2023-11-12T11:28:14+01:00typeintlen1440000width1200height1200.dat [INFO] Writing file: out/imagesensor02023-11-12T11:28:14+01:00_dx14.0dy14.0dz0.1typefloatlen1000000width1000height1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-12T11:28:14+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out /t/al_kalpha_params_figure_error_in_ratio_large.pdf \ --inPixels=false \ --title "Al Kα, 1.49 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5
-> Yeah, this brings down the inner one well, but again at a extreme cost to the outer one.
I think the optimization routine really found a good minimum. Every change leads to significant worsening in other aspects.
Let's shortly run Ti and Fe lines to see what they look like with the parameters.
FUZZ_IN=3.257544581618656 \ FUZZ_ORTH=0.2242798353909466 \ FUZZ_IN_SCALE=0.9814814814814816 \ FUZZ_OUTER_SCALE=9.22976680384088 \ FUZZ_IN_RATIO=1.083333333333333 \ ./raytracer \ --width 1200 --speed 10.0 --nJobs 32 --vfov 10 --maxDepth 10 \ --llnl --focalPoint --sourceKind skXrayFinger \ --rayAt 1.013 --sensorKind sSum --energyMin 4.50 --energyMax 4.52 \ --usePerfectMirror=false \ --ignoreWindow --sourceDistance 130.297.m --sourceRadius 0.42.mm \ --telescopeRotation 90.0 --sourceOnOpticalAxis \ --ignoreMagnet --targetRadius 40.mm
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer2023-11-12T11:31:36+01:00typeuint32len1440000width1200height1200.dat [INFO] Writing file: out/counts2023-11-12T11:31:36+01:00typeintlen1440000width1200height1200.dat [INFO] Writing file: out/imagesensor02023-11-12T11:31:36+01:00_dx14.0dy14.0dz0.1typefloatlen1000000width1000height1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-12T11:31:36+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out /t/ti_kalpha_params_figure_error.pdf \ --inPixels=false \ --title "Ti Kα, 4.51 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5
This is very close to the "PANTER model"!
and Fe:
FUZZ_IN=3.257544581618656 \ FUZZ_ORTH=0.2242798353909466 \ FUZZ_IN_SCALE=0.9814814814814816 \ FUZZ_OUTER_SCALE=9.22976680384088 \ FUZZ_IN_RATIO=1.083333333333333 \ ./raytracer \ --width 1200 --speed 10.0 --nJobs 32 --vfov 10 --maxDepth 10 \ --llnl --focalPoint --sourceKind skXrayFinger \ --rayAt 1.013 --sensorKind sSum --energyMin 6.39 --energyMax 6.41 \ --usePerfectMirror=false \ --ignoreWindow --sourceDistance 130.297.m --sourceRadius 0.42.mm \ --telescopeRotation 90.0 --sourceOnOpticalAxis \ --ignoreMagnet --targetRadius 40.mm
[INFO] Writing buffers to binary files. [INFO] Writing file: out/buffer2023-11-12T11:34:39+01:00typeuint32len1440000width1200height1200.dat [INFO] Writing file: out/counts2023-11-12T11:34:39+01:00typeintlen1440000width1200height1200.dat [INFO] Writing file: out/imagesensor02023-11-12T11:34:39+01:00_dx14.0dy14.0dz0.1typefloatlen1000000width1000height1000.dat
./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-12T11:34:39+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out /t/fe_kalpha_params_figure_error.pdf \ --inPixels=false \ --title "Fe Kα, 6.51 keV, 5x5 mm, figure errors, source optical axis" \ --xrange 2.5
Almost a perfect match to the LLNL Panter model too!
[X]Finally, a look at the images in 3x3 mm comparing with the slides. -> They look good, I think.[ ]Implement these fuzz values as default in code. -> Note that these are still dependent on
const ImperfectVal = pow(0.000290887991795424, 1.2)
| Al Kα (1.49 keV) | |||
|---|---|---|---|
| 50% (HPD) | 80% circle | 90% circle | |
| Point source (perfect mirrors) | 168 arcsec | 270 arcsec | 313 arcsec |
| Point source (figure errors) | 206 | 387 | 568 |
| PANTER data | 206 | 397 | 549 |
| PANTER model | 211 | 391 | 559 |
| TrAXer (perfect mirrors) | 183.19 | 304.61 | 351.54 |
| TrAXer (figure errors) | 184.22 | 305.53 | 352.86 |
| Ti Kα (4.51 keV) | |||
| 50% (HPD) | 80% circle | 90% circle | |
| Point source (perfect mirrors) | 161 | 259 | 301 |
| Point source (figure errors) | 202 | 382 | 566 |
| PANTER data | 196 | 380 | 511 |
| PANTER model | 206 | 380 | 559 |
| TrAXer (perfect mirrors) | 174.84 | 288.54 | 333.75 |
| TrAXer (figure errors) | 175.59 | 289.40 | 335.49 |
| Fe Kα (6.41 keV) | |||
| 50% (HPD) | 80% circle | 90% circle | |
| Point source (perfect mirrors) | 144 | 233 | 265 |
| Point source (figure errors) | 184 | 350 | 541 |
| PANTER data | 196 | 364 | 483 |
| PANTER model | 185 | 348 | 516 |
| TrAXer (perfect mirrors) | 160.38 | 257.79 | 296.79 |
| TrAXer (figure errors) | 161.40 | 256.37 | 298.43 |
12. Likelihood method
TODO: merge this with 6.
12.1. Clusters after likelihood method
The code for the following plots is: ./../../CastData/ExternCode/TimepixAnalysis/Plotting/plotBackgroundClusters/plotBackgroundClusters.nim based on the files:
- ./../../CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_no_tracking_eff_0.8_whole_chip.h5
- ./../../CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2018_no_tracking_eff_0.8_whole_chip.h5
which are created from the likelihood with commands as:
./likelihood /mnt/1TB/CAST/2017/DataRuns2017_Reco.h5 \ --h5out ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_no_tracking_eff_0.8_whole_chip.h5 \ --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crAll
The plots 81 and 82 show the cluster centers after the logL method without any vetos at an ε = 80%.
Finally, in fig. 99 is the combined data.
#+begincenter
#+endcenter t
12.1.1. TODO investigate the noisy pixel in 2017!
I think the reason it's in the data is that at some point when comparing with MarlinTPC we took out a cluster size cut. However, it's a bit weird that these pass the logL cut, because they should be outside of the lowest energy (do we have such a thing?) / shouldn't match a cluster geometry (except they are almost perfectly round…).
Not a problem for gold region, but should be removed.
12.2. Likelihood distribution after cuts used to build method
An interesting thought that came up while talking to Dongjin is what is the effect of the cuts used to build the likelihood method in the first place on the distribution of the likelihood points itself. In theory it should plainly cut on the tail and leave the main peak untouched. We'll quickly create a plot comparing the two cases.
We'll do this by reading the raw data itself first and then comparing the
it to everything that passes buildLogLHist.
import ggplotnim, nimhdf5, strutils, os, sequtils import ingrid / [tos_helpers, ingrid_types] # custom `buildLogLHist` that only applies cuts, nothing else proc buildLogLHist*(h5f: H5File, dset: string): seq[bool] = var grp_name = cdlPrefix("2018") & dset # create global vars for xray and normal cuts table to avoid having # to recreate them each time let xrayCutsTab = getXrayCleaningCuts() var cutsTab = getEnergyBinMinMaxVals2018() # open h5 file using template let energyStr = igEnergyFromCharge.toDset() logLStr = igLikelihood.toDset() centerXStr = igCenterX.toDset() centerYStr = igCenterY.toDset() eccStr = igEccentricity.toDset() lengthStr = igLength.toDset() chargeStr = igTotalCharge.toDset() rmsTransStr = igRmsTransverse.toDset() npixStr = igHits.toDset() let energy = h5f[grp_name / energyStr, float64] logL = h5f[grp_name / logLStr, float64] centerX = h5f[grp_name / centerXStr, float64] centerY = h5f[grp_name / centerYStr, float64] ecc = h5f[grp_name / eccStr, float64] length = h5f[grp_name / lengthStr, float64] charge = h5f[grp_name / chargeStr, float64] rmsTrans = h5f[grp_name / rmsTransStr, float64] npix = h5f[grp_name / npixStr, float64] # get the cut values for this dataset cuts = cutsTab[dset] xrayCuts = xrayCutsTab[dset] result = newSeq[bool](energy.len) for i in 0 .. energy.high: let # first apply Xray cuts (see C. Krieger PhD Appendix B & C) regionCut = inRegion(centerX[i], centerY[i], crSilver) xRmsCut = rmsTrans[i] >= xrayCuts.minRms and rmsTrans[i] <= xrayCuts.maxRms xLengthCut = length[i] <= xrayCuts.maxLength xEccCut = ecc[i] <= xrayCuts.maxEccentricity # then apply reference cuts chargeCut = charge[i] > cuts.minCharge and charge[i] < cuts.maxCharge rmsCut = rmsTrans[i] > cuts.minRms and rmsTrans[i] < cuts.maxRms lengthCut = length[i] < cuts.maxLength pixelCut = npix[i] > cuts.minPix # add event to likelihood if all cuts passed if allIt([regionCut, xRmsCut, xLengthCut, xEccCut, chargeCut, rmsCut, lengthCut, pixelCut], it): result[i] = true let cdlPath = "/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5" let h5cdl = H5open(cdlPath, "r") let xtab = getXrayRefTable() var df = newDataFrame() for idx, dset in xtab: let dsetLogL = cdlGroupName(dset, "2018", "likelihood") let cX = h5cdl[cdlGroupName(dset, "2018", "centerX"), float] let cY = h5cdl[cdlGroupName(dset, "2018", "centerY"), float] let dfLoc = toDf({ "Dset" : dset, "logL" : h5cdl[dsetLogL, float], "cX" : cX, "cY" : cY, "pass?" : buildLogLHist(h5cdl, dset) }) .mutate(f{float -> bool: "inSilver" ~ inRegion(idx("cX"), idx("cY"), crSilver)}) .filter(f{`logL` < 50.0}) df.add dfLoc echo df discard h5cdl.close() ggplot(df, aes("logL", fill = "pass?", color = "pass?")) + facet_wrap("Dset", scales = "free") + geom_histogram(position = "identity", bins = 200, alpha = some(0.5), hdKind = hdOutline) + ggtitle("Comparison of CDL logL data of events *passing* and *not passing* pre processing cuts.") + ggsave("/home/basti/org/Figs/statusAndProgress/logL_distributions_cut_passing_vs_non_passing_comparison.pdf", width = 1920, height = 1080) # first take *all* data in `df` var dfCombined = df.drop(["pass?"]).mutate(f{"Type" ~ "No cuts"}) # now add all data *passing* all cuts dfCombined.add df.filter(f{idx("pass?") == true}) .drop(["pass?"]).mutate(f{"Type" ~ "Cuts applied"}) # finally add only data passing *silver* cut dfCombined.add df.filter(f{`inSilver` == true}) .drop(["pass?"]).mutate(f{"Type" ~ "Only silver"}) ggplot(dfCombined, aes("logL", fill = "Type", color = "Type")) + facet_wrap("Dset", scales = "free") + geom_histogram(position = "identity", bins = 200, alpha = some(0.5), hdKind = hdOutline) + ggtitle("Comparison of raw CDL logL data with logL data remaining after preprocessing cuts are applied & silver *only* cut") + ggsave("/home/basti/org/Figs/statusAndProgress/logL_distributions_cut_comparison.pdf", width = 1920, height = 1080)
This code results in two different plots. First a rather confusing one, namely the comparison between the distributions from events that do and that do not pass the preprocessing cuts:
So all good in the end.
13. MarlinTPC vs. TimepixAnalysis output
UPDATE : See the note in sec. 10 for the likely solution.
One of the major remaining problems of TPA is the difference in the background rate for the 2014/15 dataset compared to the Marlin TPC result. See fig. 85 for the comparison as of (git commit: https://github.com/Vindaar/TimepixAnalysis/commit/970ed6ba07cac0283a5353ce896327848eb09abb).
The data files used to generate the plots are currently stored in: ./../../../../data/CAST/Backup/BackgroundCompareAug2019/.
It can be seen that differences in the low energy range and near \(\sim\SI{8}{\keV}\) are visible. The reason for these differences will be studied in this section.
I finished the extraction of the Marlin events. They are now stored in the TPA repository as a JSON file in: ./../../CastData/ExternCode/TimepixAnalysis/resources/marlinTestEvents/marlinEvents.json
For these events the corresponding Run numbers and event numbers are:
Run number of idx 1803795 : 437 and eventNumber 55827
Run number of idx 1325475 : 333 and eventNumber 38624
Run number of idx 1906755 : 456 and eventNumber 34270
which we can then extract from the data directories. Hmm, that doesn't seem to match what we think it does…
Update: investigating by running raw_data_manipulation on the
directory and looking at the number of hits, we find the correct
number (435) of hits at lines 55827 for run 437.
It turns out what the Marlin code calls the "eventNumber" is not the event number, but actually the event number without any empty frames! It's the event counter basically. Thus we have to determine the real event numbers from these indices above. :/
Run 437: Index: 55827, Event Number: 59424 Run 333: Index: 38624, Event Number: 57178 # why such large discrepency compared to Run 437 and 456? Run 456: Index: 34270, Event Number: 34772
These 3 files are now also found in ./../../CastData/ExternCode/TimepixAnalysis/resources/marlinTestEvents/.
NOTE: there is a one-off error in the x coordinates of all events
from Marlin TPC! The y coordinate matches what's written in the raw
TOS data files.
NOTE: Marlin TPC filters out the pixel
167 200 *
by default, since it's the noisy pixel that's visible in every of its files.
For now we just take that pixel out of the data for the test cases and see where it leads us. Will have to remove it always for oldVirtex data in TPA at some point I fear. Possibly could be done via an additional raw data manipulation step where we can apply a filtering mask via a toml file or something? Would make it more usable.
In the script we currently create DataFrames both for the events as
they are reconstructed by MarlinTPC and by TimepixAnalysis. As of
and commit hash
b70bc06b9b2bbc6dbba9a5507f9786ab0f46b84c, we generate the following
data frames and plots:
var dfReco = DataFrame() dfReco.bindToDf(reco.cluster) var dfExp = DataFrame() dfExp.bindToDf(expEvents[i].cluster) let df = bind_rows([dfReco, dfExp], id = "origin") # work around issue in ggplotnim ggplot(df, aes("x", "y", color = "from")) + geom_point() + facet_wrap(~origin) + ggtitle("Is event " & $i & " with clusters: " & $reco.cluster.len) + ggsave(&"recoed_cluster_{i}.pdf")
where bindToDf is:
proc bindToDf[T](df: var DataFrame, clusters: seq[ClusterObject[T]]) = var dfs = newSeq[DataFrame]() for i, cl in clusters: let ldf = toDf({ "x" : cl.data.mapIt(it.x), "y" : cl.data.mapIt(it.y), "ch" : cl.data.mapIt(it.ch)}) dfs.add ldf df = bind_rows(dfs, id = "from") if "from" notin df: df["from"] = toVector(toSeq(0 ..< df.len).mapIt(%~ "-1"))
That is a facet wrap of the two differnt reconstruction frameworks,
TPA and Marlin, where each cluster is colored differently.
For some reason Marlin sometimes wrongly classifies clusters. Consider
the event in fig. 86. It's obvious that the
clustering clearly does not allow a spacing of 50 pixels. Even if
for some reason the search radius of 50 pixels was set differntly
for this, it's still wrong in case of the red pixel at
(x: ~200, y: ~120). For a smaller search radius it should become
part of the green cluster and not the red one.
This becomes even more obvious if we reconstruct with a search radius of \(\SI{25}{pixel}\) in TPA. That gives us fig. 87.
Christoph used the ./../../../../data/tpc18/home/src/MarlinTPC/krieger/reconstruction/pixelbased/src/TimePixSpecialClusterFinderProcessor.cc, see from here ./../../../../data/tpc18/home/src/MarlinTPC/XrayReco.xml especially line 96, the search radius.
A little further investigation into this: I added a simple plot option into the Marlin event extraction tool to more quickly find such events with more than one cluster found: https://github.com/Vindaar/TimepixAnalysis/commit/5c4445a628669aa3f85244497301744bf62f4958
This shows that quite a few events showcase the same behavior. Another example in fig. 88.
For now in
./../../CastData/ExternCode/TimepixAnalysis/Tests/reconstruction/tInGridGeometry.nim
(as of
https://github.com/Vindaar/TimepixAnalysis/commit/2e5e183cc39199ed3a6edfb2929a2a9d722b3709)
we restrict ourselves in our test cases to single clusters. If in
either of the two frameworks more than 1 cluster was found, we skip
it (we will extend it soon to TPA.numClusters ==
Marlin.numClusters).
In those cases we compare all geometrical properties.
Take the following event, fig. 89:
Both frameworks find only a single cluster in this background event. The number of pixels is the same and the pixel content is the same too (not the value though, since we have charge values for our Marlin expectation, but so far raw ToT values from TPA). There are two small differences in the input clusters:
- TPA would normally include the "noisy pixel" mentioned above
- all
xcoordinate values for MarlinTPC are off by one, for some reason. Theyvalues however are correct.
Keeping this in mind, the table tab. 16 compares the two events.
| Property | TimepixAnalysis | MarlinTPC | Difference |
|---|---|---|---|
| hits | 218 | 218 | 0 |
| centerX | 7.622798165137616 | 7.567798137664795 | 0.05500002747282107 |
| centerY | 6.574266055045872 | 6.574265956878662 | 9.816720947242175e-08 |
| rmsLongitudinal | 4.374324961805871 | 4.374318599700928 | 6.362104943313796e-06 |
| rmsTransverse | 3.151177831860436 | 3.15118670463562 | 8.872775183910164e-06 |
| eccentricity | 1.388155539042776 | 1.388149619102478 | 5.919940298415582e-06 |
| rotationAngle | 3.000219815810449 | 2.997750043869019 | 0.002469771941430388 |
| skewnessLongitudinal | 0.3448385630734474 | 0.3436029553413391 | 0.00123560773210829 |
| skewnessTransverse | 0.2743112358858994 | 0.2803316414356232 | 0.006020405549723828 |
| kurtosisLongitudinal | -1.440002172704371 | -1.439019083976746 | 0.0009830887276247591 |
| kurtosisTransverse | -0.5166603621509731 | -0.5109220147132874 | 0.005738347437685754 |
| length | 13.60058820384985 | 13.61675834655762 | 0.01617014270776806 |
| width | 13.15926696041458 | 13.16585159301758 | 0.006584632602999463 |
| fractionInTransverseRms | 0.1880733944954129 | 0.1880733966827393 | 2.187326458846783e-09 |
| lengthDivRmsTrans | 4.316033219813605 | 4.321152512647504 | 0.005119292833899003 |
As we can see from the table, most values are very close (as can
reasonably be expected given different algorithms and
implementations. However especially centerX is obviously a little
different, which is due to the "one-off" error mentioned above. The
other properties with larger differences are all those, which depend
on the rotationAngle. This makes sense, because the rotationAngle
is the most involved calculation, making use of non-linear
optimization algorithms. See 13.4 below for
the implementation details. In short: the function being minimized is
exactly the same, but the algorithm used is TMinuit2 for Marlin and
BOBYQA for TPA.
Let's check that by modifying the raw x values before reconstruction
(added in
https://github.com/Vindaar/TimepixAnalysis/commit/5e0366a97cb5e3540b780b7654eb54606b7b1306
as an option) the center positions actually come closer. The output of
the above values is now instead, see
tab. 17
| Property | TimepixAnalysis | MarlinTPC | Difference |
|---|---|---|---|
| hits | 218 | 218 | 0 |
| centerX | 7.567798165137615 | 7.567798137664795 | 2.74728204630037e-08 |
| centerY | 6.574266055045872 | 6.574265956878662 | 9.816720947242175e-08 |
| rmsLongitudinal | 4.374324961805872 | 4.374318599700928 | 6.362104944201974e-06 |
| rmsTransverse | 3.151177831860438 | 3.15118670463562 | 8.872775182577897e-06 |
| eccentricity | 1.388155539042776 | 1.388149619102478 | 5.919940297971493e-06 |
| rotationAngle | 3.000219815795619 | 2.997750043869019 | 0.002469771926600473 |
| skewnessLongitudinal | 0.344838563066004 | 0.3436029553413391 | 0.001235607724664911 |
| skewnessTransverse | 0.274311235922031 | 0.2803316414356232 | 0.006020405513592231 |
| kurtosisLongitudinal | -1.440002172698533 | -1.439019083976746 | 0.0009830887217865403 |
| kurtosisTransverse | -0.516660362116625 | -0.5109220147132874 | 0.005738347403337674 |
| length | 13.6005882039472 | 13.61675834655762 | 0.01617014261042016 |
| width | 13.15926696045436 | 13.16585159301758 | 0.006584632563217951 |
| fractionInTransverseRms | 0.1880733944954129 | 0.1880733966827393 | 2.187326458846783e-09 |
| lengthDivRmsTrans | 4.316033219844496 | 4.321152512647504 | 0.005119292803008157 |
The difference in the values based on rotationAngle are still more
or less the same.
13.1. :
As of
https://github.com/Vindaar/TimepixAnalysis/commit/fd7073c92ab966ecc170e3960bb1052d55b3b919
we now compare all values using almostEqual allowing for a certain
epsilon, which the floats have to satisfy. These epsilons for each
property are shown in tab. 18.
| Property | Epsilon | Note |
|---|---|---|
| hits | 0 | |
| centerX | 1-e5 | |
| centerY | 1-e5 | |
| rmsLongitudinal | 1-e5 | |
| rmsTransverse | 1-e5 | |
| eccentricity | 1-e5 | |
| rotationAngle | 1-e2 | Most affected by non-linear opt algorithm |
| skewnessLongitudinal | 1-e2 | Deviate more than remaining |
| skewnessTransverse | 1-e2 | "" |
| kurtosisLongitudinal | 1-e3 | |
| kurtosisTransverse | 1-e3 | |
| length | 1-e3 | |
| width | 1-e3 | |
| fractionInTransverseRms | 1-e5 | |
| lengthDivRmsTrans | 1-e3 |
The next step is to add a couple more events for certainty (best would be a whole run, but that's problematic data wise in the CI, since we don't want to store all that data in the repo. Will have to find a good solution where to store those to be able to download) and then add checks for all events with same number of clusters found.
13.2.
An option was added in: https://github.com/Vindaar/TimepixAnalysis/commit/a7426b6f1e78ddfdd01ad8bcbbec1533827f52a9 to automatically copy the needed data files from the directory. Otherwise manually finding all files that correspond to MarlinTPC's "eventNumber" is annoying.
Changed the number of events to extract to 15. That should be enough
as a first start to have a decent understanding of geometric
differences.
Not commited at the moment, since it blows up the repo size too much.
13.3.
Based on the now 15 events, which can now be found in the TPAresources repository, which is embedded as a submodule into TimepixAnalysis (in the resources directory), we investigated what happened with a few more events.
With the changes as of
https://github.com/Vindaar/TimepixAnalysis/commit/c7563fad8e5513026c0f086c21cd772d4f7e4788
we now generate two different plot types. Either or both of the two
frameworks produces more than 1 cluster in the cluster finding stage,
a file recoed_cluster_{idx}.pdf is generated, which shows the two
frameworks side by side similar to fig. 86,
e.g. fig. 90.
On the other hand if both clusters only have a single cluster found,
we generate a non facet_wrap plot, which highlights missing pixels
in said cluster, if any. This arose due to a bug in the test case,
which caused more than the single noisy pixel to be filtered, which
was fixed here, like e.g. fig. 91.
Of the 15 events we check right now, we skip 3, due to them having
a different number of clusters in TPA than in Marlin, these are:
data0347721021331054.txt
data0430201232312198.txt
data0539081050952120.txt
On the other hand, some examples have much smaller differences in the rotation angle than the first example shown in the section before. For instance the event shown in tab. 19, or fig. 92.
| Property | TimepixAnalysis | MarlinTPC | Difference |
|---|---|---|---|
| hits | 290 | 290 | 0 |
| centerX | 1.936379310344827 | 1.936379313468933 | 3.124106084939626e-09 |
| centerY | 11.66151724137931 | 11.66151714324951 | 9.812980117374082e-08 |
| rmsLongitudinal | 0.9789389659115215 | 0.9789389371871948 | 2.872432669498437e-08 |
| rmsTransverse | 0.8146316514450767 | 0.8146316409111023 | 1.053397435946124e-08 |
| eccentricity | 1.201695225289835 | 1.201695203781128 | 2.150870681560946e-08 |
| rotationAngle | 1.071629641164285 | 1.071630239486694 | 5.983224089511907e-07 |
| skewnessLongitudinal | -0.43160063409701 | -0.4316010773181915 | 4.432211814786591e-07 |
| skewnessTransverse | -0.4530571933494556 | -0.4530574083328247 | 2.149833691067471e-07 |
| kurtosisLongitudinal | 0.1300158103087616 | 0.130016103386879 | 2.930781173582364e-07 |
| kurtosisTransverse | 0.03049568566315575 | 0.03049567155539989 | 1.410775585589108e-08 |
| length | 5.276106472381047 | 5.276106357574463 | 1.148065837952572e-07 |
| width | 4.3103265261156 | 4.310326099395752 | 4.267198479013246e-07 |
| fractionInTransverseRms | 0.296551724137931 | 0.2965517342090607 | 1.007112970796697e-08 |
| lengthDivRmsTrans | 6.476677481192575 | 6.476677424011602 | 5.718097284557189e-08 |
Comparing the behavior of the difference in the rotation angle dependent variables on the difference in the rotation angle, we end up with the following table 20 and the following (ugly, sorry) plot fig. 93
For these numbers we simply take the absolute value of the difference in rotation angle and the mean value of the absolute differences of each property.
The numbers in the table and the corresponding difference plot were created at commit: https://github.com/Vindaar/TimepixAnalysis/commit/16235d917325502a29eadc9c38d932a734d7b095
| eventIndex | eventNumber | rotAngDifference | meanPropertyDifference |
|---|---|---|---|
| 0 | 59424 | 0.000696276 | 0.000390437 |
| 1 | 57178 | 0.00246977 | 0.00523144 |
| 3 | 36747 | 3.61235e-05 | 6.33275e-05 |
| 4 | 31801 | 8.19876e-05 | 4.99778e-05 |
| 5 | 38770 | 0.00260109 | 0.00147491 |
| 6 | 55899 | 2.18814e-06 | 3.06651e-06 |
| 7 | 53375 | 2.36981e-05 | 2.68843e-05 |
| 8 | 11673 | 5.98322e-07 | 1.96771e-07 |
| 9 | 57233 | 0.00457722 | 0.00503872 |
| 10 | 25415 | 4.30135e-07 | 4.86432e-07 |
| 12 | 69254 | 1.2369e-05 | 1.3467e-05 |
| 14 | 74237 | 0.000127007 | 0.000158893 |
The event with the largest deviation is the event with index 9. It
looks like. fig 94.
By now this largely explains the plot, fig. 95, shown in a meeting several months back. Although there are still certain outliers that have a much larger difference than the test events used here. That is still open to discussion. It needs to be investigated if the larger deviations in properties derived from the rotation angle correspond to those events where the roration angle is off significantly.
13.4. How to calculate rotation angle
In case of MarlinTPC this is done via TMinuit2, see ./../../../../data/tpc18/home/src/MarlinTPC/krieger/reconstruction/pixelbased/src/GridPixXrayObjectCalculatorProcessor.cc for the implementation, and ./../../../../data/tpc18/home/src/MarlinTPC/XrayReco.xml to see that actually this processor is called.
The implementation in Marlin explcitily is thus:
// this is actually a Class definition and the `()` operator is used to // call this object... excentricity(const std::vector<double>& xInput, const std::vector<double>& yInput, const double& posX, const double& posY, const double& pitchXInput, const double& pitchYInput) : X(posX),Y(posY),x(xInput),y(yInput),pitchX(pitchXInput),pitchY(pitchYInput){ } double operator() (const std::vector<double>& p) const { double sumX(0.); double sumY(0.); double sumXSqr(0.); double sumYSqr(0.); for(int i = 0; i < static_cast<int>(x.size()); i++){ double newX = std::cos(p[0])*(x[i]-X)*pitchX - \\ std::sin(p[0])*(y[i]-Y)*pitchY; double newY = std::sin(p[0])*(x[i]-X)*pitchX + \\ std::cos(p[0])*(y[i]-Y)*pitchY; sumX += newX; sumY += newY; sumXSqr += newX*newX; sumYSqr += newY*newY; } double rmsX = std::sqrt((sumXSqr/static_cast<double>(x.size())) - \\ (sumX*sumX/static_cast<double>(x.size()) / \\ static_cast<double>(x.size()))); double rmsY = std::sqrt((sumYSqr/static_cast<double>(y.size())) - \\ (sumY*sumY/static_cast<double>(y.size()) / \\ static_cast<double>(y.size()))); double exc = rmsX / rmsY; return -exc; }
which is then called further below in processEvent:
_excentricity = new excentricity(xVec,yVec,posX,posY,pitchX,pitchY); _minuit->SetMinuitFCN(_excentricity); _minuit->SetParameter(0,"rotAngle",rotAngleEstimate,1., -4*std::atan(1.),4*std::atan(1.)); _minuit->SetPrintLevel(-1); _minuit->CreateMinimizer(); int minimize = _minuit->Minimize();
Here we can see the start parameters used and the fact that TMinuit2 is used.
The Nim implementation in TimepixAnalysis is based directly on this. It is found here:
proc eccentricity[T: SomePix](p: seq[float], func_data: FitObject[T]): float = let fit = func_data let c = fit.cluster let (centerX, centerY) = fit.xy var sum_x: float = 0 sum_y: float = 0 sum_x2: float = 0 sum_y2: float = 0 for i in 0..<len(c): let new_x = cos(p[0]) * (c[i].x.float - centerX) * PITCH - sin(p[0]) * (c[i].y.float - centerY) * PITCH new_y = sin(p[0]) * (c[i].x.float - centerX) * PITCH + cos(p[0]) * (c[i].y.float - centerY) * PITCH sum_x += new_x sum_y += new_y sum_x2 += (new_x * new_x) sum_y2 += (new_y * new_y) let n_elements: float = len(c).float rms_x: float = sqrt( (sum_x2 / n_elements) - (sum_x * sum_x / n_elements / n_elements)) rms_y: float = sqrt( (sum_y2 / n_elements) - (sum_y * sum_y / n_elements / n_elements)) # calc eccentricity from RMS let exc = rms_x / rms_y result = -exc
It's basically the same code just ported to Nim. However, we do not
use TMinuit2 to minimize the problem, but NLopt with the gradient free
LN_BOBYQA algorithm, see here:
https://github.com/Vindaar/TimepixAnalysis/blob/master/Analysis/ingrid/reconstruction.nim#L569-L613
13.5. Comparing Marlin / TPA by using (partial) Marlin results
In order to compare the impact of each difference found for Marlin vs. TPA we'll extract certain information from the Marlin results and use them instead of our own calculations. Then we simply perform the whole analysis chain and at the end we can compare the final background rate to the TPA result. By doing this for several different properties we can gauge the impact each has and maybe explain where the differences come from.
13.5.1. Comparing Marlin / TPA by clusters found in Marlin
As the (easiest) way to start, we use the previously found bad cluster
finding of Marlin. This means that instead of calling our own
findSimpleClusters proc, we will inject replacement code, which
instead reads the data from the
./../../CastData/ExternCode/TimepixAnalysis/resources/background_splitted.2014+2015.h5
file. This will be achieved by using a sort of monkey patching by
making use of NimScript's patchFile and a macro we add to the
./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/private/geometry.nim file, which performs body replacement. This has the
advantage that we leave (almost, beside the 4 line macro we add) our
normal TPA code as is. This avoids introducing bugs in the code, but
mainly prevents spoiling the code base with unrelated BS.
The patchFile proc simply replaces a certain file from a nimble
module with any other file. Module name and file paths are given. It
must be called from any of the read config.nims files during
compilation. Those files may lie in ./../../.config/,
./../../src/nim/nim_git_repo/config/config.nims (the Nim source tree
config directory) and the folder in which the file being compiled is
located.
For our attempt we will replace the local config.nims file relative
to reconstruction.nim.
Thus, the full injection works as follows:
./../../CastData/ExternCode/TimepixAnalysis/Tools/CompareMarlinTpa/UseMarlinRotAngle/
is a directory, which contains:
ls ~/CastData/ExternCode/TimepixAnalysis/Tools/CompareMarlinTpa/UseMarlinRotAngle/
where I removed some temp files etc.
runHackedAnalysis.nim is a simple script, which copies the
config.nims file, which in this dir is called
runHackedAnalysis.nims to the reconstruction.nim base directory,
perform the compilation with the correct flags (including
-d:activateHijack) and the two files being replaced, the
geometry.nim containing the injected code and a replaced dummy
threadpool. This is required, because our code cannot be thread safe
anymore, since we have to access global memory in order to make the
injection work without an actual rewrite.
The script is:
import shell, strutils, os let recoPath = "../../../Analysis/ingrid/" var undoBackup = false if fileExists($recoPath / "config.nims"): shell: cp ($recoPath)/config.nims ($recoPath)/config.nims_backup undoBackup = true shell: cp runHackedAnalysis.nims ($recoPath)/config.nims shell: nim c "-f --threads:on -d:danger -d:activateHijack" ($recoPath)/reconstruction.nim if undoBackup: shell: cp ($recoPath)/config.nims_backup ($recoPath)/config.nims else: shell: rm ($recoPath)/config.nims shell: "../../../Analysis/ingrid/reconstruction ../../../Tests/run_245_2014.h5" "--out" testfile.h5
The nims file is just two patchFile calls:
patchFile("ingrid", "geometry", "../../Tools/CompareMarlinTpa/UseMarlinRotAngle/geometry") patchFile("threadpools", "threadpool_simple", "../../Tools/CompareMarlinTpa/UseMarlinRotAngle/threadpool_simple")
and the threadpool dummy is just:
type ThreadPool* = object discard FlowVar*[T] = object when T isnot void: v: T proc sync*(tp: ThreadPool) = # dummy discard proc newThreadPool*(): ThreadPool = result = Threadpool() template spawn*(tp: ThreadPool, e: typed{nkCall | nkCommand}): untyped = when compiles(e isnot void): type RetType = type(e) else: type RetType = void FlowVar[RetType](v: e) proc read*[T](v: FlowVar[T]): T = result = v.v proc `^`*[T](fv: FlowVar[T]): T {.inline.} = fv.read()
where we make sure to export all procs, which are actually used in the
ingrid module.
The data storage in the background Marlin file is pretty ugly in my opinion. All data for each type (read background, sunrise tracking etc.) are a single 1D dataset for each property. This means all runs are part of the same dset. This means there's barely any structure in the data and every run / event or whatever specific data access has to be done via the indirection of filtering on another dataset (also meaning we have to read the whole freaking run / eventNumber datasets each time we need only a subset, since we have no idea where the data is! Well, one can read batches by "guessing" but that's…).
This means we first parse the Marlin data into a combination of the following types:
type MarlinCluster = object data: Cluster[Pix] globalIdx: int rotAngle: float MarlinEvent = object eventNumber: int clusters: seq[MarlinCluster] MarlinRuns = object run: int events: seq[MarlinEvent]
Which is done by reading the run dataset, splitting it by runs and the corresponding indices and then reading each run / all clusters of that run.
While doing this a weird behavior was uncovered.
Yesterday () I stopped working being under the
impression that for some reason the Run 245 in the background file
contained O(>130,000) events (according to the event number, not the
actual number of clusters), while the raw data file only contains
96340 files and the last event number being 123691. This does not
make any sense whatsoever.
I continued by investigating if this was some bug yesterday or it is true.
NOTE: it appears that Marlin does actually count only non empty frames indeed. However, the raw data I have only contains non empty frames in the first place. So the numbers given in the filenames should actually match the given clusters…
Ok for some stupid reason this whole numbering of Marlin does not make any sense. Aside from what I explained above, sometimes events are completely dropped after all. For instance run 245 has the following files as the first ~50 files:
-rwxrwxrwx 1 root 12 Jul 13 2018 data000000_1_071309274.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000001_1_071310277.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000002_1_071311281.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000003_1_071312284.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000004_1_071313288.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000005_1_071314291.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000006_1_071315295.txt* -rwxrwxrwx 1 root 60 Jul 13 2018 data000007_1_071316298.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000008_1_071317302.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000009_1_071318310.txt* -rwxrwxrwx 1 root 22 Jul 13 2018 data000010_1_071319309.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000011_1_071320313.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000012_1_071321316.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000013_1_071322320.txt* -rwxrwxrwx 1 root 1.3K Jul 13 2018 data000014_1_071323323.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000015_1_071324327.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000016_1_071325330.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000017_1_071326334.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000018_1_071327337.txt* -rwxrwxrwx 1 root 34 Jul 13 2018 data000019_1_071328341.txt* -rwxrwxrwx 1 root 1.5K Jul 13 2018 data000020_1_071329344.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000021_1_071330348.txt* -rwxrwxrwx 1 root 552 Jul 13 2018 data000022_1_071331351.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000023_1_071332355.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000024_1_071333359.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000025_1_071334366.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000026_1_071335365.txt* -rwxrwxrwx 1 root 2.0K Jul 13 2018 data000027_1_071336369.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000028_1_071337372.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000029_1_071338376.txt* -rwxrwxrwx 1 root 13 Jul 13 2018 data000030_1_071339379.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000031_1_071340383.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000032_1_071341386.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000033_1_071342390.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000034_1_071343393.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000035_1_071344397.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000036_1_071345400.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000037_1_071346403.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000038_1_071347407.txt* -rwxrwxrwx 1 root 288 Jul 13 2018 data000039_1_071348410.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000041_1_071350417.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000042_1_071351421.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000043_1_071352424.txt* -rwxrwxrwx 1 root 23 Jul 13 2018 data000044_1_071353428.txt* -rwxrwxrwx 1 root 2.6K Jul 13 2018 data000045_1_071354432.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000046_1_071355435.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000047_1_071356439.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000048_1_071357442.txt* -rwxrwxrwx 1 root 688 Jul 13 2018 data000049_1_071358446.txt* -rwxrwxrwx 1 root 11 Jul 13 2018 data000050_1_071359454.txt* -rwxrwxrwx 1 root 21 Jul 13 2018 data000051_1_071400453.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000052_1_071401456.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000053_1_071402460.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000054_1_071403464.txt* -rwxrwxrwx 1 root 12 Jul 13 2018 data000055_1_071404471.txt*
HOWEVER aside from dropping all events from the ROOT (now H5) file, which have less than 3 lines, it completely makes some more disappear.
> Building run 245 > for @[863985, 863986, 863987, 863988, 863989, 863990, 863991, 863992, 863993, 863994, 863995, 863996, 863997, 863998, 863999, 864000] > EventNumbers: @[7, 14, 20, 22, 27, 27, 27, 27, 39, 44, 44, 48, 57, 58, 59, 59] > Last event number: 96330 at index 880491 For run 245
where we see that the events included in the freaking H5 file are the
event numbers given there! See that event 45 should be the one after
39 from the file size of data000045_1_071354432.txt? For some reason
it's suddenly 45!!! What the heck.
I have no fucking idea what's going on here.
I'll just use all of the raw data from Marlin instead of reading it ourselves… Ugh. Not sure if that introduces any other problems, but I don't see a better way. Cross reference the commit: 6c51ee2fe51d8b491d8c4e628395f1dac5e5683b of TPA for the state that reproduces the numbers above..
Hah. Except I can't do that! The only reason I did this hacky shit is because I don't have the ToT values corresponding to the Marlin found clusters, since it only. contains. the. charge. values.!
13.5.2. Update:
Over the weekend I played some more with this. The approach is working (at least sort of) taking this into account:
- the only noisy pixel is (167, 200)
some events (from multiple clusters) have a different number of pixels despite describing the same events, because a few pixels can be far away from the other clusters, be reconstructed as their own cluster but then be dropped, because they're too small. See for example: ./../../src/MarlinTPC/XrayReco.xml From the "MyTimepixMapHandlerProcessor":
<!--number of pixels in a cluster in order for it to be seperated--> <parameter name="clusterSizeToSep" type="int">2 </parameter>
and from "MyTimePixSpecialClusterFinderProcessor":
<!--minimum number of pixels in one cluster--> <parameter name="minNoPixel" type="int">3 </parameter>
Which means: a cluster which is 2 pixels and far away will be split from the rest of the cluster if it's further away than 50 pixels, but will then be dropped! TODO: verify this with an example to show. Included a "small mismatch" plot in the hijacked geometry.nim. Result should give us plots for these! These events we just take as is, ignoring the small mismatch (we use a cut off of 5 pixels max).
If we find larger diffrences than this, we ignore the event and take the one afterwards, probably because an event disappeared as stated before this update. The next one should be the real event then. These events typically contain only 3 pixels in our raw data, e.g. fig. 96.
Figure 96: Example of event mismatch, because of 3 raw pixel event, which completely disappears in MarlinTPC (lost event number as described above). we also filter out pixels with a ToT value larger than X. X currently is not exactly known, but events like ./../../../../mnt/1TB/CAST/2014_15/CalibrationRuns/256-Run150623_12-58-49/data007600_1_130158448.txt:
0 166 298 1 166 752 2 166 984 3 166 1136 4 166 766 5 166 309 6 166 179 7 166 1333 0 165 33 2 165 913 3 165 1282 4 165 466 5 165 567 6 165 1493 7 165 1335
do not appear in Marlin. The reason has to be that their ToT values are too large (for some pixels), those being dropped and the remaining pixels are too few to count as a valid cluster (<= 3). Based on this a cut off of 400 ToT was chosen. For one run (256, calibration) it seems to work.
That seems all. With this, we'll attempt to reconstruct all of the calibration runs first.
TODO fill me in!!! See fig. 97.
NOTE: In the plots below which were included previously, there was a bug, which caused the TPA values to be switched. The ones in the fit plot were the mean values and vice versa. This is fixed now. It was only a minor bug in the ./../../CastData/ExternCode/TimepixAnalysis/Tools/CompareMarlinTpa/compareGasGainMarlinTpa.nim script which is why the records were not kept.
13.6. Comparison of Marlin & TPA charge calibration
At some point I was unsure whether the actual calibration function used in Marlin vs. the one I use is actually analytically the same.
So I simply implemented both the way they are used and calculated the charge values for all ToT values in \([0, 11810]\) and made a plot:
import ggplotnim import math, seqmath, sequtils const a = 0.3484 const b = 58.56 const c = 1294.0 const t = -12.81 const conversionFactor = 50.0 proc marlinChargeCalib(x: int): float = let p = (b - x.float - a * t) / a let q = (t * x.float - b * t - c) / a result = conversionFactor * (-p / 2.0 + sqrt(p * p / 4.0 - q) ) func calibrateCharge(totValue: int): float = # 1.sum term let p = totValue.float - (b - a * t) # 2. term of sqrt - neither is exactly the p or q from pq formula let q = 4 * (a * b * t + a * c - a * t * totValue.float) result = (conversionFactor / (2 * a)) * (p + sqrt(p * p + q)) let tots = arange(0, 11810) let mCh = tots.mapIt( marlinChargeCalib(it) ) let tpaCh = tots.mapIt( calibrateCharge(it) ) let maxDiff = block: var res = 0.0 for i in 0 ..< mCh.len: let diff = abs(mCh[i] - tpaCh[i]) if res < diff: res = diff res echo "Max difference is: ", maxDiff var df = toDf({ "tot" : tots, "marlinCharge" : mCh, "tpaCharge" : tpaCh }) df = df.gather(["tpaCharge", "marlinCharge"], key = "from", value = "charge") echo df.pretty ggplot(df, aes("tot", "charge", color = "from")) + geom_line() + ggsave("charge_calib_compare.pdf")
As one can see both given the maximum difference \(\mathcal{O}(1e-10)\) and the plot in fig. 102 shows two lines exactly on top of one another, this thought can be put aside.
Note: I also cross checked the resulting ToT calibration factors in
use in TPA again in the ingridDatabase.h5 file. They are exactly the
same as the ones used in the code above (whereas those are the ones
taken from ./../../../../mnt/1TB/CAST/2014_15/xmlbase/getGasGain.C!).
The values in the database simply have more digits after the ones used by Christoph.
13.7. TODO write about calculation of gas gain from mean of histo
Instead of mean of distribution of the data, we used that of the fit. BAD. Write about!
\clearpage
13.8. Gas gain calculation contained major bug
Finally found a major bug on
idxFix appended to files which have it.
In
./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/calibration.nim
in the calcGasGain proc, the following lines contain the bug:
let passIdx = cutOnProperties(h5f, group, crSilver, ("rmsTransverse", cut_rms_trans_low, cut_rms_trans_high)) let vlenInt = special_type(uint16) # get all ToT values as `seq[seq[uint16]]` and flatten let totsFull = totDset[vlenInt, uint16].flatten let tots = passIdx.mapIt(totsFull[it])
We first flatten the seq[seq[T]] before we apply the indices,
which map to events. Thus we filter using event indices on a
seq[T], which contains pixels.
let passIdx = cutOnProperties(h5f, group, crSilver, ("rmsTransverse", cut_rms_trans_low, cut_rms_trans_high)) let vlenInt = special_type(uint16) # get all ToT values as `seq[seq[uint16]]` let totsFull = totDset[vlenInt, uint16] # use `passIdx` to get the correct events and then flatten let tots = passIdx.mapIt(totsFull[it]).flatten.mapIt(it)
Which is what we were supposed to do.
After fixing this in commit 480e5b0094d88968170669b7ce33c2d6e2824920, the whole reconstruction was rerun (the raw data files were kept; yay for having those now).
The gas gains (fit and mean of data) are shown in figs. 103, 104. We can see that the agreement is almost perfect now.
The fit to the gas gain vs charge calibration factor fit, see fig. 106, in comparison to fig. 107 (Krieger PhD) and the previously wrong curve in fig.

However, unfortunately this does not fix the background rate below \(\SI{2}{\keV}\)! See fig. 108 for the comparison.
This results in a polya fit as shown in fig. 109. Compare this to the previos, broken plot of TPA from the same run, fig. 99 and the correct marlin results in fig. 98.
\clearpage
13.9. Comparison of XrayReferenceFile distributions
One of the ingredients for the successful application of the LogL cuts is of
course the shape of the XrayReferenceFile.
The creation of our version of the 2014/15 files is described in detail in sec. 7.4.
The contained datasets are the target filter combinations from the corresponding
calibration-cdl.h5 file, with the X-ray reference cuts applied, namely the
ones found here:
./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/private/cdl_cuts.nim
in the func getEnergyBinMinMaxVals<Year>*(): Table[string, Cuts] procs.
The resulting cluster data sets are then binned using the binning information
that was used in Krieger thesis, which is stored in:
./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/private/hdf5_utils.nim
in the func cdlToXrayBinning<Year>Map(): Table[InGridDsetKind, tuple[bins: int,
min, max: float]] procedures.
The resulting histograms are the XrayReferenceFile. As long as the histogram
function works correctly, the cuts are chosen in the same way and the input data
is correct, the histograms have to match between 2014/15 Marlin and TPA.
Variability is of course expected, because the reconstruction introduces
differences, thus changing the input data.
The shape of the distributions for the 2014/15 results are visible in Krieger thesis in the "Reference dataset" appendix C starting on page 183 (document, not PDF).
Those datasets plotted using ggplotnim as a ridgeline plot to allow shape
comparison between the different datasets, is shown in
figs. 110,
111 and
112 (sec. 13.9.1). The data file for that plot is:
./../../../../mnt/1TB/CAST/CDL-reference/XrayReferenceDataSet.h5.
The code to generate the plots here was added to the likelihood program in
commit:
41cfa60e91984dde3de352fd8fcfe56b91405f70
The same plot for the XrayReferenceFile for 2014/15 data reconstructed with
TPA is shown in the corresponding plots in sec. 13.9.2 and the 2017/18 data (CDL Feb 2019) in
sec. 13.9.3
The related files are located in:
- 2014/15 TPA: ./../../../../mnt/1TB/CAST/2014_15/CDL_Runs_raw/XrayReferenceFile2014.h5
- 2019 CDL: ./../../../../mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5
as is also visible by the year in the filename. In addition there is an attribute in the H5 file on the ROOT group mentioning the framework used for reconstruction.
\clearpage
13.9.1. XrayRefercenceFile datasets, Marlin 2014
XrayReferenceFile.h5 of 2014.XrayReferenceFile.h5 of 2014.XrayReferenceFile.h5 of 2014.\clearpage
13.9.2. XrayRefercenceFile datasets, TPA 2014
XrayReferenceFile.h5 of 2014.XrayReferenceFile.h5 of 2014.XrayReferenceFile.h5 of 2014.\clearpage
13.9.3. XrayRefercenceFile datasets, TPA 2019
XrayReferenceFile.h5 of 2019.XrayReferenceFile.h5 of 2019.XrayReferenceFile.h5 of 2019.\clearpage
13.9.4. Things of note
Note that the height of each ridge in the plots depends on the relative count in a dataset compared to the others. For the shown plots the maximum allowed overlap was set to \(\num{1.75}\). This means all others show relative counts to the largest entry.
One thing is striking from immediately. Comparing the fraction within transverse RMS plot of 2014 Marlin to 2014 TPA. The relative height of this dataset is much lower in the TPA reconstructed dataset.
The same behavior is visible in the other properties.
The likely cause is that the TPA reconstructed code is still using the same charge cuts for the filtering of raw CDL data to reference datasets as the Malrin dataset.
They were not changed after performing the spectrum fits using
cdl_spectrum_creation, as is described in sec. 17.2.1!
This is possibly one of the most important reasons why the background is still this much lower for the TPA reconstructed rate than for Marlin!
Need to check this tomorrow .
Finally, of course even then the question remains why the charge seems to be so different. Is the charge calibration / the gas gain fit still the issue?
13.9.5. Compare CDL data for lower most spectra
In order to understand why such a difference in number of events happens when comparing the CDL based reference dataset, it seems like a good idea to compare the raw CDL spectra of Marlin and TPA and additionally add the CDL cuts still being used in TPA for 2014 data to the plot of the Marlin data (both from Krieger thesis as well as reading both spectra into one plot).
So let's write a small plotting script, which reads both:
- ./../../../../mnt/1TB/CAST/CDL-reference/calibration-cdl.h5 2014 Marlin
- ./../../../../mnt/1TB/CAST/calibration-cdl-2014.h5 2014 TPA
files and plots the contained spectra including the charge cut values.
Script is now live here:
- ./../../CastData/ExternCode/TimepixAnalysis/Plotting/compareCdlMarlinTpa/compareCdlMarlinTpa.nim
- https://github.com/Vindaar/TimepixAnalysis/blob/master/Plotting/compareCdlMarlinTpa/compareCdlMarlinTpa.nim
TODO: Need to fix the bin ranges in some plots (use percentiles?) and actually compare them.
- have way less statistics in the first place, only 1 run for C 0.6 kV???
- still less in Cu 0.9 kV 23000 to 28000 roughly
UPDATE : As it turns out as of commit
1d963c412ed45c0294146d5597e127e76c01aa46 the file containing the the CDL runs
from 2014 is simply missing a bunch of runs. :O
./../../CastData/ExternCode/TimepixAnalysis/resources/cdl_runs_2014.html.
Why are those runs missing? See section 7.4. There we
generated said file from the directory
/mnt/1TB/CAST/CDL-reference, which wastaken from Christoph's backup on
tpc00. However, that must be wrong andmissing some runs for some reason, given that
- the
xlsxfile mentions more runs as "ok" - the 2014 Marlin
calibration-cdl.h5file literally contains data from the missing runs!
NOTE: Update after reading the above linked section again. No, the issue is not that the runs are missing from the directories. I didn't finish generating the file in the first place and instead used the file created by Hendrik. Never trust other people. :'(
Thus, in order not to be fooled again, Thus, we are going to regenerate the
CDL 2014 file from the existing calibration-cdl.h5 file!
import nimhdf5, strutils, sugar, sequtils, algorithm import ingrid / cdl_spectrum_creation const path = "/mnt/1TB/CAST/CDL-reference/calibration-cdl.h5" const header = "| Run # | Type | FADC? | Target | Filter | HV / kV |" const sepLine = "|-------+--------------+-------+--------+--------+---------|" let h5f = H5file(path, "r") var file = open("cdl_runs_2014.org", fmWrite) file.write(header & "\n" & sepLine & "\n") var runs: seq[(float32, seq[string])] for grp in h5f: let tfKindStr = dup(grp.name, removePrefix("/calibration-cdl-apr2014-"), removeSuffix("kV")) let r = h5f[grp.name & "/RunNumber", float32].deduplicate runs.add zip(r, repeat(tfKindStr.split('-'), r.len)) for (r, tf) in runs.sortedByIt(it[0]): file.write("| " & concat(@[$(r.int), "rtXrayFinger", "n"], tf).join("|") & " |\n") file.close() discard h5f.close()
Updated run list live with commit c17bb697b88fe25a2ce2595af650d366c6093272.
UPDATE : It turns out there is one run, which is
definitely missing in our data, namely run 8 (the first Cu Ni 15 kV
run. According to the excel file that run stopped after about 5 minutes and had
6005 events. Should not matter too much.
13.9.6. Determine lower pixel cut used for Marlin CDL reconstruction
It seems like first and foremost, after doing the above we still have less
clusters in the TPA calibration-cdl-2014.h5 file (~16000 vs. 22000).
Let's quickly write a script to plot a histogram of the NumberOfPixels dataset
in the Marlin file:
import nimhdf5, ggplotnim const path = "/mnt/1TB/CAST/CDL-reference/calibration-cdl.h5" let h5f = H5file(path, "r") let hits = h5f["/calibration-cdl-apr2014-C-EPIC-0.6kV/NumberOfPixels", float32] let df = toDf(hits) ggplot(df, aes("hits")) + geom_histogram(bin_width = 1.0) + xlim(0.0, 25) + ggsave("/tmp/hits_marlin_C_EPIC_0.6k.pdf")
Thus, the smallest number of pixels allowed is 3.
In our code (reconstruction or rather technically in geometry.nim) the cut
is set to 6 pixels (check is done for > cutoff_size!).
TODO: From here we need to reconstruct the whole CDL data. Then
- create
calibration-cdl-2014.h5again - check number of entries of the datasets, specifically C EPIC 0.6kV
- fix up the charge cuts for the reference file
- recreate
XrayReferenceFile.h5 - replot the plots comparing:
totalChargeand properties of the raw uncut CDL data- comparison of XrayReference histograms
13.10. Comparison of background / signal lnL distributions
Cu-EPIC-0.9kV line is way too low (compare with (g) in 124). This is interesting as this is the main bin which shows largest difference between background rate of Marlin vs. TPA! NOTE: Part of the reason for the difference is explained in the section below.IMPORTANT:
The plots of the lnL distributions created by TPA shown above
(fig. 119 and fig. 120) are stacked!!
I knew that, but due to not looking at the plots for a long time I
forgot and thought I set position = "identity" in the mean time. Yet
I did not!
The plots below fig. 125 and fig. 126 show the same plot with identity placement using frequency polygons for better visibility.
13.10.1. UPDATE:
There was indeed a bug in the determination of the ROC curves, which is partly the explanation why the ROC curves look differently in the figures 123 and 124.
The fix is done in commit
d57b18761b36097b707a32e52cabb4d02bdd3cd1. The problem was that we
accidentally used the already filtered background data, that is the
data without any logL = Inf values. This obviously reduces the
apparent background rejection.
The fixed plot is shown in fig. 127. Note however,
that this is still different compared to the Marlin plots above. For
instance the Cu-EPIC-0.9kV case has a background rejection of
\(\SI{78}{\percent}\) while the same case for Marlin (subplot (g)) still
has a background rejection of \(\SI{84}{\percent}\).
Cu-EPIC-0.9kV line is still too low (compare with (g) in 124). This is interesting as this is the main bin, which shows largest difference between background rate of Marlin vs. TPA!13.10.2. Create ROC curves using TPA created cdl 2014 file
Since the curves in fig. 127 still look different
from the Marlin ones, the next attempt is to look at the curves
created by using the TPA calibration-cdl2014.h5 file (generated from
./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/cdl_spectrum_creation.nim).
The explanation as to how this file is generated is given in section 7.4.
This is done with the following command:
./likelihood /mnt/1TB/CAST/2014_15/DataRuns2014_Reco.h5 \ --createRocCurve \ --altCdlFile /mnt/1TB/CAST/2014_15/CDL_Runs_raw/calibration-cdl-2014.h5 \ --altRefFile /mnt/1TB/CAST/2014_15/CDL_Runs_raw/XrayReferenceFile2014.h5
The result is the following ROC curve plot, fig. 128.
calibration-cdl.h5 and reference dataset generated by cdl_spectrum_creation (TPA). Very obvious that something is different and likely wrong!It's very obvious that something is amiss. The wiggliness of the lowest line is clearly unexpected. Either we have some binning artifact at play here or we unexpectantly have very little statistics.
Let's look at the distributions for the signal like data of the logL values in fig. 129 compared with the same plot using the CDL file created by Marlin in 130.
We'll create a similar plot as the above two with both datasets in one plot for easier visibility of the differences. We'll do this by dumping the data frame of the input of each plot to a CSV file, adding it to this repository and write a small script to plot it in this file.
We can then compare the number of entries in each target filter combination (maybe we lose statistics due to something we do / maybe we have too little data to begin with - missing runs - or whatever).
IDEA: how to check for likelihood: Use Marlin CDL file, and calculate logL value for each cluster from TPA and compare!
- TODO Is bad line shape due to bad charge cut values?
During calculation of the XrayReferenceFile we need precise values to cut to the real main peak of each target + filter combination.
I think I didn't update the charge values for those cuts, which probably leads to very bad behavior, because essentially the XrayReferenceFile partly misses data.
\clearpage
14. General investigations
14.1. Chi Sq of gas gain fit
With the recent changes of commit PUT HASH HERE AFTER MERGE we now show the reduced \(\chi^2 / \text{d.o.f.}\) in the title of the plot. However, we can see that the result often seems much too small. Interestingly this is mainly the case for chips other than 3!
See for instance fig. 131 for a polya for chip 3 and compare it to the fit for chip 4 in fig. 132 we see a super small value. But at least the fit works even better than chip 3. On the other hand the same plot for chip 6 fig. [[ shows a fit that is clearly broken (although possibly only outside the fitting range!! Investigate range!) and also has a tiny \(\chi^2 / \text{d.o.f.}\).
\clearpage
14.2. Comparison of Fe spectrum pixel fits Python/Nim
While rewriting the fitting code of TPA recently () in order to both get rid of Python dependencies and make use of the fitting macro written for the CDL fits, I started to write comparison tests for what I replaced.
First of all for the fit to the Fe spectrum for the pixel data.
While doing that I noticed some really weird behavior of the different
fitting routines. Python's scipy.curve_fit gave a mostly reasonable
result, although also there a small bump is visible
The test file is: ./TimepixAnalysis/Tests/reconstruction/tFitFe.nim
NOTE: Prior to 68bb07c6245736f6aeeef26aa453b84e563eb04c the new Nim based fitting routines
ignored the defined bounds, because there was a
var result = ...
line in the proc that returns the bounds, which caused us to never
return anything, since we shadowed the result variable, but didn't
say result alone in the last line. This was a bug introduced when
the bounds proc was made a proc. It was a template before that.
Both fits were changed to use the exact same binning (which is checked in the test; previously the Nim fit used a bin width of 3 instead of 1!). A small bug in the calculation of the binning for the Nim code was fixed at that time too.
Both fits also just use essentially an error of 1.0. In case of Python
this means we don't actually provide an error, but it will just use an
error of 1 by default.
For scipy:
https://github.com/scipy/scipy/blob/v1.4.1/scipy/optimize/minpack.py#L460-L463
this is the function that will be used in the optimizer. If
transform is none (defined in the calling scope based on curve_fit
sigma argument) it simply doesn't have any transformation of the
function.
The declared fit function for is (in Python it's manually defined):
declareFitFunc(feSpectrum): ffExpGauss: "Mn-Kalpha-esc" ffExpGauss: name = "Mn-Kbeta-esc" eN = eN("Mn-Kalpha-esc") * p_ar[14] # p_ar[14] is an additional fit parameter emu = emu("Mn-Kalpha-esc") * 3.5 / 2.9 # lock to relation to `Mn-Kalpha-esc` arg es = es("Mn-Kalpha-esc") # lock to `es` of `Mn-Kalpha` ffExpGauss: "Mn-Kalpha" ffExpGauss: name = "Mn-Kbeta" eN = eN("Mn-Kalpha") * p_ar[14] emu = emu("Mn-Kalpha") * 6.35 / 5.75 # lock to relation to `Mn-Kalpha` arg es = es("Mn-Kalpha") # lock to `es` of `Mn-Kalpha`
where p_ar[14] is an additional free parameter, which is used to fix
the K-beta line amplitdes with respect to the K-alpha amplitdes.
scipy.curve_fit with bounds (calls scipy.least_squares with trf (trust region fit) algorithm) vs. the fit with nlopt from Nim. We see that the Python fit has a weird bump near 170 pixels, but the mpfit fit suffers from a kink on the LHS of the main photo peak. The \(\chi^2/\text{d.o.f.}\) for the mpfit fit is \(\num{13.84}\). For Python it is not known.
#+CAPTION The \(\chi^2/\text{d.o.f.}\) for the nlopt fit is
The fit parameters for the curve_fit and mpfit fits are as
follows:
Nim: P[0] = -1.087688911915764 +/- 1.020700318722243 P[1] = -0.03301370388533077 +/- 0.0583324151869608 P[2] = 10.12406261365524 +/- 0.3217326542192619 P[3] = 135.9964525615895 +/- 0.3265645977010108 P[4] = 9.400429767716425 +/- 0.3688217584104574 P[5] = 0.0 +/- 0.0 P[6] = 0.0 +/- 0.0 P[7] = -2.141722770607383 +/- 0.4279572434041407 P[8] = 0.01571410561881723 +/- 0.002074000869198006 P[9] = 70.33982513756375 +/- 0.239498233291062 P[10] = 270.0442719089232 +/- 0.1211042551739025 P[11] = 17.38469606218239 +/- 0.1061788189141645 P[12] = -0.07737684791538217 +/- 0.0 P[13] = 0.002361728445329643 +/- 0.0 P[14] = 0.02658955973285415 +/- 0.007043296423750531 Python: p_0 = -3.8091112579898607 +- 664.0685776777244 p_1 = -0.2543209679305682 +- 149.9177486702038 p_2 = 9.827137587248982 +- 1.1551248129979157 p_3 = 135.83010652988253 +- 1.1361264737583983 p_4 = 8.687212459716198 +- 1.152672293912876 p_5 = -4.071919592698672e-15 +- 2.33793705312362e-09 p_6 = -3.271059822613274e-15 +- 1.2607718060554034e-08 p_7 = -6.872960496798991 +- 14066.635730347338 p_8 = -0.07057917088580651 +- 655.669585354836 p_9 = 67.62206233455022 +- 1.9885162481017526 p_10 = 269.82499185570555 +- 0.318570739199299 p_11 = 15.935582326626962 +- 0.3989026220769561 p_12 = -0.8415886895272773 +- 1.2624092861023852 p_13 = 0.00884604223679859 +- 0.006014451627636819 p_14 = 0.10175893815442262 +- 0.027813638900144932
The fits for the energy calibration were also moved over to Nim. The resulting fit parameters are also tested in ./../../CastData/ExternCode/TimepixAnalysis/Tests/reconstruction/tFitFe.nim and match well with the Python results (which is no surprise of course, given the fact that it's a linear fit).
The plot for the run used for testing is shown in fig. 136.
Parameters of calibration fit:
Nim: a^-1 = 21.332694254459806 +- 0.028456349644216023 Python: a^-1 = 21.31800535054976 +- 0.009397812699508474
\clearpage
14.2.1. Fe charge spectrum
The charge spectrum was also converted to use mpfit from Nim. Here the fit is much simpler, which results in almost perfect agreement of the two libraries. See fig. 137.
The declared fit function for the charge fit is:
ffGauss: "Mn-Kalpha-esc" ffGauss: "Mn-Kalpha" ffGauss: name = "Mn-Kbeta-esc" gN = gN("Mn-Kalpha-esc") * (17.0 / 150.0)# lock to Kalpha escape peak gmu = gmu("Mn-Kalpha-esc") * (3.53 / 2.94) gs = gs("Mn-Kalpha-esc") # lock to Kalpha escape peak ffGauss: name = "Mn-Kbeta" gN = gN("Mn-Kalpha") * (17.0 / 150.0) # lock to Kalpha escape peak gmu = gmu("Mn-Kalpha") * (6.49 / 5.90) gs = gs("Mn-Kalpha") # lock to Kalpha escape peak
Which means that the K-beta lines are fixed with respect to the K-alpha lines.
| Parameter | Meaning | Mpfit results | Mpfit errors | scipy.curvefit (trf) | curvefit errors |
|---|---|---|---|---|---|
| 0 | NKα-esc | 8.768296054521993 | 0.2683507496720102 | 8.768286883183828 | 0.8780793171060238 |
| 1 | μKα-esc | 427.6354202844113 | 1.685120722694063 | 427.6354634703872 | 5.514192772380387 |
| 2 | σKα-esc | 48.21096938527347 | 1.721099489439823 | 48.211058213503755 | 5.63194902213723 |
| 3 | NKα | 63.8798536616741 | 0.2120196952745237 | 63.87985873512651 | 0.6937700015058812 |
| 4 | μKα | 859.0423007834373 | 0.2641305903574854 | 859.0422957628506 | 0.8642828475288842 |
| 5 | σKα | 70.36757123264431 | 0.2754759027072539 | 70.36756104039033 | 0.9014067701994025 |
Finally the energy calibration of the charge spectrum is shown in fig. 138.
Here the fit parameters are essentially the same for Python and Nim.
Nim: a^-1 = 6.867026670389081 +- 0.0009881714159211403 Python: a^-1 = 6.86702663955951 +- 0.00210501332154937
14.3. DONE General removal of Python & Plotly in TPA
After the work mentioned above, I also went ahead and removed all usage of Python in TimepixAnalysis (which was mainly the Fe spectra fits anyways now) and the remaining Plotly plots.
Same as with the python dependencies, which slow down the run time significantly, each call to a Plotly plot also takes a long time, especially because we have to open a plot in the browser in order to save it.
Finally, the code is a lot clearer now in regards to that.
- ./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/calibration.nim:
All procedures, which involve calibration steps. These parts are
involved in the actual data reading and writing of results. They
call into:
- ./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/calibration/calib_fitting.nim: contans the procedures, which get prepared data, perform a fit and return the fit results.
- ./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/calibration/calib_plots.nim:
contans the procedures, which get prepared data and fit results
and create a
ggplotnimplot - ./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/calibration/fit_functions.nim:
contans the actual fitting routines, used in
calib_fitting.nim.
This work was done over the weekend - .
14.4. Reconstruct all 2017/2018 data again
It is time to reconstruct all data from 2017 and 2018 again to see the impact of all fixes (especially the total charge fix mentioned 13.8) on the background rate and also on the variation of the peak positions.
This was attempted on in the evening. After fixing a few minor bugs. Mainly:
- start gas gain fit from 2 ToT clock cycles, due to crazy noise on chip 4 in run 108 with many ToT counts of 1.
- fix minor ingrid database opening / closing bug
- fix \(\chi^2\) used in Nlopt fits. Instead of returning and thus minimizing the reduced chi square, stick with normal chi square. This had the effect that we'd divide the \(\chi^2\) for the gas gain polya fits twice by the number of degrees of freedom.
14.4.1. 2017 / beginning 2018 (Run 2)
After these fixes the code ran without trouble for the 2017 / beginning 2018 data.
The resulting gas gain versus energy calibration fit is shown in fig. 139 (and the 2014/15 comparison in fig. 140).
At the same time the variation of the peak positions of the spectra against time in those months is shown via fig. 141.
The remaining plots were all generated using ./../../CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/plotData.nim as follows:
./plotData /mnt/1TB/CAST/2017/CalibrationRunsRuns2017_Reco.h5 --backend=ggplot --runType calibration --no_occupancy --no_fadc ./plotData /mnt/1TB/CAST/2017/DataRuns2018_Reco.h5 --backend=ggplot --runType background --no_occupancy --no_fadc ./plotData /mnt/1TB/CAST/2018_2/CalibrationRuns2018_Reco.h5 --backend=ggplot --runType calibration --no_occupancy --no_fadc ./plotData /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 --backend=ggplot --runType background --no_occupancy --no_fadc
which generates the plots in a specific directory under figs/.
Remember that the file type for the output can be set in the
./../../CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml file as filetype = "pdf" # or png, svg....
The corresponding plot for the charge distribution is shown in fig. 142.
Finally, the same plot with a smaller binnign (a 30 minutes) for each is shown in fig. 143 and 144.
- TODO Investigate temperature relation of this
It appears that the majority of CAST runs don't have their temperature log files stored. Due to a bug the fallback temperature location in
TOS/log/was apparently selected.At this point it is possible that the TOS directory on the actual PC that took the data at CAST still has a useful
temp_log.txtfile in its log directory. However, due to COVID-19 restrictions access to that PC isn't possible right now.Alternatively all shift logs have an entry for the InGrid temperature at the beginning of the shift. However, this value was never put into the e-log. That means also here someone would have to access CAST and take a look at them.
See 14.5 for a study on this.
\clearpage
14.4.2. 2018 end of the year (Run 3)
After successfully running through the Run 2 dataset, I attempted to do the same for Run 3. However, I was soon greeted by a Fe pixel spectrum fit failing, because it apparently only had 14 elements in it (resulting in NaNs during calculation, causing the fit to fail).
This was run 305, marked as rtCalibration in
./../data_taking_2017_runlist.html:
| Run # | Type | DataType | Start | End | Length | # trackings | # frames | # FADC Backup? | Backup? | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| 305 | rtCalibration | rfNewTos | 0 days 13:58 | 32655 | 25702 | y* |
To investigate the crash, I inserted a few ggplotnim calls into the code before the Fe index filtering happens to look at the distributions of the following variables:
- posx
- posy
- ecc
- rmstrans
- hits
to get an idea what the run looks like.
In order to make a proper plot for that though, I had to fix
facet_wrap in ggplotnim:
https://github.com/Vindaar/ggplotnim/pull/67
and this run also became a recipe here:
https://github.com/Vindaar/ggplotnim/blob/master/recipes.org#facet-wrap-for-data-of-different-ranges
This resulted in the following facet plot, fig. 145. Compare this to the fig. 146, which is from a real calibration run. The eccentricity and hit datasets especially are very obvious take aways.
After this and a few other minor changes / fixes, it was finally possible to produce the equivalent plots of the variation of pixel positions. See figures 147, 148, 149, 150.
And lastly the plot for the fit of the gas gain vs. charge calibration factors in fig. 151
14.5. Dependency of calibration peak position on temperature
While at CERN I copied over all temperature records from the shift forms, because the log files might be lost for good. I need to check the hard drive both at CERN and at Uni in detail again. Some information might still be found in some archive of the TOS directory for instance.
The data for the temperature is found in both:
- ./../data_taking_2017_runlist.html
- ./../../CastData/ExternCode/TimepixAnalysis/resources/cast_2017_2018_temperatures.csv
Given the temperature data that we have, while only being one data point per day (not even, because the temperature readout was broken for the 2018 part of Run 2), we can normalize the peak position of the Fe calibration data by the absolute temperature.
By comparing this to the raw peak position with both normalized to 1, we can check whether a correlation seems likely.
A script to perform this calculation was written here:
~/CastData/ExternCode/TimepixAnalysis/Tools/mapSeptemTempToFePeak.nim
In it we perform a simple linear interpolation for a temperature for each calibration run based on the two closest temperatures according to their date.
The result is shown in fig. 152 and 153.
PeakPos) of all calibration runs in Run 2 and 3 to the position normalized by the absolute temperature (PeakNorm) calculated from the nearest data points. Behavior between the two cases is very comparable, putting the hypothesis of a strong temperature correlation to the peak position into question.PeakPos) of all calibration runs in Run 2 and 3 to the position normalized by the absolute temperature (PeakNorm) calculated from the nearest data points. Behavior between the two cases is very comparable, putting the hypothesis of a strong temperature correlation to the peak position into question.14.5.1. TODO investigate statistical distributions of comparisons
We should calculate the mean, median and variance of the raw positions and the temperature normalized one. Then we have a better understanding of how large the impact of the temperature normalization is.
14.5.2. TODO in addition investigate temperature variation within CDL data
For the CDL data we do have temperature data. That means there we can in principle go and investigate how the peak position of those peaks (remember we had large variation there as well!) on the temperature.
14.6. Calculate total charge of background data over time
The idea is the assumption that the first effect, even visible in the background data, on the variation of the detector performance (gas gain, possibly other things?) is the total charge that the detector measures in a given time interval during background data taking.
This has one single assumption: The background rate of cosmics is, on average over the chosen time span, constant.
To do this, we're going to write a small script that takes the
DataRuns*_reco.h5 files as input and calculates:
- relatively easy to do. Just read all data. Can't do it conveniently using dataframes though I think. In any case, just walk timestamps, until
ΔT = x min, calc average of those events.
- do same, but applying a filter, e.g.
totalCharge > 1e5or whatever- do same, but don't do any averaging, just sum up
Use pdfunite to combine all resulting plots into one PDF!
A detailed study of this is found in 15.
14.7. Debug software efficiency discrepancy
NOTE: This section started from the evaluation of the systematic uncertainty of the software efficiency in section 24.1.7.4.
Things that were initially forgotten:
- filter to silver region for the data
- apply X-ray cuts
Initially running a simple evaluation of the effective software efficiency from the calibration data (using only an rms transverse cut of <= 1.5 and cuts of energy around 1 keV range around escape & photo peak) yielded the following two things of note:
- for the photo peak we seem to have a software efficiency above 80% by quite a bit, often 87% or so
- for the escape peak we have efficiencies around 40 % (!!!). What the heck is going on here?
Two ideas to debug the escape peak behavior:
- create histogram of likelihood data. Can we see multiple distributions, i.e. likely that input data already has tons of non photons?
- generate fake low energy photons from photo peak photons by taking out pixels randomly
Regarding 1: Comparing the histograms of the escape and photo peaks seems to show that the escape peaks have a lot more entries at higher L values than the escape peak. That could either just indicate that this is what such energies typically look like or it could be indicative of background.
Let's first look at option 2 and then evaluate.
Computation of the software efficiency based on fake data at different energies (see code above), called like:
./calibration_software_eff_syst ~/CastData/data/CalibrationRuns2018_Reco.h5 --fake --energies 0.5 --energies 1.0 --energies 1.5 --energies 2.0 --energies 3.0 --energies 4.0 --energies 4.5 --energies 0.25 --energies 0.75 --energies 0.923
The 1 keV data looks as it should. Between 1 and 4.5 keV it's pretty much too low everywhere. But in particular below 1 keV it drops like a rock. Why?
Comparisons of fake data at different energies vs. the reference
histograms from the CDL data for all the relevant parameters going
into the likelihood method.
It is of note that the distributions look surprisingly
realistic. However, in particular the rmsTransverse distribution
looks a bit off especially at low energies. That could explain why the
software efficiency is too low there maybe.
But then again, if that's the case then we should be able to see a
difference in the combined property distributions that actually go
into the likelihood method!
Or maybe not? What happens if the joint distributions look fine, but the way these 3 are combined is wrong?
[X]We should plot the 3 properties as a scatter plot for each data point
with x, y, color (one property on each) and compare the scatters of the fake data & the real CDF data. Do we see a difference there?
[X]In addition look at the likelihood histograms of real vs. fake data. Only minor annoyance as there's no likelihood dataset in theXrayReferenceFileso we have to load the data from thecalibration-cdlfile and filter it according to the cuts applied for the method (we havebuildLogLHistor whatever it's called for that, no?)[ ]look at distribution of data for CDL when only using Xray cuts vs. Xray + Reference cuts. Do Ref cuts remove elements in the peak of the distribution ? Could explain why eff. is higher for 5.9 keV photopeak in CAST data[X]include silver region cut on the data preselection![X]compute efficiency of CDL raw data after being filtered by
buildLogLHistcuts (i.e. Xray cuts + reference cuts). It should reproduce 80% for the 5.9 keV data. However, what it gives is:Efficiency of logL cut on filtered CDL data (should be 80%!) = 0.7454049775745317
Efficiency of logL cut on filtered CDL data (should be 80%!) = 0.9459327812956648
All of the above points were valid investigations, and partially the reason for the discrepancy, but not remotely the real reason.
First let's look at the scatter plots of the different properties though (they are pretty and still interesting!):



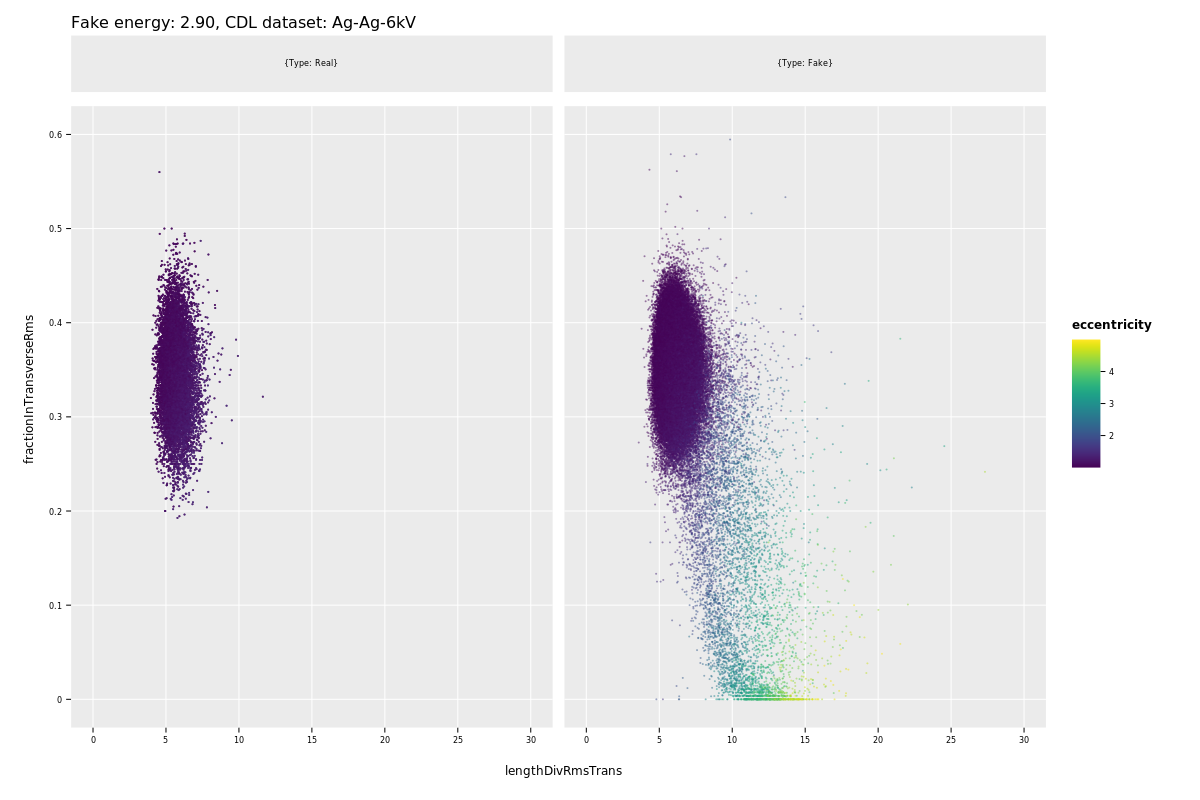

The effect of the CDL cuts on the CDL data is of course extreme! After seeing this the first reaction might be "oh of course the efficiency is so low".

Given that the efficiencies were still at the 40%s, at least at low energy, Klaus and me agreed that it would be a good idea to also use the X-ray cuts to filter out preliminary stuff (the idea being that the software efficiency measures the percentage of real photons passing the logL cut. By applying the cuts we're very unlikely to remove real photons, as they are still loose, but very likely to remove background stuff and other weird events (double photons etc)).
Therefore we included the 'X-ray cuts' for the CAST data as well with the following effect:


With these cuts now the computed efficiencies were as follows:
Dataframe with 2 columns and 32 rows:
Idx Escapepeak Photopeak
dtype: float float
0 0.7814 0.9802
1 0.7854 0.9758
2 0.7388 0.9748
3 0.7954 0.979
4 0.7396 0.9759
5 0.7913 0.9758
6 0.781 0.9715
7 0.7356 0.97
8 0.7709 0.9685
9 0.75 0.9742
10 0.8238 0.9771
11 0.7667 0.976
12 0.7399 0.9718
13 0.7506 0.9774
14 0.7461 0.977
15 0.7644 0.973
16 0.7967 0.9778
17 0.7445 0.9788
18 0.7378 0.9724
19 0.7645 0.9718
20 0.7401 0.972
21 0.7745 0.969
22 0.7417 0.969
23 0.7469 0.9756
24 0.7765 0.977
25 0.7732 0.9713
26 0.7391 0.967
27 0.7309 0.9687
28 0.7102 0.9732
29 0.7553 0.9661
30 0.7387 0.9712
31 0.7639 0.9735
so the numbers were now much more realistic (around 76% for escape) but still way too high for the photo peak.
"What gives?" was the question…
So, I started looking into this and at some point noticed that when
applying the logL cut the numbers seemed to be different than when I
compute the logL cut in my code. This eventually made me realize that
there's a nasty bug in the code that computes the logL cut values. The
histograms from which each cut value was determined were not correctly
assigned the right indices for the table storing the datasets! Thanks
to the values iterator for Table to return data in an arbitrary
order…
So, after fixing this, the efficiencies finally made sense!
One issue that does affect the computation of the software
efficiency outside the code that normally computes the logL cut values
when trying to determine the efficiency later, is that the the logL
cut is determined purely on the set of CDL data. Each target/filter
dataset of course is not a sharp line, but rather somewhat wide,
(ref. figure
), which means that
when computing the effective software efficiency for each CDL line
afterwards using the precomputed logL cut values and each cluster's
energy, some clusters will be placed in a different target/filter
bin. That changes the logL cut required for the cluster, which has a
direct effect on the software efficiency.
14.8. Effective efficiency for Tpx3 data…
(… but also confusion affecting Tpx1)
I extracted the effective efficiency code to compute it for the Tpx3 data in ./../../CastData/ExternCode/TimepixAnalysis/Tools/determineEffectiveEfficiency.nim and extended it to compute means & shift around the data.
This lead to the realization that the efficiencies were first too high. Then I changed the calculation of the logL values so that they are computed on the fly and then the values were too low.
Digging into the logL distributions and the reference data files used before in ./../../CastData/ExternCode/TimepixAnalysis/Tools/compareLogL.nim lead to the following plots:
where Local means the data files that were stored on my laptop as of
and Void for the desktop files.
It is clearly visible that the reference distributions look identical (comparing newly computed - the new default - with the actual files), but the likelihood values are sharply different for the local file.
Even retracing the same computation conceptually leads to a very different result.
At least the VoidRipper data agrees more or less. I have absolutely no idea why the laptop file looks the way it does, but honestly I don't give a fuck.
15. Detector behavior against time
The code to produce the plots in this section is found in
- ./../../CastData/ExternCode/TimepixAnalysis/Plotting/plotTotalChargeOverTime/plotTotalChargeOverTime.nim
- https://github.com/Vindaar/TimepixAnalysis/blob/master/Plotting/plotTotalChargeOverTime/plotTotalChargeOverTime.nim
One of the first things to understand about detector performance is its stability against time. Only if the detector is stable or at least its instability is understood, does it make sense to perform a full analysis.
In order to have a measure that is as simple as possible and is least likely to suffer from artifacts / bugs in the code, we decided to take a look at the raw charge if it is binned against time. The idea being that both for background as well as calibtration data the mean charge collected over a certain - long enough - time window should be constant. Every variation around that value should be purely dependent on detector properties, particularly the gas gain.
Thus I wrote the above code to plot such plots. The code simply walks the sorted timestamps of each cluster and sums each the values of different variables one might want to look at until a window of time is reached. Then it just normalizes by (or calculates) one of 3 things:
- number of clusters in time window
- number of hit pixels in time window
- median of the values seen in the time window.
Below we will discuss the different things we looked at so far and what is visible in each and possible steps to take to solve different issues.
15.1. Energy behavior against time
This section outlines the study of the time dependency of our detector.
It contains multiple related topics and more or less follows the order in which the conclusions were drawn, which might not necessarily be the best way to later understand the final conclusion. For that reason section 15.1.7 gives a brief summary of the learned lessons.
15.1.1. Mean "cluster" charge vs. time
As mentioned above, the mean cluster charge is both the simplest value we can look at as well as the one that should be directly correlated to the gas gain. If taking the mean cluster charge directly the values one sees should in principle be constant in time, however they will also depend on the energy spectrum of the physical processes one looks at. For the background data this is the spectrum of cosmic radiation (or rather their mean ionization energy in our detectors) and for the calibration data it is the spectrum of the source.
In order to better be able to visualize the calibration and background data is one plot instead of normalizing the total charge in a time window by the total number of clusters in the window, we normalize by the total number of hit pixels. This then should in principle be a direct measure for the gas gain. It is essentially what we already do to calculate the gas gain for each run, except with a different time binning.
Fig. 163 shows said distributions, split into the three data taking periods we had at CAST. Namely end of 2017, beginning of 2018 and end of 2018. Keeping all in one plot results in too much wasted space. The time binning window is \(\SI{100}{\minute}\) long in this case. Previously we looked at a window of only \(\SI{10}{\minute}\), but there was too much statistical fluctuation still to see.
The most important take away visible from the figure is though that there is a very strong correlation of the background behavior with the calibration behavior. Each dip or increase is visible in both datasets. However, there is a distinct offset between the two. The calibration does systematically have more charge than the background. This needs to be investigated, see sec. 15.1.1.1.
It is quite evident that a strong time dependence of the gas gain is visible. This is not unexpected, because the same was already seen in the actual gas gain values calculated for each run, but variations on shorter times are visible here.
This variation by itself is not necessarily too problematic. The question is simply: How does a variation in gas gain
- affect the energy calibration?
- affect the stability of the geometric properties used for the log likelihood method?
These two things are partially studied in the following sections.
15.1.2. Median cluster energy vs. time
The first question is whether the variation seen in gas gain above has an impact on the energy calibration. Given that the energy calibration is done in a pretty complicated matter with respect to the time scales involved, it is expected that variations in gas gain will have an effect on the resulting energy calibration.
See sec. 14.4 for some of the plots that show the gas gain vs. energy calibration factor plots and the corresponding fit and sec. 21 for a detailed explanation on the procedure (TODO!).
Fig. 164 shows the median cluster energy of all clusters within \(\SI{100}{\minute}\) bins. A similar variation in energy is visible to the charge per pixel variation as above, even if maybe not quite as extensive.
However, given that the energy is a scalar property it is in principle possible to correct the variation perfectly and achieve a flat mean / median cluster energy.
Variations in energy are problematic, because they can essentially cause clusters to be moved from one CDL energy bin to another, which have very different distributions and thus different cut values. Also see sec. 22 for a discussion on how to mitigate that. If a cluster is moved to a "wrong" energy bin it follows that the efficiency drops drastically (or a cluster that should have been classified as signal is not anymore).
Possible solutions to approach a more flat energy behavior when binned in time are two fold, sec. 15.1.3 and [BROKEN LINK: WONTFIX 2. Change energy calib to use closest two calibration runs].
15.1.3. 1. Bin background by time for more gas gain values
Each run is long enough to suffer from the time variation in the
charge as seen in from the last meeting.
This means that the gas gain varies too much to assign a single
value for the gas gain for a whole run, resulting in a definite
problem for the variation.
Possible solution: change the way the gas gain is calculated in
the reconstruction. Instead of calculating the polya for each
run, bin it also by time (have to look at different binning times
to find the shortest possible time which still gives us good
enough statistics!) and then calibrate each of these intervals
individually based on the energy calibtration function.
Current approach:
- DONE fix the remaining bug of accessing indices for charge / timestamps
- write each polya fit to a dataset that comprises of interval length and index + the start and end times as attributes
- DONE take a very long background run, raw data manip + reconstruct it
- run gas gain calculation on it with:
- TODO varying interval times
- DONE write out each gas gain w/ timestamp to a file
- DONE compare plots / polya fit datasets once merged
- TODO can create overlapping histos / ridgelines of different histograms
- TODO find optimal value for interval that is short enough for enough statistics
- DONE run on all background data and reproduce plots from last week
As it stands most of the above things have been implemented now. However, the finer details regarding:
- a comparison of different interval times
- optimizing for the best time interval regarding enough statistics
have not been done. In addition the resulting plot in fig. 165 brings up a few more questions.
The figure shows a few interesting things.
First, the variation of the background data is mostly unchanged along its "gaussian" behavior, but the larger variation across time has become a lot smaller (compare directly with fig. 164).
Second, the mean value of the median cluster energies is slightly lower now. Before it frequently fluctuated above \(\SI{2}{\kilo\electronvolt}\), while it is almost exclusively at and below \(\SI{2}{\kilo\electronvolt}\) now.
Third, the subplot on the RHS corresponding to Run 3 (end of 2018) features slightly higher median energy values. The reason for this is that the energy calibration factors used (the ones derived from the "gas gain vs. energy calbration factors" fit) were the ones used for Run 2 (2017 / beginning 2018), which leads to a slight offset in energy (too high), but should not lead to a change in the fluctuation. To avoid a similar issue in the future the information about each run period was now added to the InGrid database.
Fourth, the variation of the calibration data is almost completely unchanged, see section [BROKEN LINK: STARTED Understand variation in calibration data] below.
- DONE Compute time binned median cluster energy with \(\SI{30}{\minute}\) bins
To make sure we're not seeing some weird Moiré like pattern due to binning the gas gain by 30 minutes and the plot by 100 minutes, we should create the plot with 30 minute binning as well.
Doing this results in the following fig. 166
Figure 166: Behavior of median cluster energy in \(\SI{30}{\minute}\) bins against time, filtered to only include non noisy events. Gas gain was binned in \(\SI{30}{\minute}\) intervals. End 2018 energies were calculated based on 2017 gas gain vs. energy calibration factor fits though (accidentally). Also the gas gain fit is still unchanged, because so far we only have one fit per calibration run. As can be seen now the variation of cluster energies for the background data increase a bit. This might just be due to the variations in background rate showing up in the data now? The more important thing is the varition towards low values that can still be seen. However, the good thing is that it does not seem like the number of "bad" points has increased by a factor of 3, which might mean that the outliers towards low values do have some reason to be understood by looking at the individual gas gain bins, which are binned by 30 minute intervals in the shown data.
NOTE: It is important to note that there is no guarantee that the exact same 30 minute binning intervals will be chosen for the plot here compared to the binning of the gas gain! The gas gain performs certain cuts on the input data, which moves the start and stops of the 30 minute intervals slightly.
- Find out where outliers in background come from
The code used in this slide is:
- ./../../CastData/ExternCode/TimepixAnalysis/Plotting/plotGasGainIntervals/plotGasGainIntervals.nim
- plotGasGainIntervals on Github
To understand where the outliers in the background come from, we will look at the computed gas gain values used for the computation of the energy for plot fig. 166.
Fig. 167 shows the gas gain behavior of the background data of the 30 minute gas gain slices.
The gas gain is shown as used for the energy calibration ("Gain" = mean of the data), fit result (GainFit) and mean of the distribution described by the fit. The values shown for the fits do not include the dynamic fit range of the polya fit from sec. 15.4 yet!
As can be seen there are still many outliers. The outliers that are worse than \(\SI{80}{\percent}\) of the maximum gas gain value observed are written to a CSV file. From the current computation they are shown in tab. 22.
Table 22: All gas gain slices that are higher than \(\SI{80}{\percent}\) of the maximum gas gain observed. CalcType GasGain Run timestamp Date RunPeriod SliceIdx Gain 5890 79 1509769378 2017-11-04T05:22:58+01:00 30/10/2017 17 Gain 5884 92 1510816897 2017-11-16T08:21:37+01:00 30/10/2017 21 Gain 5801 92 1510920516 2017-11-17T13:08:36+01:00 30/10/2017 78 Gain 5988 97 1511511714 2017-11-24T09:21:54+01:00 30/10/2017 31 Gain 6474 109 1512422366 2017-12-04T22:19:26+01:00 30/10/2017 9 Gain 5901 112 1512639062 2017-12-07T10:31:02+01:00 30/10/2017 39 Gain 5792 112 1512695549 2017-12-08T02:12:29+01:00 30/10/2017 70 Gain 6250 112 1512855077 2017-12-09T22:31:17+01:00 30/10/2017 158 Gain 5741 113 1512908433 2017-12-10T13:20:33+01:00 30/10/2017 13 Gain 6002 115 1513081031 2017-12-12T13:17:11+01:00 30/10/2017 37 Gain 6600 121 1513331076 2017-12-15T10:44:36+01:00 30/10/2017 27 Gain 7125 127 1513738968 2017-12-20T04:02:48+01:00 30/10/2017 17 Gain 6117 164 1520306085 2018-03-06T04:14:45+01:00 17/02/2018 58 Figure 167: Behavior of gas gain in \(\SI{30}{\minute}\) bins against time. The gas gain is shown as used for the energy calibration ("Gain" = mean of the data), fit result (GainFit) and mean of the distribution described by the fit. The values shown for the fits do not include the dynamic fit range of the polya fit from sec. 15.4 yet! Taking the slice from run 164 listed and looking at the plot we see fig. 168 in comparison to the slices before fig. 169 and after 170.
Figure 168: Slice 58 of run 164 of chip 3 where the gas gain is estimated way too high. Left fit range is determined automatically, but right side is not. There is a large background (noise?) visible in the data, which leads to a deviation to higher values for the mean of the data. The fit seems more reasonable, but suffers from the same issue that it takes too much of the right flank into account. The slices before and after look fine, see fig. 169 and fig. 170. See fig. 186 for a follow up after this investigation showing the same slice. Figure 169: Slice 57 of run 164 of chip 3. The slice before the one shown in fig. 168, which is estimated too high. This slied looks normal. Figure 170: Slice 59 of run 164 of chip 3. The slice after the one shown in fig. 168, which is estimated too high. This slied looks normal again. - DONE Occupancy of the bad slice
I've hacked in a way to plot occupancies (both in charge values and raw counts) into the gas gain interval plotting code used for the above section.
A comparison of the time slices 57 and 58 from above are shown in fig. 171, 172 for counts and fig. 173 and 174 for the charge.
Structure is visible in the charge occupancy for the slice 58, which is not understood. Need to take a look at the individual clusters found in the time slice.
There are two main features visible for the slice 58.
- On the raw counts occupancy in fig. 172 we can see a slight mesh signature in the upper half of the chip.
- There is a large cluster like signature visible on the left side of the charge occupancy fig. 174. It seems unlikely to be the result of events which are full frames, since that would result in a shape that covers the full width of the chip.
Figure 171: Occupancy (in raw counts) of slice 57 of run 164 of chip 3. This is the non noisy slice from fig. 169. There is one noisy pixel, but otherwise it looks relatively homogeneous. Figure 172: Occupancy (in raw counts) of slice 58 of run 164 of chip 3. This is the noisy slice from fig. 168 There is some weird grid like behavior visible on most of the chip. The noisy pixel stands out the most still. Figure 173: Occupancy (in charge) of slice 57 of run 164 of chip 3. This is the non noisy slice from fig. 169. The noisy pixel is still visible, but a region near the edge of the chip is more visible, possibly a region with small sparks. Figure 174: Occupancy (in charge) of slice 58 of run 164 of chip 3. This is the noisy slice from fig. 168 In charge there is clear structure visible now. The orientation is unclear but the large cluster on the left side does not seem to resemble events with 4095 pixels. - DONE Look at individual events in bad slice 58
From looking at the occupancies of the noisy slice 58 I hacked some more to output event displays as scatter plots from the noisy slice as well as the one before.
A few things are immediately recognizable:
- most events look fine
- there is an explanation for the mesh like signature discussed in the last section
- the cluster visible on the left side in the charge occupancy is the result of a single event.
There are 3 or 4 events which are noisy (full frames, hits:
>=4095), which however are not active pixel to pixel, but rather have some gaps. These gaps produce the mesh like signature. It is to note that such events then always have the same charge values for all pixels. At the same time these events would possibly not be filtered out from thermsTransversecut. But this needs to be verified, see section below.One of those events is event 45 of the slice, shown in fig. 175.
Figure 175: One of the events which explans the mesh like signature visible on the raw counts occupancy of slice 58 in fig. 172. Together with 3 or 4 other events like it the mesh signature makes sense. The event producing the large cluster is event 15, shown in fig. 176. It seems excessively large even for an \(\alpha\) particle. A typical \(\alpha\) in my opinion should rather look like event 156 in fig. 177.
Also the event is already at the \(\num{4096}\) pixel threshold.
Figure 176: The event which is the reason for the cluster structure found on the left hand side of the charge occupancy in fig. 174. The ionization is so high that most pixel in the center have charge values fitting more to 2 or even more electrons in origin. Also the event has most likely some lost pixels, since it's already at the \(\num{4096}\) pixel threshold (even if that is not visible). For an \(\alpha\) it seems like too much ionization, compare with fig. 177 for a possible \(\alpha\) event. Figure 177: A possibly (in my opinion) more typical \(\alpha\) event. It shows reasonable ionization in terms of density and thus charge per pixel of \(\mathcal{O}(\text{gas gain})\). The event 15 raises the question what is visible on the LHS. Jochen was pretty certain in our group meeting on that the event looks like some ionizing particle and not like a spark. By that assumption the event should be extended towards the chip on the left.
Note: the actual event number is
44674in run number 164.The full septem event is shown in fig. 178.
With the combination of the results from this section and the one below about run 109 it is pretty evident that the low energy values / high gas gain values can be attributed to bad clusters being used for the gas gain calculation. This is why we added a cut to the number of hits per cluster to be used for the gas gain computation in:
- https://github.com/Vindaar/TimepixAnalysis/commit/5dc8e50a1d734820f732ace2ae9ee37f9fc0268e
- https://github.com/Vindaar/TimepixAnalysis/blob/master/Analysis/ingrid/calibration.nim#L430
Figure 178: The event which is the reason for the cluster structure found on the left hand side of the charge occupancy in fig. 174 seen as a full septemboard event. It is well visible that it is indeed just an extremely highly ionizing track. - DONE Run 109, Slice 9 - Look at another noisy slice
Next we're going to look at one of the other noisy slices, shown in tab. 22. We pick the one with the highest gas gain in the table:
Gain 6474 109 21512422366 2017-12-04T22:19:26+01:00 30/10/2017 9 Using ./../../CastData/ExternCode/TimepixAnalysis/Plotting/plotGasGainIntervals/plotGasGainIntervals.nim in combination with a slight hack to only plot slice 9 from run 109 and using a lot of fixes to the
ggplot_utilsas well asggplotnimitself.Fig. 179 shows the charge occupancy of the time slice. It's very visible that it shows the same kind of behavior as slice 58 of run 164, namely a mesh like structure (only visible on count occupancy) and some very large events in charge.
The events most visible is the 2 track structure and a blob in the bottom left. It turns out that the two tracks are actually from two different events about 5 minutes apart! Fig. 180 and 181 show these two events. Fig. 182 shows the blob in the bottom left.
Figure 179: Occupancy (in charge) of slice 9 of run 109 of chip 3. This is another noisy slice from the median energy cluster In charge there is clear structure visible now. The orientation is unclear but the large cluster on the left side does not seem to resemble events with 4095 pixels. Figure 180: Event 7106of run 109, slice 9. The first of two events, which show up as a 2 track "event" together on the occupancy in fig. 179.Figure 181: Event 7191of run 109, slice 9. The second of two events, which show up as a 2 track "event" together on the occupancy in fig. 179.Figure 182: Event 7095of run 109, slice 9. An event, which shows up as the spherical blob in the bottom left on the occupancy in fig. 179.TODO include polya?
- DONE verify if noisy events pass gas gain cuts!
In the code used above
plotGasGainIntervals, which creates the occupancies for the gas gain slices, no filtering similar to the gas gain cuts used for the gas gain as well as in the binned vs time plotting script are performed.Check whether applying those cuts, as done in
plotTotalChargeVsTime, removes the mesh like signature. And make sure the big cluster mentioned above stays (if it doesn't we still don't understand why the slice is noisy!).NOTE: Short check using hacky way for run 109, slice 9. The gas gain cuts as expected do not remove the two tracks, cause their centers are too far in the center and their rmsTransverse is "healthy". Need cut on number of hits for sure. The spherical event in the bottom left is removed indeed. In fact only about 90 of ~200 events survive the cut.
UPDATE: Indeed, including the cut on hits of 500 pixels per cluster for the gas gain data, gets rid of the broken behavior. In combination with the above (90 / 200) events remaining after filtering in
plotGasGainIntervals.nimmeans that indeed many noisy cuts did pass the cuts. - DONE Make the right hand side of the fit dynamic, understand why data is bad
We should make the RHS of the polya fit dynamic as well to see whether this fixes the issue seen in the gas gain (and thus for the energy calibrations).
If we make the fit range dynamic on the RHS we have to make sure that the mean of the data is only computed in that range as well!
Finally also we should understand why we have such behavior. What to the events look like there?
This is solved and described in sections 15.1.3.2.3 and 15.1.3.2.2 (the previous sections). Essentially the problem was there were clusters with way too much charge per pixel (extremely ionizing tracks), which skewed the gas gain data. A cut on the number of hits per cluster was implemented in https://github.com/Vindaar/TimepixAnalysis/commit/5dc8e50a1d734820f732ace2ae9ee37f9fc0268e.
- DONE Reevaluate gas gain & energy behavior against time after 500 hits cut
After having introduced a cut on the cluster size of 500 pixels in:
we need to reevaluate whether this actually got rid of our original problem, namely the fact that we saw time slices with very large gas gains in fig. 167.
The corresponding figure with the included cut is shown in fig. 183, which was created after:
- making sure the
chargedataset was removed, because it countained all the attributes, which slowed down running over the data. Added aconfig.tomloption in reconstruction to delete thechargedatasets when running--only_charge - recomputed gas gains after charge was readded
- plotted using
plotGasGainIntervalafter fixing the code for new gas gain intervals in dataset
Note: this means the file was not reconstructed from scratch from the raw datafile.
For the distribution of the gas gain values against time (for the 3 different computational methods) one can easily see now (cf. fig. 167) that the data is much more stable in time. The variations of the mean of the data (points in red) in particular do not deviate to extremely large values anymore.
The difference between mean of data / fit results is larger again for the end of 2018 data than the other two intervals. Because of this, I looked at two different polya distributions of these gain slices to get an idea how the difference can be explained. This comparison is shown in fig. 184 and 185. The former shows a polya from run period 2 where the values match much closer (~100) than the latter from run period 3 where we observe a difference of ~500.
Looking at the distributions in detail we can see the effect of the threshold. The distribution from run period 2 in fig. 184 has a much more prominent cutoff on the left side (the threshold is clearly visible), whereas the latter plot shows a much more gradual decay. Having more data on the left hand side of the distribution means the mean of the gas gain is going to be shifted to lower values in comparison, whereas the fit does not include that range (see the stroked line in the fit).
Aside from that there are still some outliers, which need to be investigated, possibly the cuts have to be taken a bit more stringent even?
Figure 183: Distribution of the gas gain values against time (for the 3 different computational methods) after the cut of a minimum of 500 pixels in each cluster for the input to each gas gain slice was implemented. As one can easily see (cf. fig. 167) that the data is much more stable in time. The variations of the mean of the data (points in red) in particular do not deviate to extremely large values anymore. The difference between mean of data / fit results is larger again for the end of 2018 data than the other two intervals. Aside from that there are still some outliers, which need to be investigated, possibly the cuts have to be taken a bit more stringent even. Figure 184: A polya distribution of run 186 (run period 2, slice 10 of chip 3), which shows a fit where the gas gain determined from the mean of the data is only about ~100 lower than the fit parameter. This is in comparison to fig. 185 of run period 3 where the data determined value is ~500 lower. See the text for a discussion. Figure 185: A polya distribution of run 283 (run period 3, slice 10 of chip 3), which shows a fit where the gas gain determined from the mean of the data is ~500 higher than the fit parameter. This is in comparison to fig. 184 from run period 2, in which the difference is much lower on average. See the text for a discussion. In addition it is interesting to look at the same polya distribution of run 164 as shown in fig. 168, which started the investigation into the individual events etc. It is shown in fig. 186. Indeed the distribution now looks as expected!
Figure 186: Reevaluation of slice 58 of run 164 of chip 3 where the gas gain previously way too high. Having removed the large "noise" (which was due to highly ionising events + some real noise) by cutting on cluster sizes < 500 hits, the distribution now looks as expected. See fig. 168 for what it looked like before. Finally, we need to recompute the energy of the clusters an find out if the situation has improved there over fig. 166 where we saw the massive number of extreme outliers to very low energies in the background data.
- Gas gain vs. energy calibration factors of 2017/18
Fig. 187 shows the new gas gain vs. energy calbration factors using the 500 pixel cut per cluster data and binned gas gain computation. The gas gain used for the fit is the mean of all gas gain slices (30 min)!
It is visible that the agreement of data and fit is much better now than it was in fig. 139. Take note that the y values remain unchanged from the previous values, but the gas gain values change. Thus, some points "switch" places.
Figure 187: Gas gain vs. energ calibration fit with the cut of 500 pixels per cluster required. This uses the mean of all gas gain slices (30 min) in each run. Compared with the previous fit in fig. 139 the data is much more stable resulting in a better fit. Note that the y values (calibration factors) are the same in both plots (they only depend on the Fe fit), but the gas gain values (x) are different, resulting in some points "switching places". For instance the 2nd data point in the old plot is now the 1st in this plot and fits much better now. Sanity check:
To make sure that running the energy calibration on the existing file actually works as expected now (the dataset is overwritten in
calibration.nim, so that's not a real worry in principle), we will do a quick sanity check of comparing the energy distributions for background and calibration (without any cuts) before and after re-calculating the energy based on the above new fits.These plots were created using
karaPlot, with \(\num{500}\) bins between \(\SIrange{0}{15}{\keV}\).TODO: The same needs to be done for when we switch over to
- Fe spectrum based energy calibration factors for the above plot based on the same time slices as used for the gas gain
- the energy computed by taking a linear interpolation between the closest two calibration runs for each cluster
Energy histograms for calibration data before new gas gain based energy computation in fig. 188 and for the background data in fig. 189.
Figure 188: Histogram of energies computed from the charge for the calibration data in run 2 (2017/18) to be used as a comparison with the energies after they are computed from the new gas gain vs. energy calibration factors, based on the mean of the gas gains of all time slices found in each calibration run. The values in the plot are still computed with the gas gain values using all clusters in silver region and 0.1 < rmsTransverse < 1.5.Figure 189: Histogram of energies computed from the charge for the background data in run 3 (end of 2018) to be used as a comparison with the energies after they are computed from the new gas gain vs. energy calibration factors, based on the mean of the gas gains of all time slices found in each calibration run. The values in the plot are still computed with the gas gain values using all clusters in silver region and 0.1 < rmsTransverse < 1.5. - Gas gain vs. energy calibration factors of Run 3 (20182)
This is the same comparison as in the previous section, but this time for the Run 3 (end of 2018) data.
Fig. 190 shows the new gas gain vs. energy calbration factors using the 500 pixel cut per cluster data and binned gas gain computation. The gas gain used for the fit is the mean of all gas gain slices (30 min)! Again the data and fit match much better now. The error bars now wouldn't even need to be increased by a factor of 100 anymore (meaning that one source of systematic errors has been removed!).
Figure 190: Gas gain vs. energ calibration fit with the cut of 500 pixels per cluster required. This uses the mean of all gas gain slices (30 min) in each run. Compared with the previous fit in fig. 151 the data is much more stable resulting in a better fit. Note that the y values (calibration factors) are the same in both plots (they only depend on the Fe fit), but the gas gain values (x) are different, resulting in some points "switching places". Sanity check:
See the previous section for the meaning of sanity check in this context.
Energy histograms for calibration data before new gas gain based energy computation in fig. 191 and in fig. 192 for the background.
Figure 191: Histogram of energies computed from the charge for the calibration data in run 3 (end of 2018) to be used as a comparison with the energies after they are computed from the new gas gain vs. energy calibration factors, based on the mean of the gas gains of all time slices found in each calibration run. The values in the plot are still computed with the gas gain values using all clusters in silver region and 0.1 < rmsTransverse < 1.5.Figure 192: Histogram of energies computed from the charge for the background data in run 3 (end of 2018) to be used as a comparison with the energies after they are computed from the new gas gain vs. energy calibration factors, based on the mean of the gas gains of all time slices found in each calibration run. The values in the plot are still computed with the gas gain values using all clusters in silver region and 0.1 < rmsTransverse < 1.5. - Should the mean of the data for gas gain use data range as fit?
This is a question that I asked myself looking at the plots in the previous section. Given that we see effects of the threshold having such a strong impact, why is it a good idea to use the full range of the data for the mean? Shouldn't one have a "real" distribution without threshold effects to get the gas gain from the mean of the data?
While cutting away more of the data on the left hand side moves the mean of the distribution to the right and thus increases the gas gain determined. However, in principle the absolute value of the gas gain does not matter. All that matters is that it's "stable" over time! Or rather which when changing changes proportionally to the derived energy calibration factor.
UPDATE: In the last meeting we put this question to rest. The interpretation for the behavior seen in the gas gain values based on the raw data vs. the fit parameters is most likely indeed this one. But as established, the absolute gas gain values do not matter actually. The goal is just to calibrate out variations in gas gain such that each time slice can be calibrated correctly, which does work as seen in the previous section (especially compare the gas gain vs. energy calibration plots with single values for each calibration run and those with individual fits; the fit is essentially the same).
- TODO Investigate the remaining outliers?
We should look at the remaining outliers to find out if it's still related to individual events behaving badly.
- DONE calculate time binned median cluster plot w/ position cuts
Finally it is a good idea to compute the same plot as fig. 165, but with cuts on the position. So instead of taking every cluster into account, we should only use those which are close enough to the center, e.g. in the gold region. That should in principle eliminate some possible geometric causes.
Fig. 193 shows the behavior of the cluster energy agains time if only clusters are considered, which are in the silver region and conform to
0.1 < rmsTransverse < 1.5. That is the same cut as applied to the gas gain computation itself.As can be seen the behavior is almost exactly the same as for the uncut data, with the exception that the values are higher. The latter makes sense, because some clusters were cut off on the sides of the chip, which leads to a smaller median energy values.
Figure 193: Behavior of median cluster energy in \(\SI{30}{\minute}\) bins against time, filtered to only include non noisy events only including events in the silver region and cut to 0.1 < rmsTransverse < 1.5(the same cuts used for the calculation of the gas gain itself). Gas gain was binned in \(\SI{30}{\minute}\) intervals. End 2018 energies were calculated based on 2017 gas gain vs. energy calibration factor fits though (accidentally).
- making sure the
- DONE Optimize for best interval times (enough stats) + compare polyas of different times
Comparison of different polyas depending on energy might be interesting.
In principle could have some simple script that performs the current analysis on a single run with different parameters. Just replace values in the toml file, write the resulting dataset for the first (or whatever) poyla to a CSV file and finally create a plot of the different distributions. UPDATE: : This is somewhat what we ended up doing at the end of Dec 2020.
The code we use is ./../../CastData/ExternCode/TimepixAnalysis/Tools/optimizeGasGainSliceTime.nim, which performs the steps outlined in the last paragraph (but only for the background datasets the new gas gains are computed).
We computed gas gains for the following intervals (in minutes):
const timeIntervals = [45, 60, 90, 120, 180, 300]
Of course this is not comparable to a proper optimization, but more of a high-level determination of a suitable range!
The CSV files were written to ./../../CastData/ExternCode/TimepixAnalysis/Tools/out/ but are now available in: ./../../CastData/ExternCode/TimepixAnalysis/resources/OptimalGasGainIntervalStudy/ TODO: add github link.
The source file contains some documentation on what the "optimization" script does. The major part is the following:
On a practical side:
- We have to read the `config.toml` file each time and replace it by the version
containing the interval we wish to compute.
- Then run the reconstruction with `–onlygasgain`, which creates a dataset
`gasGainSlices<IntervalLength>`.
- Recompute the energy using `–onlyenergyfrome`, which overrides the existing
energies. This means we have to start with computing the median values and write to CSV for the existing 30 min stored in the files first.
- Compute the median cluster energies according to `plotTotalChargeVsTime`.
- Store DF as CSV.
NOTE: All plots shown in the rest of this section show the energy with a factor \(\num{1e6}\) missing!
In order to get an idea of what the time dependency looks like, we computed histograms of all median cluster values computed in each run period for each of the computed gas gain interval times. The idea is that the optimal lenght should be that, which is still gaussian and has the smallest sigma.
A ridgeline plot of each histogram is shown in figs. 194, 195 and 196.
From these already, in particular the first run period, a likely candidate for an optimal length is \(\SI{90}{\minute}\), because those distributions still look mostly gaussian, but are less wide than the shorter time scales. Beyond that the distributions look less gaussian and do not get much smaller.
Figure 194: Comparison of all computed gas gain slice lengths in a single ridgeline plot for the first run period at the end of 2017. Each ridge is the histogram of all median cluster energies in that period binned by the specified time in min. In this run period it is visible that a decent time length is about \(\SI{90}{\minute}\) as for shorter lengths the distribution just becomes wider and for longer times it becomes first and foremost less gaussian. Figure 195: Comparison of all computed gas gain slice lengths in a single ridgeline plot for the first run period at the end of 2018. Each ridge is the histogram of all median cluster energies in that period binned by the specified time in min. Figure 196: Comparison of all computed gas gain slice lengths in a single ridgeline plot for the secon drun period at the end of 2018. Each ridge is the histogram of all median cluster energies in that period binned by the specified time in min. Figure 197: Comparison of all computed gas gain slice lengths in a facet plots with all plots for each run period in a single plot. Essentially all plots fig. 194, 195 and 196 in one. NOTE: The plots showing the explicit time dependency show multiple data points at one time, due to a simplified calculation!
The plots for all data points against time can be found in ./../Figs/statusAndProgress/binned_vs_time/optimize_gas_gain_length/. The equivalent plots for 30 minutes are shown elsewhere in this section. Here we only compare the plot for 90 and 120 minutes to highlight the choice of 90 minutes.
Fig. 198 shows the behavior against time for the \(\SI{90}{\minute}\) calculation. Aside from maybe the first couple of bins in the 2017 window (top left) there is no significant time dependence visible. In contrast fig. 199 shows the same plot for the \(\SI{120}{\minute}\) interval length. Here we start to see some time dependent behavior. This is the major reason to use 90 minutes for the remaining data analysis.
Figure 198: Calculation of all median cluster energies based on gas gains binned by \(\SI{90}{\minute}\). There is essentially no time dependent behavior visible yet. Figure 199: Calculation of all median cluster energies based on gas gains binned by \(\SI{120}{\minute}\). Here slight time dependent behavior is visible. - STARTED Understand variation in calibration data
To understand why the median energy variation is unchanged between figures 164 and 165 it is first important to note how the energy is calculated for the calibration data.
One might assume that each calibration run is computed by its own fit to the \(^{55}\text{Fe}\) spectrum. But that is not what is done.
Energy calibration of calibration runs is done exactly in the same way as for the background runs. That means it is a 2 phase process.
- The charge and gas gain of each calibration run is calculated.
- The fit to the charge spectrum of the \(^{55}\text{Fe}\) run is performed, determining the position of the photopeak and escape peak.
- The linear fit to the two positions in charge space is performed, which maps a number of charge values to an energy value. The slope is then a conversion factor between a charge and an energy.
- This is done for each calibration run.
- A scatter plot of each calibration run's gas gain vs the aforementioned energy calibration factor is done. A linear fit is performed to finally have a linear function mapping a gas gain to a conversion factor to be used to map charge values to energy values.
- A second pass over the calibration data is performed. The fit function from 5 is applied to every cluster's total charge found in the calibration run.
The fit from point 5 is the one shown e.g. in fig. 151.
So to an extent the variation in the calibration data is expected is it reflects the calibration runs which lie further away from the actual fit of the fit from point 5 above.
On the other hand: The median cluster energy of the calibration runs is a somewhat special value. That is because in an \(^{55}\text{Fe}\) spectrum there are essentially only 2 different kinds of clusters. Namely the photopeak and the escape peak. That means the median value of these will strongly depend on the the shape of the spectrum and in particular:
- how much background does the spectrum have
- what is the ratio between photo and escape peak?
In principle it is possible to have small geometric effects that change the amount of escape photons visible. If the emission of the source is slightly more to the side of the detector there is a larger chance for an escape photon to actually escape, thus increasing the escape peak. The effect this can have in the CAST setup (source is \(\sim\SI{0.5}{\meter}\) away from the detector and the window is pretty small) is questionable, but should be inveistigated.
- DONE look at occupancy of calibration runs
This is interesting to get a feeling for how large an effect the geometric things mentioned in the above paragraph might play.
As can be seen both in fig. 200 (for the full run period 2) and fig. 201 (for only a typical \(\sim\SI{3}{\hour}\) calibration run) the occupancy is very flat, with the exception of the window strongback being somewhat visible.
A cut to the silver region is probably advisable for the time dependent energy plot to remove the drop off towards the edges.
Figure 200: Occupancy of the raw pixel events without any full events of of all runs in the run period 2 (2017 / beginning 2018). It's visible that most of the chip is lit up pretty well Figure 201: Occupancy of the raw pixel events without any full events of of a single run in run period 2 (run 126 - it is a typical run at \(\sim\SI{3}{\hour}\).) The data is clamped to the \(95^{\text{th}}\) quantile. - STARTED calculate ratio of escape peak and photo peak amplitudes
Creating a scatter plot of these ratios / a histogram / a timeseries might shed some light on whether the variation can be an intrinsic to the individual calibration runs and not the energy calibration itself. Even if that would not explain which effect plays a major role (be it geometric or something else)
Fig. 202 shows the behavior looking at the ratio of photo and escape peak via their position, i.e. the mean charge of the peak.
Fig. 203 looks at the ratio of the photo and escape peak via their amplitude.
Figure 202: Comparison of the ratio of the photo peak divided by the escape peak position in charge. Figure 203: Comparison of the ratio of the photo peak amplitude divided by the escape peak amplitude in charge. There is a strong change in the variation of the peak ratios, up to 25%! And one might imagine to see an inverse relationship between the median cluster energy of the binned calibration data.
15.1.4. WONTFIX 2. Change energy calib to use closest two calibration runs
UPDATE: This has been deemed unnecessary, as the main reason we wanted to do this was to remove systematic detector behaviors if there were such during the data taking periods at CAST. But as we saw in section 15.1.3.3 the detector properties might change in time systematically, but not in such a way that there is no linear dependence of the gas gain and the resulting fit parameters for the energy calibration of each calibration run. With that this is going on ice for the time being.
Change the energy calibtration to not use all runs and perform the "gas gain vs energy calibration slope fit". Instead only look at the weighted mean of the energy calibrations of the two closest calibtration runs, i.e. linear interpolation. Then the gas gain won't be needed at all anymore.
15.1.5. DONE 3. Change gas gain vs. energy calib fit to perform 55Fe fits for each gas gain slice
So that we use the same binning for the fitting of the "gas gain vs. energy calibration factors" function, we should read the gas gain slice information and then perform one Fe spectrum fit for each of these time slices.
UPDATE:
Two ways have been implemented (an additional one has been put on
ice, namely the above) based on a new enum:
GasGainVsChargeCalibKind, which is set using the config.toml field,
excerpt from the current TOML file:
[Calibration] # the gas gain vs energy calibration factor computation to use # - "": the default of just one gas gain per calibration run based on # one gas gain interval for each run # - "Mean": use the mean of all gas gain time slices # - "Individual": use the factors obtained from one Fe fit per gas gain slice gasGainEnergyKind = "Individual"
which is mapped to the following type:
type GasGainVsChargeCalibKind* = enum gcNone = "" gcMean = "Mean" gcIndividualFits = "Individual"
gcNone: is a backward compatible to compute things, namely for the case of having reconstructed and computed the gas gains for a file before gas gain slicing was introduced; a fallback modegcMean: refers to computing the mean gas gain value of all gas gain time slices found in a calibration run and using mapping that mean value to the energy calibrationgcIndividualFits: refers to the actual implementation of the topic of the section, namely fitting individual Fe spectra to each time slice, such that individual gas gain values are mapped to individual calibration factors. If the data in calibration runs suffers from actual time dependent behavior, this should yield a better fit function, which maps better over a wide range of gas gain values.
We will now compare the results from gcMean and
gcIndividualFits. Note that the results from gcMean were already
discussed in section 15.1.3.3 to an extent. In particular in terms of the
comparison of gcMean with the old computation. The main difference
between those is that gcMean includes the fixes to the computation
of the gas gain (filtering of large clusters and region filter).
First in fig. 204 we see the result of
gcMean. The fit includes error bars, which have been enlarged by a
factor of 100. The errors represent numerical errors on the linear fit
to the peak positions of each Fe spectrum. The problem is that the
numerical error is very small. It mostly depends on the uncertainty
of the Fe fit. The variation of the detector is larger than the actual
numerical errors. However, by enlarging the errors instead of taking a
large error for all data points we keep the information of the
likely good fits to the Fe spectra. If a fit to the Fe spectra is
relatively bad, this should lead to a larger error on the linear
fit. We wish to assing a lower weight to such points.
This first fit describes the data very well already.
In comparison the second fig. 205
shows the same for individual Fe spectrum fits to each time slice in
which a gas gain is computed. Note that here the error bars are not
increased by a factor of 100. The calibration values (y axis) however
are wrong by a factor of \(\num{1e6}\).
Aside from the visual difference due to different errors bars and
significantly more data points the fit is very similar to the gcMean
case. This is reassuring, because it implies that possible variations
during the calibration runs are possibly more statistical than
systematic in nature.
gcMean).gcIndividual). The calibration factors are wrong by a factor of \(\num{1e6}\). Other than the title suggests, the error bars here are not enlarged by a factor of 100!15.1.6. Replace mean/median etc. computation in plotTotalChargeOverTime.nim
To simplify the code and to more easily add additional computations for the
15.2, the calculateMeanDf implementation in
plotTotalChargeOverTime was changed.
While implementing this a few issues (presumably in the new implementation) were uncovered.
Instead of walking over the timestamps of the data for each run and binning manually by N minutes, we use the existing gas gain slices. This has the advantage that we don't have to perform a somewhat complicated computation and compute essentially a running mean (and need variables for each variable). Instead we can work on the full subset of each gas gain slice.
Note that when using the gas gain slices, one needs to take care to apply the slice start / stop (which correspond to indices in the individual chip's indices) not to the indices of the full dataset, but rather of the reduced dataset by the gas gain cuts.
In addition there is an issue with those gas gain slices, which are at the end of a run. Those are shorter than the N minutes we want. Some end up significantly shorter then. These have too little statistics and thus result in a worse energy calculation! It would be important to probably include these into the second to last slice (unless they are ~80% or so of a slice interval length). Those short slices are included in fig. 207, which explains the outliers seen (compare with fig. 208 in which those slices are simply excluded). (UPDATE: fixed, plots in this section do not use that fix!).
TODO: include plot including short slices
The old computation in fig. 206 yields different results for that reason. While we did apply a filtering, the N minute intervals were not aligned with the gas gain computations! All in all the results are very compatible, even if the first run period does look maybe a tiny bit more variable in time.
The comparison of the time difference (i.e. the length) of the considered intervals from the "direct mapping" (by walking timestamps and creating an interval after each N seconds) and the usage of the gas gain slices directly is shown in fig. 209.
The code to generate the plot is:
import ggplotnim, sequtils, sugar let df1 = readCsvTyped("/tmp/nov2017_0.csv") let df2 = readCsvTyped("/tmp/nov2017_mean_0.csv") var diffs1 = newSeq[float]() var diffs2 = newSeq[float]() for i in 0 ..< df1["timestamp", int].size.int: let t = df1["timestamp", int] if i == 0: continue diffs1.add (t[i] - t[i-1]).float for i in 0 ..< df2["timestamp", int].size.int: let t = df2["timestamp", int] if i == 0: continue diffs2.add (t[i] - t[i-1]).float var dfP = toDf({"gainSlices" : diffs1, "directMapping" : diffs2}) dfP["idx"] = toSeq(0 ..< dfP.len) dfP = dfP.gather(["gainSlices", "directMapping"], "key", "val").dropNull("val", convertColumnKind = true) echo dfP.pretty(-1) ggplot(dfP, aes(idx, val, color = key)) + geom_point(alpha = some(0.5)) + ylim(0, 1e4) + ylab("Time difference in s") + margin(top = 1.75) + ggtitle("Length of each time slice in 2017/18 data (direct mapping vs gain slices)") + ggsave("/tmp/timediff.pdf")
where in the plotTotalChargeOverTime.nim we introduced a hacky
output for the CSV files read above. In each proc which generates the final
DF that contains the mean / median / etc values introduce something like:
let df2 = result.mutate(f{float -> int: "timestamp" ~ `timestamp`.int}) var num {.global.} = 0 df2.writeCsv(&"/tmp/nov2017_{num}.csv") inc num
- DONE fix last gas gain slice in each run
See https://github.com/Vindaar/TimepixAnalysis/issues/50.
: this has been fixed in commit
f177d378021368cdc9eeb866ba650e5bd58e2397: https://github.com/Vindaar/TimepixAnalysis/commit/f177d378021368cdc9eeb866ba650e5bd58e2397There is now a
minimumGasGainIntervalconfig field in theconfig.tomlfile. If a slice would be shorter than this amount of time in minutes, it will be merged into the previous slice. The default for that variable is set to 25 minutes.The plots in this whole section do not take this fix into account!
15.1.7. Summary & conclusion of energy behavior
In summary, by applying the lessons learned in this section we arrive at a mostly flat median cluster energy over time, which is one of the most fundamental detector properties.
The lessons learned are as follows:
- do not perform the gas gain computation by run, but rather by time intervals. The optimal time lenght was determined to be $\SI{90}{\minute}
- when computing the gas gain for each time slice filter out events with \(> \num{500}\) active pixels and clusters with their center outside the silver region
With this we manage to get from the original time dependency in fig. 210 to fig. 211.
We can see a huge improvement in the stability of the energy over time. A gaussian distribution with a width of \(\mathcal{O}(\SI{10}{\percent}\) remains as a systematic error to be included in the final limit calculation.
15.1.8. Addendum: outlier in mean charge plot
:DATE:
While attempting to recreate the mean charge plot of 100 min
intervals, similar to
using:
nim c -d:danger plotTotalChargeOverTime && \ ./plotTotalChargeOverTime ~/CastData/data/DataRuns2017_Reco.h5 \ ~/CastData/data/DataRuns2018_Reco.h5 \ --interval 90 \ --cutoffCharge 0 \ --cutoffHits 500 \ --calibFiles ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --calibFiles ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --applyRegionCut --timeSeries
after a small rewrite to again produce the mean + sum charge plots, it resulted in the following plot:
Especially in the bottom left pane (beginning 2018) there is one very strong outlier at values over 4000.
Digging into where this comes from via:
echo "Last length was: ", tStartIdx - i, " and res ", result[j] if n == "totalCharge" and result[j].float > 4000.0: echo "length: ", tStart, " to ", i, ", = ", tStartIdx - i echo "data ", data[tStartIdx ..< i] echo "length: ", tStart, " to ", i, ", = ", tStartIdx - i let df = toDf({ "tstamp" : tstamps[tStartIdx ..< i], "ch" : data[tStartIdx ..< i] }) ggplot(df, aes("tstamp", "ch")) + geom_point() + ggsave("/t/test.pdf") if true: quit() # ... and if data[i] > 1e7: echo "INDEX ", i, " is ", data[i] echo pretty(df[i .. i], precision = 8)
in the code to find the index & slice in which the mean charge is so large and within that if it is a single event or an average large value.
Its output was:
Last length was: -291 and res 3617.678852842497 Last length was: -236 and res 3598.673882329947 Last length was: -264 and res 3641.173822604179 Last length was: -280 and res 3605.069529817737 INDEX 237826 is 26316762.40340682 DataFrame with 15 columns and 1 rows: Idx rmsTransverse eventNumber centerY fractionInTransverseRms hits centerX timestamp eccentricity totalCharge passIdx energyFromCharge lengthDivRmsTrans sliceNum runNumber runType dtype: float float float float int float int float float int float float int float constant 0 0.1927173 10848 10.803683 0.017994859 389 4.9118959 1519185676 17.454171 2.63167624e+07 3092 184.13387 57.936507 4 152 background
Last length was: -284 and res 4254.708882948279 length: 1519181784.0 to 237906, = -284 ???? data Tensor[system.float] of shape "[284]" on backend "Cpu" 1.68539e+06 205137 201441 206790 622698 303646 342420 291833 604419 596356 319416 670859 432885 452852 304175 846901 565319 352798 466391 377176 397090 293363 162558 526110 207020 345741 319893 301364 1.04708e+06 390639 1.0254e+06 346379 494977 716693 1.20072e+06 610760 256509 173341 426503 284683 814225 569041 739574 314467 266767 361373 1.48926e+06 726477 411304 275435 437679 439111 532635 263605 612498 256107 260832 451291 494898 158837 645821 290485 413361 414344 552745 752441 511996 247317 752622 426083 274576 560313 248534 82960.4 349464 406526 269853 246654 324982 125291 453567 353351 70229.5 105742 355082 312163 218937 515320 224887 540044 688247 152195 363122 430243 435442 547016 649466 414965 453190 366832 286729 868068 311237 819931 505166 334158 196456 534143 229294 366923 351357 837295 521709 486612 370174 323139 163335 471073 564746 482601 309848 1.57928e+06 1.06362e+06 442279 800070 218032 690609 422052 700527 976659 499942 474658 359143 387098 306101 295397 171737 531351 247587 441442 618648 264360 429737 218503 808569 1.02645e+06 580813 344111 1.73806e+06 250116 729248 237528 1.03964e+06 304583 388759 298463 279643 460584 431884 436135 204595 238155 357546 525117 523138 382828 548200 373566 872981 964381 1.05226e+06 475805 439081 158074 658351 461735 776620 864060 464007 279170 100585 230106 297941 227093 422738 1.10164e+06 147949 772283 1.29533e+06 543220 668338 419537 592404 352563 421508 833071 570334 176963 1.13414e+06 358889 612626 272287 884650 1.11804e+06 2.63168e+07 1.51075e+06 656776 426873 949940 1.19846e+06 94669.7 93799 327250 449350 486857 624928 700651 430752 254255 310247 1.31443e+06 599228 1.00603e+06 394776 469805 449809 336905 193892 1.01557e+06 997765 946682 169701 462733 400066 1.5937e+06 382060 127537 265382 442016 462554 494394 721419 312335 1.0488e+06 332989 825896 1.5551e+06 523187 313771 856802 195173 696572 849662 527441 377369 931255 397475 214142 420752 580139 220324 883653 827271 368623 1.04245e+06 195019 169403 140307 389981 811432 684180 640685 626364 355756 424588 477057 862284 682780 1.01364e+06 296700 548328 184223 495866 329890 length: 1519181784.0 to 237906, = -284
The plot /t/test.pdf from this is the following:
where we can clearly see that the individual event 10848 is the
culprit with a charge value over 2e7 from run number 152.
Writing a simple script to just plot a single event number ./../../CastData/ExternCode/TimepixAnalysis/Plotting/plotEvent/plotEvent.nim yields
./plotEvent -f ~/CastData/data/DataRuns2017_Reco.h5 --run 152 --eventNumber 10848 --chip 3
i.e. one very weird event that is probably some kind of spark. Kind of pretty though!
In any case, this is proof again (that I had forgotten!) that it's very important to use the median in this context.
15.2. Time behavior of logL variables
NOTE:: All plots about the mean, variance, skewness and kurtosis do not contain data from "short" gas gain slices (shorter than 100 clusters) and still contain the calibration data which is binned by 30 min instead of 90!, ref https://github.com/Vindaar/TimepixAnalysis/issues/50
15.2.1. TODO redo plots with 90 minute gas gain slices for calibration and fixed last slice
15.2.2. Median of variables used for logL vs. time
Behavior of median eccentricity in \(\SI{100}{\minute}\) bins against time, filtered to only include non noisy events. This variable is one of the likelihood inputs. Thus it should be as stable as possible, otherwise it affects the efficiency of the likelihood method and will mean that our \(\SI{80}{\percent}\) software efficiency will be a pipe dream very soon for the background data at least.
Here the variation is still visible. This is important, because the energy calibtration does not enter the calculation in any way! Need to understand this behavior. Why does it fluctuate? How does it fluctuate in time? This should be as flat as possible. Variations in gas gain seem to play a role. Why? Either means noisy pixels are active sometimes that distort the geometry or we have more multi hits which affect the calculations.
NOTE: maybe this could be visible if we did take into account the charge that each pixel sees. Currently we just treat each pixel with the same weight. In principle each computation could be weighted by its charge value. Problematic of course, due to influence of gaussian statistics of gas gain!
See also section 15.3 for related discussions.
15.2.3. Mean of clusters for logL variables vs. time
15.2.4. Variance of clusters for logL variables vs. time
15.2.5. Skewness of clusters for logL variables vs. time
15.2.6. Kurtosis of clusters for logL variables vs. time
15.3. CDL distributions against background data
The plots shown here are created using the: https://github.com/Vindaar/TimepixAnalysis/blob/master/Plotting/plotCdl/plotCdl.nim code.
It compares the CDL reference distributions (for each of the 3 log
likelihood input variables) with the same distributions for
background. This is done by walking over all background clusters and
classifying each cluster based on the energy into one of the CDL
intervals in the same way as it is done in likelihood.nim.
The plots shown in fig. 227, 228 and 229 show the full distributions of all background data (no distinction if 2017 or 2018 nor if tracking or not).
Each plot looks pretty reasonable in terms of their separation power. More interesting is how this will look like in terms of time dependency.
15.3.1. TODO Check time dependent behavior of background behavior against time
A more detailed way to study this is to look at the behavior of the properties against time. A simplified version of this was already shown in 15.2.2.
We can calculate these distributions as shown in fig. 227 for background also binned according to time. Using a test to compare each time sliced distribution to the "true" full background spectrum (possibly separate for the separate Run 2 and 3?):
Three approaches come to mind (and there are infinitely more):
- compare using \(\chi^2\) test, but problematic because depends strongly on individual bin differences
- better maybe: Kolmogorov-Smirnov test Klaus worry about it (he's probably right): I'd probably implement that myself. Is it hard to implement? Figure out and maybe do or use nimpy+scipy: https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.kstest.html also see:
- Łukaszyk–Karmowski metric a measure for the "distance" of two distributions. That seems to be interesting, even if not for this particular problem, because we care less only about the distance, than the shape similarity!
Related term: Inverse distance weighting https://en.wikipedia.org/wiki/Inverse_distance_weighting
15.4. Determine polya fit range dynamically
See the following issue on the TPA repository: https://github.com/Vindaar/TimepixAnalysis/issues/47 The below is the text of the issue.
We use a hardcoded lower bound for the polya fit, which by default is at 1200 e⁻:
The reason for a lower bound in the first place are threshold effects showing up in the data, which make the polya not fit very well. This leads to a bad prediction of the gas gain (NOTE: does not directly matter for the following analysis, since we use the mean of the data anyway), which slows down the fitting significantly. Each fit can take up to 1 s instead of ~(20 ms).
Example 2 and 3 here are cases where the left edge would significantly cause a bad fit, due to a sharper than real drop off, because the pixel threshold of the is visible.
We need to dynamically determine the lower bound. Essentially find the maximum of the data and then walk to the left up to maybe once more than 1 consecutive bin (to avoid stat fluctuations mess things up) to be lower than maybe 60% of the max or so.
Finally, also show the actual range of the fit and the extended range that is plotted for the fit line here in the plots. Just draw two geomline or add a new column before plotting with a label "fit / extended" or something like this.
UPDATE:
The dynamic polya bound estimation was finished INSERT COMMIT / CODE HERE
## Our heuristics are as follows: ## - Find 95% of max of counts. ## - Take mean of all indices larger than this ## - walk left from mean until we reach 70% of max ## -> minimum value
See fig. 234 for an example
where the determination of the range works well, but the fit is still
bad. The gas gain is probably extremely overestimated, compare the gas
gain from the fit G_fit = 4323.3 vs. from the mean of the data G =
6467.5. Maybe an argument to use the fit after all?
TODO: is such an example the reason for the outliers in the energy vs. time behavior seen in fig. 165?
A good example for chip 3 is shown in fig. 233.
15.4.1. DONE also limit the upper data range?
UPDATE: This has been superseeded by the discussion of
Given fig. 234, maybe a good idea to also limit the upper data range to only fit along the falling edge and not on the "background" seen in the above plot?
16. All \(^{55}\text{Fe}\) spectra on a grid
This shows all \(^{55}\text{Fe}\) spectra in two single grids (look at original plots!). One for pixels and one for charge.
These plots were created using:
https://github.com/Vindaar/TimepixAnalysis/blob/master/Plotting/plotTotalChargeOverTime/plotTotalChargeOverTime.nim
when running with the --createSpectra command line option.
17. 95% CLs limit calculation
I started the calculation of the 95% confidence limit.
For that I first of all ported the TLimit / mclimit code from ROOT, now found here: ./../../CastData/ExternCode/mclimit/ https://github.com/SciNim/mclimit
With this and an additional ROOT tool found here: ./../../CastData/ExternCode/mclimit/tools/calc_with_root.cpp we are now able to write a non linear optimization problem to find the limit for us.
That is part of TPA and now found here: ./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/limitCalculation.nim
The important thing I learned while writing this tool is the usage of non linear equality constraints when working with such problems. Previously I would always try to treat the function to be minimized as a loss function and hope that the algorithm would fine the global minimunm, but in a case like this we'd almost always end up in some bizarre local minimum (there are a lot of those here…). By using an inequality constraint it works flawlessly:
Instead of:
proc calcCL95(p: seq[float], data: FitObject): float = var obsCLs: float obsCLb: float runLimitCalc(p[0], data, obsCLs, obsCLb) result = pow(abs(obsCLs - 0.05), 2) ... proc main = ... var opt = newNloptOpt[FitObject](LN_COBYLA, 1, @[(l: 1e-12, u: 1e-8)]) let varStruct = newVarStruct(calcCL95, fitObj) opt.setFunction(varStruct)
We should instead do:
proc calcCL95(p: seq[float], data: FitObject): float = var obsCLs: float obsCLb: float runLimitCalc(p[0], data, obsCLs, obsCLb) result = obsCLs proc constrainCL95(p: seq[float], data: FitObject): float = var obsCLs: float obsCLb: float runLimitCalc(p[0], data, obsCLs, obsCLb) result = abs(obsCLs - 0.05 - 1e-3) - 1e-3 echo result, " at a CLs: ", obsCLs, " and CLb ", obsCLb, " for param ", p proc main = ... var opt = newNloptOpt[FitObject](LN_COBYLA, 1, @[(l: 1e-12, u: 1e-8)]) let varStruct = newVarStruct(calcCL95, fitObj) opt.setFunction(varStruct) var constrainVarStruct = newVarStruct(constrainCL95, fitObj) opt.addInequalityConstraint(constrainVarStruct)
which makes it work nicely!
The limitCalculation program has a compile time option:
-d:useRoot
which will then try to call the ROOT helper script mentioned above.
Compile said script using:
g++ -Wall --pedantic `~/opt/conda/bin/root-config --cflags --glibs` -o calcLimit calc_with_root.cpp ../tests/mclimit.cpp ../tests/mclimit.h
The limit calculation program will dump the current iteration flux to
/tmp/current_flux.pdf and the data to /tmp/current_data.csv (which
is given as a command line arg to the ROOT script if that option is
selected.
The result using the 2014/15 data (in a broken and fixed version) in fig. 237, 238 and 239.
17.1. Axion flux scaling
Total flux in units of \(\si{\year^{-1}\centi\meter^{-2}}\) given:
is \(f_{\text{tot}} = \SI{7.783642252184622e+20}{\year^{-1}\cm^{-2}}\).
From this we can calculate the number of axions, which entered the magnet bore within the time frame of the solar tracking.
Further we have to take into account the "weights" of each axion we simulate (\(N_{\text{sim}}\)). Weight in this context implies the product of all possible losses, that is:
- conversion probability (depends on \(g_{a\gamma}^2\)!)
- telescope inefficiency
- window transmission
- gas absorption
By calculating a histogram based on all simulated axions and using the weights as weighting, we should get a binning, which describes the scaled number of photons expected given a time corresponding to: Number of total simulated axions divided by the number of axions per year: \(t_{\text{sim}} = N_{\text{sim}} / f_{\text{tot}} * A_{\text{bore}}\) as a fraction of a year
Assumption: Taking:
Run 2 / 3 (2017,18) Background data: 3526.3 h Tracking data: 180.29 h
and 2014/2015 total background data: 14772495.26004448 / 3600 = 4103.47
Resulting in roughly: 209 h
17.2. Limit calculations
Using background of 2017 and 2018 data, added up. Only no tracking data. A tracking candidate set is sampled using Poisson statistics.
Background data is scaled down to correct tracking time before candidates are drawn.
- gaγ = 1e-12
Final limit is below gae times this gaγ.
Rather arbitrary selection of systematic errors:
- "Tel" -> Telescope efficiency
- "Window" -> detector window
- "Software" -> software efficiency of the logL method
- "Stat" -> fake statistical errors
For these limit calculations, the following files were used:
Run 2:
- ./../../../../mnt/1TB/CAST/2017/CalibrationRuns2017_Reco.h5 for energy calibration
- ./../../../../mnt/1TB/CAST/2017/DataRuns2017_Reco.h5 as basis for which to calculate non tracking data
Run 3:
- ./../../../../mnt/1TB/CAST/2018_2/CalibrationRuns2017_Reco.h5 for energy calibration
- ./../../../../mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 as basis for which to calculate non tracking data
All steps desrcibed below were done at commits:
8d1583fa88c865689e7d0593e3b6ce658dbfd957
and the more important commit containing all relevant changes:
1d32139c7743956f30490d9e7d286ad880c62740
or easier the git tag: SPSC_Sep_2020.
The git tag was updated to the reflect the git commit containing the bug fixed mentioned here:
WARNING: The aforementioned commit and all results presented below, are wrong. They include an incorrect scaling of the total background time! Instead of O(3500 h) it was scaled down to O(350 h)! This means all coupling constants etc. below cannot be taken at face value! The relevant commit to fix the bug: +https://github.com/Vindaar/TimepixAnalysis/commit/ba5176f32d697a6bfbaa629ec10e24a899e5a6f4 This commit does not exist anymore. It contained the following lines:
let lhGrp = h5f["/likelihood".grp_str] result = lhGrp.attrs["totalDuration", float] - let trackingTime = h5Cands.mapIt(it.readDuration).sum / 10.0 + let trackingTime = h5Cands.mapIt(it.readDuration).sum echo "Total tracking time ", trackingTime / 3600.0, " h" let secondsOfSim = N_sim.float / totalFluxPerYear * 86400 * 365 ... let scale = trackingTime.float / secondsOfSim / (100 * 100) * areaBore echo &"Scale = {scale}" let gaeDf = readAxModel(axionModel, scale)
where the trackingTime later would then be used to scale the signal
hypothesis. See warning 2 below.
The effect of this bug was a scaling of the signal hypothesis and not the background data (as we did for the hypothetical case of scaling the background down by a factor of 10). That's why the result is quite a big improvement.
WARNING2: Turns out past me once again was not as stupid as the not
as past me thought. Instead of removing that factor of 10 outright,
I have to modify it of course. That factor is the
ratio of tracking to non tracking time! My mind yesterday read
trackingTime as backgroundTime. Of course we need to scale the the
signal hypothesis to the tracking time! By performing the fix shown
above, we essentially scale it to the background time. This leads to a
~20 times larger value for the input signal hypothesis. Of course,
that means the optimization varying \(g_{ae}\) will then have to scale
the parameter down even further resulting in a much lower limit (which
is then wrong of course).
17.2.1. Filtering by non tracking data
First we add the tracking information using the LogReader tool. This was done via:
./cast_log_reader ../resources/LogFiles/tracking-logs --h5out /mnt/1TB/CAST/2017/DataRuns2017_Reco.h5 ./cast_log_reader ../resources/LogFiles/tracking-logs --h5out /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5
which adds the tracking information to each run in the files.
Then we perform the likelihood method. For that we need the
calibration-cdl and XrayReferencFile.
- Generate the CDL raw H5 files:
raw_data_manipulation /mnt/1TB/CAST/CDL_2019/ --runType back --out /mnt/1TB/CAST/CDL_2019/CDL_2019_raw.h5
- Reconstruct those runs:
reconstruction /mnt/1TB/CAST/CDL_2019/CDL_2019_raw.h5 --out /mnt/1TB/CAST/CDL_2019/CDL_2019_reco.h5 reconstruction /mnt/1TB/CAST/CDL_2019/CDL_2019_reco.h5 --only_fadc reconstruction /mnt/1TB/CAST/CDL_2019/CDL_2019_reco.h5 --only_charge reconstruction /mnt/1TB/CAST/CDL_2019/CDL_2019_reco.h5 --only_gas_gain reconstruction /mnt/1TB/CAST/CDL_2019/CDL_2019_reco.h5 --only_energy_from_e
which gives us a fully reconstructed H5 file.
- Calculate CDL spectra using simple cuts and perform all fits using mpfit (instead of previously nlopt - for latter we need better constraints on the solution I feel):
cdl_spectrum_creation CDL_2019_reco.h5 --dumpAccurate
which generated the following fit parameters file: ./../../CastData/ExternCode/TimepixAnalysis/resources/archive/fitparams_accurate_1600446157.txt
- Based on these new fit parameters the cuts, which cut to the main
peaks we are interested in of each CDL target + filter combination
has to be updated. The cuts are inserted here:
./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/private/cdl_cuts.nim
lines: 98-156 in
getEnergyBinMinMaxVals2018. This is currently hardcoded and has to be changed manually. I opened an issue to change this in the future, see here: https://github.com/Vindaar/TimepixAnalysis/issues/46 Based on the above file, the values were changed to:
let range0 = replace(baseCut): minCharge = 0.0 maxCharge = calcMaxCharge(3.52e4, 1.31e4) minRms = -Inf maxRms = Inf maxLength = 6.0 let range1 = replace(baseCut): minCharge = calcMinCharge(4.17e4, 1.42e4) maxCharge = calcMaxCharge(4.17e4, 1.42e4) maxLength = 6.0 let range2 = replace(baseCut): minCharge = calcMinCharge(7.76e4, 2.87e4) maxCharge = calcMaxCharge(7.76e4, 2.87e4) let range3 = replace(baseCut): minCharge = calcMinCharge(1.34e5, 2.33e4) maxCharge = calcMaxCharge(1.34e5, 2.33e4) let range4 = replace(baseCut): minCharge = calcMinCharge(2.90e5, 4.65e4) maxCharge = calcMaxCharge(2.90e5, 4.65e4) let range5 = replace(baseCut): minCharge = calcMinCharge(4.38e5, 6.26e4) maxCharge = calcMaxCharge(4.38e5, 6.26e4) let range6 = replace(baseCut): minCharge = calcMinCharge(4.92e5, 5.96e4) maxCharge = calcMaxCharge(4.92e5, 5.96e4) let range7 = replace(baseCut): minCharge = calcMinCharge(6.63e5, 7.12e4) maxCharge = calcMaxCharge(6.63e5, 7.12e4)
- With the new cuts in place, calculate the
calibration-cdl.h5file:
cdl_spectrum_creation /mnt/1TB/CAST/CDL_2019/CDL_2019_reco.h5 --genCdlFile --year=2018
which generated the following file: ./../../../../mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5
- Generate the
XrayReferenceFile:
cdl_spectrum_creation /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 --genRefFile --year=2018
which generated the following file: ./../../../../mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5
- Using those files finally perform the likelihood cuts on the non tracking data only:
./likelihood /mnt/1TB/CAST/2017/DataRuns2017_Reco.h5 --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --h5out lhood_2017_no_tracking.h5 ./likelihood /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --h5out lhood_2018_no_tracking.h5
If no --tracking argument is given only non tracking data is
considered by default! The resulting files are now found here:
./../../CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_no_tracking.h5
./../../CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2018_no_tracking.h5
Finally, we can calculate the limit:
./limitCalculation ../../resources/LikelihoodFiles/lhood_2017_no_tracking.h5 ../../resources/LikelihoodFiles/lhood_2018_no_tracking.h5 \ --candFiles ../../resources/LikelihoodFiles/lhood_2017_no_tracking.h5 --candFiles ../../resources/LikelihoodFiles/lhood_2018_no_tracking.h5 \ --axionModel ../../../AxionElectronLimit/axion_gae_1e13_gagamma_1e-12_flux_after_exp_N_25000.csv
where we use a relatively simple ray tracing simulation, now living here: ./../../CastData/ExternCode/TimepixAnalysis/resources/archive/axion_gae_1e13_gagamma_1e-12_flux_after_exp_N_25000.csv and the following axion emission created from
readOpacityFile.nim: ./../../CastData/ExternCode/AxionElectronLimit/solar_model_tensor.csv and the above 2 likelihood results files. Note, that we hand the same input for the "candidates" as for the background. For the time being the candidate files are ignored and instead we draw samples from poisson statistics using the given background (which is added, Run 2 and 3) and scaling that to the tracking time we have (done via magic scaling based on the ratio of background to tracking data times shown in 17.1.
From here different cases were considered as mentioned below and the final output is compared to the real TLimit results using the ./../../CastData/ExternCode/mclimit/tools/calc_with_root.cpp by calling it after optimization finishes in the ./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/limitCalculation.nim tool.
17.2.2. Bugged results
- BUG All error
"Tel" : SystematicError(cand: 0.05, back: 0.05), "Window" : SystematicError(cand: 0.10, back: 0.10), "Software" : SystematicError(cand: 0.05, back: 0.05), "Stat" : SystematicError(cand: 0.3, back: 0.1)
file:///home/basti/org/Figs/SPSC_Sep_2020/bug_background_scaled_by_10/limit_2017_18.pdf file:///home/basti/org/Figs/SPSC_Sep_2020/bug_background_scaled_by_10/limit_2017_18_sb.pdf
Result: CLs+b = 0.0305291597439857 CLs: 0.05108029471779695 CLb: 0.59767 gae = 5.074856644868851e-10
- BUG Comparison to TLimit:
We can see that the two agree reasonably well, with some differences expected due to statistics. In this case specifically the observed
CLsseems to be a little off, but the expected one agrees fairly well (if using different RNG seeds / running RNG for each calculation the variations are pretty large!) The difference on the coupling constant is relatively small though. For a final limit one would use much more statistics of course. UPDATE: the above isn't quite what I thought. The CLs and CLs+b lines were switched for TLimit! Fits well!Variable Nim TLimit CLb 0.59809 0.59833 CLs 0.05100 0.04919 CLs+b 0.03050 0.02943 <CLb> 0.50001 0.50001 <CLsb> 0.02241 0.02093 <CLs> 0.04483 0.04186
- BUG Comparison to TLimit:
- BUG No back error, stat:
"Tel" : SystematicError(cand: 0.05, back: 0.0), "Window" : SystematicError(cand: 0.10, back: 0.0), "Software" : SystematicError(cand: 0.05, back: 0.0), "Stat" : SystematicError(cand: 0.3, back: 0.1)
Result: CLs+b = 0.03081065617552044 CLs: 0.05107274715387877 CLb: 0.60327 gae = 4.779836690425872e-10
- BUG No back error, no stat:
"Tel" : SystematicError(cand: 0.05, back: 0.0), "Window" : SystematicError(cand: 0.10, back: 0.0), "Software" : SystematicError(cand: 0.05, back: 0.0), "Stat" : SystematicError(cand: 0.0, back: 0.0)
Result: CLs+b = 0.03175752901095216 CLs: 0.05099890641061193 CLb: 0.62271 gae = 3.949640875352285e-10
- BUG Large errors
"Tel" : SystematicError(cand: 0.10, back: 0.10), "Window" : SystematicError(cand: 0.20, back: 0.20), "Software" : SystematicError(cand: 0.10, back: 0.10), "Stat" : SystematicError(cand: 0.4, back: 0.3)
file:///home/basti/org/Figs/SPSC_Sep_2020/bug_background_scaled_by_10/limit_2017_18_large_err.pdf file:///home/basti/org/Figs/SPSC_Sep_2020/bug_background_scaled_by_10/limit_2017_18_sb_large_err.pdf
Result: CLs+b = 0.02795326380881176 CLs: 0.0510049517540585 CLb: 0.54805 gae = 7.541351890563966e-10
- BUG Additional scaling of background by factor 10
This uses the default errors first mentioned in 17.2.2.1:
"Tel" : SystematicError(cand: 0.05, back: 0.05), "Window" : SystematicError(cand: 0.10, back: 0.10), "Software" : SystematicError(cand: 0.05, back: 0.05), "Stat" : SystematicError(cand: 0.3, back: 0.1)
Coupling gae: 4.126320414568255e-10
Compare coupling constant with the same case without fake scaling: gae = 5.074856644868851e-10
So essentially the gains are not really not that impressive.
Variable Nim TLimit CLb 0.97023 0.97085 CLs 0.05089 0.05142 CLs+b 0.04937 0.04992 <CLb> 0.50008 0.50029 <CLsb> 0.00537 0.00552 <CLs> 0.01075 0.01104 - BUG The same with only "stat" background errors
"Tel" : SystematicError(cand: 0.05, back: 0.0), "Window" : SystematicError(cand: 0.10, back: 0.0), "Software" : SystematicError(cand: 0.05, back: 0.0), "Stat" : SystematicError(cand: 0.3, back: 0.1)
Coupling gae: 4.060293063844513e-10
So essentially the same as above.
Variable Nim TLimit CLb 0.97094 0.97214 CLs 0.05119 0.05121 CLs+b 0.04970 0.04978 <CLb> 0.50027 0.50096 <CLsb> 0.00455 0.00454 <CLs> 0.00910 0.00906
- BUG The same with only "stat" background errors
17.2.3. Fixed results
Fixed the background time bug mentioned above.
"Tel" : SystematicError(cand: 0.05, back: 0.05), "Window" : SystematicError(cand: 0.10, back: 0.10), "Software" : SystematicError(cand: 0.05, back: 0.05), "Stat" : SystematicError(cand: 0.3, back: 0.1)
file:///home/basti/org/Figs/SPSC_Sep_2020/limit_2017_18.pdf file:///home/basti/org/Figs/SPSC_Sep_2020/limit_2017_18_sb.pdf
Result: CLb = 0.59856 CLs = 0.05100046684407774 CLs+b = 0.03052683943419117 <CLb> = 0.50001 <CLsb> = 0.02244371896890084 <CLs> = 0.04488654020699755
Coupling gae: 1.583260509545671e-10
This fits much better to the values of my master thesis as well (2.21e-22 for gae·gaγ, especially given slightly worse background rate for the old data).
- Scale background rate by a factor of 10
We do the same as above in the bugged case, and scale down the background by a factor of 10 to see the "almost no candidates left" case.
Coupling gae = 1.302047822065652e-10
CLb = 0.96994 CLs = 0.05098874895777124 CLs+b = 0.04945602716410064 <CLb> = 0.50152 <CLsb> = 0.005342676981636716 <CLs> = 0.01065296893770282
So essentially the relative improvement becomes even smaller, as one might expect.
file:///home/basti/org/Figs/SPSC_Sep_2020/limit_2017_18_background_scaled_10.pdf file:///home/basti/org/Figs/SPSC_Sep_2020/limit_2017_18_sb_background_scaled_10.pdf
17.2.4. Notes to expand on
Klaus raises a couple of interesting points. In the 2013 CAST paper they don't optimize for CLs, but effectively for CLs+b. He writes:
ich hab mir das gae-Papier von CAST ( https://iopscience.iop.org/article/10.1088/1475-7516/2013/05/010/pdf) nochmal etwas ganuer angesehen. Es ist mir nun klar, dass es mindestens zwei Effekte gibt die, deren Limit stärker machen:
- Sie benutzen eine Likelihood die von s+b abhängt und setzen ein 95%-CL dort wo Delta chi2 (= Delta 2 ln L) < 4 ist. Damit testen Sie die s+b-Hypothese und nicht wie wir die s-Hypothese. Das wäre eher vergleichbar mit unserem CLs+b < 0.05 (und nicht CLs < 0.05), und da CLs := CLs+b / CLb und CLb immer < 1 ist CLs immer schwächer als CLs+b.
- Die haben etwas „Glück“, weil deren best-fit bei negativem gae2*gag2 liegt, sie haben also weniger tracking-Kandidaten als vom background erwartet. Das führt dann zu einem besseren observed limit als das expected limit (das sie nicht angeben). Dieser „Glück“-Effekt ist für CLs+b ausgeprägter als für CLs (wegen dieser Problematik wurde CLs überhaupt erfunden).
If instead of optimizing for CLs+b as well, our limit of course improves, too. However, we still don't improve all that significantly. My answer to him was:
Ja, ich kann auch nach CLs+b optimieren. Dann sieht das Ganze aus, wie in den angehangenen Plots (Fehler erneut auf alles) mit den folgenden Ergebnissen:
Coupling gae = 6.301273529016057e-10
CLb = 0.60616 CLs = 0.08423420776925818 CLsb = 0.05105940738141354
<CLb> = 0.50001 <CLsb> = 0.03551818335221245 <CLs> = 0.07103494600550479
Wenn ich jetzt nach <CLsb> optimieren würde, wäre das Limit natürlich noch etwas niedriger. Genauer gesagt läuft es dann auf 5.68e-10 (bzw. e-22) heraus.
17.3. Different channels
17.3.1. TODO add none gold region as additional channel
17.3.2. TODO use one channel for each energy bin
This way we can assign the statistical error as systematic errors for each channel.
Possibly works better than stats option?
17.3.3. TODO investigate stats option
So far still gives weird results.
17.3.4. TODO use one channel per pixel
This way we can directly make use of the flux information for each location on the chip.
17.3.5. TODO investigate 2013 data using PN-CCD eff. and ray tracer
As mentioned in its own section 28, we should try to approximate that better.
17.4. CLs+b and limit computations
17.4.1. Understanding CLb, CLs+b and validating mclimit
The following text is the result of a lot of confusion and back and forth. The logic follows my current, hopefully less confusing, understanding.
First we start with a recap of the mathematics to understand what the method actually entails and then we cross-check our understanding with an implementation to compare analytic results with numerical ones.
- The maths behind CLs+b
The major problem with the limit calculation at the moment is the actual limit to the coupling constant when optimizing for a
CLs ~ 95%. In the sense that the results we get are of the order of~5e-22 GeV⁻¹, whereas the result of my master thesis was~2e-22 GeV⁻¹and the 2013 paper got~8e-23 GeV⁻¹. By itself these numbers of course are not suspicious, but:- the 2014/15 data analyzed in the master thesis compared to the
2017/18 data are quite comparable (the background rate is even
lower in the new data than in the old). Naively one would expect
an improvement in the limit and not a factor 2 worsening?
The code used for the analysis is different, but both use essentially
TLimit. - computing a limit based on the data from the 2013 paper (see 28) with our code yields a value close to our new numbers. A different approach should yield a different number than reported in the paper for sure, but it shouldn't be different by one order of magnitude.
This lead to a validation from the basics. First of all a check on simple 1 bin cases, which can be computed analytically for Poisson statistics and comparing them to
mclimit/TLimit.To do that, first we need to be able to compute CLb and CLs+b by hand (CLs is just the ratio) for a simple case.
Starting from the Poisson distribution:
Pois(k, λ) = exp(-λ) * λ^k / fac(k)
or in TeX:
where
λis the (real numbered) mean of the Poisson distribution andkthe (integer) number of occurrences. Then for a set of(k, λ)the Poisson distribution gives the probability to getkoccurrences if the mean isλ.What we need to realize to apply the Poisson distribution in interpreting our results as a physics result is the following: If we restrict our data to a single bin for simplicity (for easier language), then we have the following 3 numbers after our experiment is over, which characterize it and the physics is supposed to describe:
- the number of counts we measured in our background data:
b(which will make up our background hypothesis) - the number of candidates we measured in our "signal region":
d(in our case our tracking dataset / for validation purposes toy experiments based drawing from Poisson) - by including a hypothesis we test for:
sthe signal counts we expect in our experiment (time, area, … normalized) for a given interaction / process / coupling constant, etc. (depending on the kind of thing one studies - in our case the axion coupling constant).
With these three numbers, we can use the Poisson distribution to answer the following question: "What is the probability to measure the given number of candidates, given our background and signal hypotheses?" For this to make sense, we need to assume our background to be our "true" background distribution (i.e. we need enough statistics). The signal hypothesis is typically inserted from theory as a continuous distribution already, so here changing the signal
smainly changes the question (probability given this signal A vs. that signal B).In addition the result obtained for that pair of (
s, b) and a singled_i, is the probability to measure thatd_iif we could reproduce the full experiment many times (that ratio of experiments would result ind_icandidates).With these assumptions / this understanding out of the way, we can now consider the meaning of
CLs+band the derivedCLb, andCLs.The main source of truth for the discussion on
CLs+bhere is Thomas Junk's paper aboutmclimit: https://arxiv.org/pdf/hep-ex/9902006.pdf(Note to reader and self: it's helpful to read that paper, do other things and read it again a year later apparently :P).
The major bits of information for a basic understanding is all on page 2 and 3 in form of equation (1) - (6).
First let's revise those equations and then make them understandable by removing the majority of things that make them look complicated.
(Make sure to read those two pages in the paper fully afterward)
For experiments like those described above, we want some "test" statistic
Xto discriminate signal like outcomes from background like ones (read: some function mapping to a scalar that possibly gives very different results for signal- vs. background-like outcomes).If one has multiple channels
i(a total ofn) that are independent of one another, one can compute that test statisticX_ifor each channel and produce a finalXas the product:Because we are looking at experiments following the Poisson distribution (for which we know the probability density function, and as mentioned above, we assume our data to be well understood as to describe it by such a distribution), we can take the PDF as a likelihood function to describe our experiment. This allows us to define such a test statistic
X_ibased on a likelihood ratio:\[ \left. X_i = \frac{f_P(k = d_i, \lambda = s_i + b_i)}{f_P(k = d_i, \lambda = b_i)} = \frac{e^{-(s_i + b_i)} (s_i + b_i)^{d_i}}{d_i!} \middle/ \frac{e^{-b_i} b_i^{d_i}}{d_i!} \right. \]
The main takeaway of this test statistic for further understanding is simply that it is monotonically increasing for increasing
d_i. This is a very useful property as we shall see.With a test statistic
Xdefined, we can write down a statement about the probabilityPof measuring less than or equal to (\[X \leq X_{\text{obs}}\]) what our model (background hypothesisband our "new physics" signals) expects and call itCL_{s+b}:\[ CL_{s+b} = P_{s+b}(X \leq X_{\text{obs}}) \]
We call
CL_{s+b}the "confidence level" of excluding the possibility of the presence of a new "signal" within our background model.The probability \(P_{s+b}\) is now simply the sum of all poisson probabilities for our given
(s_i, b_i)pairs for which the test statisticX_iis smaller or equal to the observed statistic (the value for whichd_iis the observed number of candidates).\[ P_{s+b}(X \leq X_{\text{obs}}) = \sum_{X(\{d'_i\}) \leq X(\{d_i\})} \prod_i^n f_P(k = d'_i, \lambda = s_i + b_i) = \sum_{X(\{d'_i\}) \leq X(\{d_i\})} \prod_i^n \frac{e^{-(s_i + b_i)} (s_i + b_i)^{d'_i}}{d'_i!} \]
Here
d_irefer to the observed candidates in each channel andd'_ito any possible integer. The set ofd'_iis now restricted by the condition for the test statistic of thatd'_ito be smaller than the measuredd_iin that channel. While this would be an extremely cumbersome definition in general, thanks to the test statistic being monotonically increasing ind_i, we can directly name the set of alld'_ifor which the condition is met: \[\{d'_i \mid 0 \leq d'_i \leq d_i\}\], or in words all natural numbers smaller and equal to the measured number of candidates in the channel.This defines the required knowledge to compute
CL_{s+b}. With it we can make statements about exclusions of new physicss. Though technically, an exclusion is described by \(1 - CL_{s+b}\), becauseCL_{s+b}describes the probability to measure less or the equal number we expect (i.e. a null measurement). An exclusion makes a statement about the opposite though:"With what probability can we exclude the existence of new physics
s?".Thanks to the Poisson distribution being normalized to 1, the tail of all cases where \(X > X_{\text{obs}}\) is thus \(1 - CL_{s+b}\).
From
CL_{s+b}we can further define two related numbers,CL_bandCL_s.CL_bis simply the case for \(s = 0\).CL_sis defined fromCL_{s+b}andCL_bby taking the ratio:\[ CL_s = \frac{CL_{s+b}}{CL_b} \]
The introduction of
CL_sis mainly done due to some not-so-nice properties ofCL_{s+b}. Namely, if we have less candidates than the background,CL_{s+b}may even exclude the background itself with a high confidence level. Especially, given that typically we deal with real experiments with limited measurement time (in particular such that our original assumption of taking the measured background as a "true" background distribution) is never going to be true. Combining this with using this technique to excluding new physics, measuring less candidates than expected from background will happen thanks to statistics.The keen reader may realize that
CL_sis essentially an "implementation" of our original test statistic as the form of the probabilityP. That is because \(P_x\) is proportional tof_P(d, x)where \(x \in \{s+b, b\}\). Thus \(P_s\) is sort of proportional tof_P(d, s+b) / f_P(d, b). This is ignoring multiple channels and the sum over the possibled'_i. The significance of it still eludes me anyway, but it seems significant. :)Taking away multiple channels, the four equations reduce to 3 because in that case \(X \equiv X_i\) and become simpler.
Let's look at them again and simplify them further if possible:
\begin{equation} \label{X_test_statistic_math} \left. X = \frac{f_P(k = d, \lambda = s + b)}{f_P(k = d, \lambda = b)} = \frac{e^{-(s + b)} (s + b)^d}{d!} \middle/ \frac{e^{-b} b^d}{d!} = \frac{e^{-s} (s + b)^d}{b^d} \right. \end{equation}This simplified form makes it much easier to see the monotonic scaling in
d.Further \(P_{s+b}\) can now also be significantly simplified:
\begin{equation} \label{P_s+b_math} P_{s+b}(X \leq X_{\text{obs}}) = \sum_{X(\{d'\}) \leq X(\{d\})} \frac{e^{-(s + b)} (s + b)^{d'}}{d'!} = \sum_{d' = 0}^{d_\text{observed}} \frac{e^{-(s + b)} (s + b)^{d'}}{d'!} \end{equation}where we used the fact that we know \(X(d') \leq X(d_{\text{observed}})\) for all \(0 \leq d' \leq d_{\text{observed}}\).
With this understanding (re-)reading the paper by T. Junk should hopefully prove a lot easier.
- the 2014/15 data analyzed in the master thesis compared to the
2017/18 data are quite comparable (the background rate is even
lower in the new data than in the old). Naively one would expect
an improvement in the limit and not a factor 2 worsening?
The code used for the analysis is different, but both use essentially
17.4.2. Comparing analytical computations with numerical ones
Armed with this knowledge we can now perform some analytical
computations about what we expect for CL_{s+b} given certain (s, b,
d) cases and compare those with the results from mclimit.
The idea will be plainly just that. We will compute 1 bin cases
analytically using the previous section and compare the results both
with the mclimit implementation in Nim as well as the ROOT
implementation (we have extracted the TLimit.h/cpp files from ROOT
to look behind the scenes, but the code produces the same results if
the ROOT installation is used for these).
Let's start with a short script to implement the analytical computation from above, namely the equations \eqref{X_test_statistic_math} and \eqref{P_s+b_math}. For the test statistic equation \eqref{X_test_statistic_math} we will also check whether we made any mistakes in our simplification.
First we define a generalized exponentiation (for more mathy syntax):
import math template `^`(x, y: untyped): untyped = pow(x.float, y.float)
which allows us to write x^y where x and y don't have to be
integers.
Now we need to define the Poisson distribution:
proc poisson(k: int, λ: float): float = # use mass function to avoid overflows # direct impl: # result = exp(-λ) * λ^k / fac(k).float result = exp(k.float * ln(λ) - λ - lgamma((k + 1).float))
where we have defined it using the mass function to avoid overflows.
Now we have everything to define the test statistics. First the reduced test statistic we obtained from simplifying equation \eqref{X_test_statistic_math}:
proc testStatistic(s, b, d: int): float = result = exp(-s.float) * (s + b)^d / (b^d)
and the full test statistic is just the ratio of two Poisson distributions:
proc testStatisticFull(s, b, d: int): float = result = poisson(d, (s + b).float) / poisson(d, b.float)
With this we can now compute some numbers and compare the correctness
of our reduction. We will just draw some random integers in {0, 50}
and check if the two always give the same results:
echo testStatistic(3, 1, 2) echo testStatisticFull(3, 1, 2) #| 0.7965930938858231 | #| 0.7965930938858231 | import random for i in 0 ..< 50: let sr = rand(1 .. 50) let br = rand(1 .. 50) let dr = rand(1 .. 50) let tS = testStatistic(sr, br, dr) let tSF = testStatisticFull(sr, br, dr) doAssert abs((tS - tSF) / (tS + tSF)) < 1e-9
where we compare the results by taking into account the absolute values of the actual test statistic values. If the values are huge the absolute difference is going to be larger of course. The code holds true.
With some confidence built up, we can now implement CLs+b via
equation \eqref{P_s+b_math}:
proc CLsb*(s, b, d: int): float = for k in 0 .. d: result += poisson(k, (s + b).float)
Let's compute some random examples:
echo CLsb(1, 1, 1) echo CLsb(4, 6, 3) echo CLsb(1, 5, 2) echo CLsb(8, 2, 6) echo CLsb(12, 84, 83)
Now it's time to check whether this actually gives the same results as mclimit.
We will import the code of the analytical computation above into
another small script. We won't go over everything in detail here, as
the majority is just boilerplate to run mclimit:
import mclimit import sequtils import random # from /tmp/ import the analytical file import clsb_analytical # monte carlo samples const nmc = 100_000 # a short template to create a `Histogram` from a single integer # with 1 bin and sqrt errors template toHisto(arg: int): untyped = let counts = @[arg].toTensor.asType(float) let err = counts.toRawSeq.mapIt(it.sqrt).toTensor Histogram(ndim: 1, bins: @[0'f64].toTensor, counts: counts, err: err) # a helper to print out all information about the `mclimit` results proc print(limit: ConfidenceLevel) = echo "CLb: ", limit.CLb() echo "CLsb: ", limit.CLsb(true) echo "CLs: ", limit.CLs(true) echo "⟨CLb⟩: ", limit.getExpectedCLb_b() echo "⟨CLsb⟩: ", limit.getExpectedCLsb_b() echo "⟨CLs⟩: ", limit.getExpectedCLs_b() # a short procedure to convert integers to histograms and evaluate `mclimit` proc eval(s, b, c: int, stat: bool) = let ch = Channel(sig: toHisto s, back: toHisto b, cand: toHisto c) var rnd = wrap(initMersenneTwister(44)) let limit = computeLimit(@[ch], rnd, stat = stat, nmc = nmc) print(limit) # test the `eval` proc: eval(12, 84, 83, stat = true) echo "Analytical CLsb: ", CLsb(12, 84, 83)
Running this yields the following final output:
CLb: 0.463 CLsb: 0.18506 CLs: 0.3996976241900648 ⟨CLb⟩: 0.5244 ⟨CLsb⟩: 0.2006512017908536 ⟨CLs⟩: 0.3826300568094081 Analytical CLsb: 0.09893305736634564
Huh? Why is CLsb from mclimit about a factor of 2 larger than the
analytical computation? As it turns out, it is because we used
statistical fluctuations (using the stat argument). The code
handling this is:
if stat: var new = output[chIdx].field var old = input[chIdx].field if stat: for bin in 0 ..< new.getBins: let gaus = gaussian(0.0, old.err[bin]) var val = old.counts[bin] + rnd.sample(gaus) when redrawOnNegative: while val < 0.0: val = old.counts[bin] + rnd.sample(gaus) elif clampToZero: val = if val < 0.0: 0.0 else: val ## NOTE: without the `tpoisson` yields exactly the same numerical values as ROOT version ## but yields inf values. So better introduce and get non inf values for expected? new.counts[bin] = val
where redrawOnNegative is a compile time option I added going beyond
ROOT's implementation to handle the case where sampling on a gaussian
at low count values around 0 (i.e. sigma is large relative to count in
that bin) can yield negative values. These result in inf values for
the expected ⟨CLsb⟩ and ⟨CLs⟩ values. It is still unclear to me how to
handle this case. clampToZero is another such option. Need to learn
more statistic, but intuitively I feel like either option introduces
obvious bias.
The ROOT implementation of the same (note: in ROOT's case there is one fluctuation for signal and background histograms; in Nim this is a single template):
for (Int_t channel = 0; channel <= input->GetSignal()->GetLast(); channel++) { TH1 *newsignal = (TH1*)(output->GetSignal()->At(channel)); TH1 *oldsignal = (TH1*)(input->GetSignal()->At(channel)); if(stat) for(int i=1; i<=newsignal->GetNbinsX(); i++) { Double_t g = generator->Gaus(0,oldsignal->GetBinError(i)); newsignal->SetBinContent(i,oldsignal->GetBinContent(i) + g); } newsignal->SetDirectory(0); TH1 *newbackground = (TH1*)(output->GetBackground()->At(channel)); TH1 *oldbackground = (TH1*)(input->GetBackground()->At(channel)); if(stat) for(int i=1; i<=newbackground->GetNbinsX(); i++) newbackground->SetBinContent(i,oldbackground->GetBinContent(i)+generator->Gaus(0,oldbackground->GetBinError(i))); newbackground->SetDirectory(0);
Note that the ROOT for loop starts at 1 and goes until <= to number
of bins, due to under/overflow bins in ROOT histograms.
This fluctuation of course is supposed to account for statistical
variation, reducing the significance of the given numbers. Thus, it
makes sense that the CLs+b numbers would decrease, as there is
uncertainty.
Let's compare the numbers without these statistical fluctuations:
# compare with `stat = false` eval(12, 84, 83, stat = false) echo "Analytical CLsb: ", CLsb(12, 84, 83)
and behold:
CLb: 0.44297 CLsb: 0.09963 CLs: 0.2249136510373163 ⟨CLb⟩: 0.52996 ⟨CLsb⟩: 0.1189515328144663 ⟨CLs⟩: 0.2244537942759195 Analytical CLsb: 0.09893305736634564
The numbers match very well.
Finally, let's do the same as for the test statistics and draw some random numbers and compare if these always match:
proc compareAnalytical(s, b, c: int, stat: bool): bool = let ch = Channel(sig: toHisto s, back: toHisto b, cand: toHisto c) var rnd = wrap(initMersenneTwister(44)) let limit = computeLimit(@[ch], rnd, stat = stat, nmc = nmc) let nCLsb = limit.CLsb() let aCLsb = CLsb(s, b, c) print(limit) echo "Analytical result: ", CLsb(s, b, c) result = abs(nCLsb - aCLsb) < 1e-3#, "No was " & $nCLsb & " vs. " & $aCLsb & " for inputs " & $(s, b, c) import sets var passSet = initHashSet[(int, int, int)]() var failSet = initHashSet[(int, int, int)]() for i in 0 ..< 50: let sr = rand(1 .. 10) # small signal let br = rand(1 .. 100) # large background let dr = rand(1 .. 120) # background compatible candidates let pass = compareAnalytical(sr, br, dr, stat = false) if pass: passSet.incl (sr, br, dr) else: failSet.incl (sr, br, dr) echo "Set of passed arguments " for el in passSet: echo el echo "Set of failed arguments " for el in failSet: echo el
For some sets of arguments the two operations do not yield the same results. Hence the usage of two sets to show passed and failed arguments:
Set of passed arguments Set of failed arguments (4, 51, 34) (2, 12, 73) (1, 92, 106) (8, 52, 85) (1, 89, 72) (10, 16, 48) (6, 74, 46) (6, 11, 120) (8, 63, 12) (4, 47, 102) (5, 53, 36) (9, 29, 98) (5, 91, 71) (6, 38, 102) (6, 26, 37) (2, 8, 27) (10, 81, 104) (4, 3, 43) (6, 98, 107) (9, 13, 115) (2, 41, 102) (10, 2, 46) (7, 72, 101) (7, 28, 36) (2, 89, 40) (9, 41, 118) (1, 85, 25) (7, 22, 41) (2, 37, 34) (2, 7, 38) (10, 43, 28) (4, 9, 113) (5, 91, 112) (5, 29, 77) (7, 84, 44) (6, 76, 26) (3, 83, 45) (2, 48, 93) (5, 37, 41) (6, 94, 22) (1, 97, 19) (10, 58, 115) (3, 65, 50) (6, 68, 16) (2, 84, 94) (3, 99, 19) (7, 92, 71) (8, 17, 21) (6, 55, 99) (2, 57, 25)
Glancing at the data it seems that the arguments seem to break for the case of having a strong excess in candidates over what the background hypothesis and signal hypothesis would predict. Given that this is not a practical problem (if one had more significantly more candidates than background and signal, one would be looking at systematic errors or plain and simply a discovery after all). Let's see if this assumption is correct and compute the difference of (s+b - c) of the two sets:
echo "Diff of (s+b - c) of passed arguments " for el in passSet: echo (el[0] + el[1] - el[2]) echo "Diff of (s+b - c) of failed arguments " for el in failSet: echo (el[0] + el[1] - el[2])
The raw diff results are shown below. Looking at them it seems that
Diff of (s+b - c) of passed arguments 21 -13 18 34 59 22 25 -5 -13 -3 -59 -22 51 61 5 25 -16 47 56 41 -43 1 78 79 -47 18 58 -8 83 28 4 -38 34 Diff of (s+b - c) of failed arguments -59 -25 -22 -103 -51 -60 -58 -17 -36 -93 -34 -1 -68 -12 -29 -100 -43
that assumption is not completely wrong, but there are outliers that pass despite having a negative (s+b - c). What are they?
echo "Diff of (s+b - c) of passed arguments " for el in passSet: let res = (el[0] + el[1] - el[2]) if res < -35: echo "Passed despite negative s+b - c of ", res, ". Result was: " discard compareAnalytical(el[0], el[1], el[2], stat = false)
Let's pick out the worst offenders at -35:
Diff of (s+b - c) of passed arguments Passed despite negative s+b - c of -59. Result was: CLb: 1.0 CLsb: 0.99999 CLs: 0.99999 ⟨CLb⟩: 0.54319 ⟨CLsb⟩: 0.4205981533438428 ⟨CLs⟩: 0.7743112968645277 Analytical result: 0.9999999999999931 Passed despite negative s+b - c of -43. Result was: [0/20821] CLb: 1.0 CLsb: 0.99999 CLs: 0.99999 ⟨CLb⟩: 0.53879 ⟨CLsb⟩: 0.4255816708911221 ⟨CLs⟩: 0.7898841309065168 Analytical result: 0.9999999813194499 Passed despite negative s+b - c of -47. Result was: CLb: 1.0 CLsb: 0.99999 CLs: 0.99999 ⟨CLb⟩: 0.53656 ⟨CLsb⟩: 0.1237885973165958 ⟨CLs⟩: 0.230707837551431 Analytical result: 0.9999999241683505 Passed despite negative s+b - c of -38. Result was: CLb: 1.0 CLsb: 0.99998 CLs: 0.99998 ⟨CLb⟩: 0.53553 ⟨CLsb⟩: 0.2434893473565324 ⟨CLs⟩: 0.4546698548289217 Analytical result: 0.9999970870413577
So these are all cases where essentially both approaches yield about 1.0. I suppose with this we can consider this topic satisfied.
The ROOT code to compute the same numbers (without any of the fancy stuff):
#include <iostream> #include "TH1.h" #include "TROOT.h" #include "TSystem.h" //#include "mclimit.h" #include "TLimit.h" #include "TRandom3.h" #include "TMath.h" #include "TLimitDataSource.h" #include "TConfidenceLevel.h" using namespace ROOT; int main(){ TH1D sh = TH1D("sh", "sh", 1, 0.0, 1.0); TH1D bh = TH1D("bh", "bh", 1, 0.0, 1.0); TH1D dh = TH1D("dh", "dh", 1, 0.0, 1.0); TRandom3 rng = TRandom3(44); const int nmc = 1000000; // set the S, B, C values we want to look at sh.SetBinContent(1, 2.0); bh.SetBinContent(1, 8.0); dh.SetBinContent(1, 7.0); TLimitDataSource* dataSource = new TLimitDataSource(); dataSource->AddChannel(&sh, &bh, &dh, NULL, NULL, NULL); TConfidenceLevel* limit = TLimit::ComputeLimit(dataSource, nmc, bool (0), &rng); std::cout << " CLb : " << limit->CLb() << std::endl; std::cout << " CLsb : " << limit->CLsb(true) << std::endl; std::cout << " CLs : " << limit->CLs(true) << std::endl; std::cout << "< CLb > : " << limit->GetExpectedCLb_b() << std::endl; std::cout << "< CLsb > : " << limit->GetExpectedCLsb_b() << std::endl; std::cout << "< CLs > : " << limit->GetExpectedCLs_b() << std::endl; delete dataSource; delete limit; return 0; }
which needs to be compiled like so:
g++ -Wall -pedantic `root-config --cflags --glibs` -O3 -o tlimit_root tlimit_root.cpp mclimit.cpp mclimit.h
(if no local copy of the TLimit implementation is around, remove the
#include mclimit.h line and replace it by #include "TLimit.h"
instead. And remove the mclimit.cpp mclimit.h of the
compilation command. It produces a tlimit_root binary we can run:
./tlimit_root
CLb : 0.313241 CLsb : 0.220396 CLs : 0.703599 < CLb > : 0.591809 < CLsb > : 0.332313 < CLs > : 0.561521
(NOTE: running the ROOT code above with a bool (1) argument to the
ComputeLimit constructor results in the case of inf results for
the expected limits, that the modified fluctuation in the Nim code is
supposed to handle).
Let's compare this finally with the nim code:
discard compareAnalytical(2, 8, 7, stat = false)
CLb: 0.3109 CLsb: 0.22177 CLs: 0.7133161788356385 ⟨CLb⟩: 0.58935 ⟨CLsb⟩: 0.3307008861114531 ⟨CLs⟩: 0.5611281685101435 Analytical result: 0.2202206466016993
As we can see, all three possibilities give essentially the same result.
17.4.3. Extracting likelihood data for limit computation to play around
Let's write the background clusters passing logL to a CSV file to play around with.
File: ./../Misc/scaled_limit_calc_input_0.8eff.csv
Energy,Flux,Back,BackErr,Cand,CandErr 0,4.2785e-11,0.1022,0.0723,0,0 0.2,2.0192e-09,0.1534,0.08855,0,0 0.4,4.8349e-09,0.4601,0.1534,0,0 0.6,1.1896e-08,1.125,0.2398,1,1 0.8,1.6939e-08,1.431,0.2705,2,1.414 1,2.0612e-08,1.636,0.2892,2,1.414 1.2,2.044e-08,0.6135,0.1771,1,1 1.4,1.9855e-08,0.4601,0.1534,0,0 1.6,1.8576e-08,0.3067,0.1252,1,1 1.8,1.5655e-08,0.7157,0.1913,0,0 2,1.3166e-08,0.3067,0.1252,0,0 2.2,1.1091e-08,0.3579,0.1353,1,1 2.4,9.4752e-09,0.1534,0.08855,0,0 2.6,7.2641e-09,0.409,0.1446,0,0 2.8,5.1626e-09,0.6646,0.1843,0,0 3,4.0621e-09,1.278,0.2556,2,1.414 3.2,5.6932e-09,1.176,0.2452,0,0 3.4,4.5366e-09,1.176,0.2452,1,1 3.6,3.8188e-09,0.7669,0.198,0,0 3.8,3.0057e-09,0.5112,0.1617,0,0 4,2.3909e-09,0.1534,0.08855,0,0 4.2,1.9964e-09,0.1022,0.0723,0,0 4.4,1.8131e-09,0.1022,0.0723,0,0 4.6,1.5083e-09,0.1022,0.0723,0,0 4.8,1.322e-09,0.1022,0.0723,0,0 5,1.1509e-09,0.2556,0.1143,1,1 5.2,1.0028e-09,0.3579,0.1353,0,0 5.4,8.8352e-10,0.5112,0.1617,1,1 5.6,7.0316e-10,0.4601,0.1534,1,1 5.8,5.5092e-10,0.7157,0.1913,2,1.414 6,4.0887e-10,0.2045,0.1022,0,0 6.2,2.8464e-10,0.5624,0.1696,0,0 6.4,1.7243e-10,0.3579,0.1353,0,0 6.6,3.5943e-11,0.3579,0.1353,0,0 6.8,0,0.3579,0.1353,0,0 7,0,0.2556,0.1143,0,0 7.2,0,0.3067,0.1252,0,0 7.4,0,0.2045,0.1022,0,0 7.6,0,0.2045,0.1022,1,1 7.8,0,0.3579,0.1353,1,1 8,0,0.5112,0.1617,2,1.414 8.2,0,0.7669,0.198,1,1 8.4,0,0.9202,0.2169,2,1.414 8.6,0,0.7669,0.198,0,0 8.8,0,1.227,0.2505,1,1 9,0,1.227,0.2505,3,1.732 9.2,0,1.585,0.2847,2,1.414 9.4,0,1.125,0.2398,1,1 9.6,0,0.9202,0.2169,1,1 9.8,0,0.9202,0.2169,0,0 10,0,0,null,0,null
All three histograms scaled to the same time, corresponding to tracking time. Candidates are drawn using:
proc drawExpCand(h: Histogram): Histogram = ## given a histogram as input, draws a new histogram using Poisson ## statistics var pois: Poisson var rnd = wrap(initMersenneTwister(0x1337)) result = h.clone() for i in 0 ..< h.counts.len: let cnt = h.counts[i] pois = poisson(cnt) let cntDraw = rnd.sample(pois) result.counts[i] = cntDraw result.err[i] = sqrt(cntDraw)
"Flux" column is the solar axion flux after full ray tracing onto the detector (including window + argon absorption). Using \(g_ae = 1e-13\) and \(g_aγ = 1e-12\). To rescale the flux to another \(g_ae\):
proc rescale(flux: var Tensor[float], gae_new, gae_current: float) = echo "gae current ", gae_current flux.apply_inline(x * pow(gae_new / gae_current, 2.0))
Information on scaling of the background data:
proc computeScale(backgroundTime: Hour, trackToBackRatio: UnitLess, N_sim: float, eff: float): UnitLess = let resPath = "../../../AxionElectronLimit" let diffFluxDf = toDf(readCsv(resPath / "axion_diff_flux_gae_1e-13_gagamma_1e-12.csv")) defUnit(yr⁻¹) defUnit(m⁻²•yr⁻¹) defUnit(m²•s¹) let fluxPerYear = simpson(diffFluxDf["Flux / keV⁻¹ m⁻² yr⁻¹", float].toRawSeq, diffFluxDf["Energy / eV", float].map_inline(x * 1e-3).toRawSeq) .m⁻²•yr⁻¹ # compute signal let trackingTime = backgroundTime / trackToBackRatio echo "Total background time ", backgroundTime, " h" echo "Total tracking time ", trackingTime, " h" let secondsOfSim = (N_sim / fluxPerYear).to(m²•s¹) echo &"secondsOfSim = {secondsOfSim}" let areaBore = π * (2.15 * 2.15).cm² # area of bore in cm² echo &"areaBore = {areaBore}" # - calculate how much more time is in tracking than simulation # - convert from m² to cm² # - multiply by area of bore #let scale = totalFluxPerYear / N_sim.float * 5.0 / (100 * 100) * areaBore * (trackingTime / (86400 * 365)) result = (trackingTime / secondsOfSim * areaBore * eff).to(UnitLess) echo &"Scale = {result}"
which yields the following output:
Total background time 3318. Hour h Total tracking time 169.7 Hour h secondsOfSim = 4.052e-07 Meter²•Second areaBore = 14.52 CentiMeter² Scale = 1.751e+09 UnitLess
Finally, let's extract the raw background counts in addition: File: ./../Misc/background_counts_logL_pass_0.8eff.csv The file contains the run number of each passed cluster and its energy:
runNumber,Energy 91,3.104 91,1.727 91,1.939 91,7.836 91,4.728 79,8.036 79,5.914 79,3.615 79,3.262 79,3.472 168,9.538 168,9.25 168,9.729 168,10.69 168,9.392 168,10.57 168,10.58 ... # and so on
Compute a histogram compatible with the above already scaled histogram by:
proc toHisto(df: DataFrame): Histogram = let energy = df["Energy", float].toRawSeq let (histo, bins) = histogram(energy, range = (0.0, 10.0), bins = 50) result = toHisto(histo, bins)
With this one can play around e.g. with TLimit:
#include <iostream> #include "TH1.h" #include "TROOT.h" #include "TSystem.h" // #include "../tests/mclimit.h" #include "TLimit.h" #include "TRandom3.h" #include "TMath.h" #include "TLimitDataSource.h" #include "TConfidenceLevel.h" #include "TVectorD.h" #include "TObjString.h" // csv parser #include "csv.h" using namespace ROOT; int main(int argc, char* argv[]){ const int nbins = 51; TH1D sh = TH1D("", "", nbins, 0.0, 10.2); TH1D bh = TH1D("", "", nbins, 0.0, 10.2); TH1D dh = TH1D("", "", nbins, 0.0, 10.2); TRandom3 rng = TRandom3(44); const int nmc = 100000; if (argc < 2) { std::cout << "Please give an input file!" << std::endl; return -1; } io::CSVReader<4> in(argv[1]); // in.read_header(io::ignore_extra_column, "Flux", "Energy", "back", "cand"); in.next_line(); double sig, back, cand, energy; int k = 0; while(in.read_row(sig, energy, back, cand)){ sh.SetBinContent(k, sig); bh.SetBinContent(k, back); dh.SetBinContent(k, cand); k++; } if (k != nbins) { std::cout << "File input does not match desired number of bins. Actual bins: " << k << std::endl << "Desired bins: " << nbins << std::endl; return -1; } TLimitDataSource* dataSource = new TLimitDataSource(); Double_t backEVal[4] = {0.05, 0.1, 0.05, 0.1}; Double_t candEVal[4] = {0.05, 0.3, 0.05, 0.1}; TVectorD backErr(4, backEVal); TVectorD candErr(4, candEVal); TObjArray names; TObjString n1("Software"); TObjString n2("Stat"); TObjString n3("Tel"); TObjString n4("Window"); names.AddLast(&n1); names.AddLast(&n2); names.AddLast(&n3); names.AddLast(&n4); dataSource->AddChannel(&sh, &bh, &dh, &candErr, &backErr, &names); TConfidenceLevel* limit = TLimit::ComputeLimit(dataSource, nmc, bool (0), &rng); if (argc < 3){ std::cout << " CLb : " << limit->CLb() << std::endl; std::cout << " CLs : " << limit->CLs() << std::endl; std::cout << " CLsb : " << limit->CLsb() << std::endl; std::cout << "< CLb > : " << limit->GetExpectedCLb_b() << std::endl; std::cout << "< CLsb > : " << limit->GetExpectedCLsb_b() << std::endl; std::cout << "< CLs > : " << limit->GetExpectedCLs_b() << std::endl; } else{ std::cout << limit->CLs() << std::endl; std::cout << limit->CLb() << std::endl; } delete dataSource; delete limit; return 0; }
Compile again with:
g++ -Wall -pedantic `root-config --cflags --glibs` -O3 -o comp_limit_root comp_limit_root.cpp csv.h
Note that the csv.h file is required!
17.5. Investigate signal limit calculation
We need to check and compare what the count numbers in the "signal" (signal hypothesis for limit calculation) look like in three different cases:
- our current
limitCalculation.nimcode - the code from my M.Sc. thesis
- the 2013 pn-CCD paper
Essentially the questions we need to ask are:
- how many counts are found in the signal histogram in the final, optimized limit? i.e. at the 95% CLs or equivalent
- how are these counts computed starting from some form of a theoretical description of the solar flux? This latter question will be hard to answer for the pn-CCD paper for sure. If we can even get the counts there!
17.5.1. 2013 pn-CCD paper
We will take a look at the 2013 CAST axion-electron paper:
https://iopscience.iop.org/article/10.1088/1475-7516/2013/05/010/pdf
or:
~/org/Papers/cast_axion_electron_jcap_2013_pnCCD.pdf
First we will present their data and their limit calculation method, then try to follow their computations to reproduce their result. Finally, we will ask a few questions about their methods and results and try to answer them.
- Data and limit method
The data analyzed in the paper is from the 2004 data taking campaign using a pn-CCD detector behind the (then single) X-ray telescope.
In consists of \SI{1890}{\hour} of background data and \SI{197}{\hour} of tracking data. In the tracking dataset 26 candidates were measured. The X-ray telescope is said to focus the \SI{14.5}{\centi\meter\squared} coldbore area onto an area of \SI{9.3}{\milli\meter\squared} on the CCD.
The data is shown in fig. 240 and tab. 23, where the background data was scaled to the tracking time.
import ggplotnim let ratio = 1890.0 / 197.0 let df = readCsvTyped("/home/basti/org/Misc/2013_pnCCD_data.csv") .mutate(f{"FullBackground" ~ `Background` * ratio}, f{float: "BackErr" ~ sqrt(`FullBackground`) / ratio}) .gather(@["Candidates", "Background"], key = "Type", value = "Counts") echo df let binWidth = 0.2857 ggplot(df, aes("binCenter", "Counts", color = "Type")) + geom_point() + geom_errorbar(data = df.filter(f{`Type` == "Background"}), aes = aes(yMin = f{`Counts` - sqrt(`BackErr`) / 2.0}, yMax = f{`Counts` + sqrt(`BackErr`) / 2.0})) + geom_errorbar(data = df.filter(f{`Type` == "Candidates"}), aes = aes(yMin = f{`Counts` - sqrt(`Counts`)}, yMax = f{`Counts` + sqrt(`Counts`)})) + xlab("ω [keV]") + ggtitle("2004 pn-CCD data of 2013 CAST g_ae paper") + ggsave("/home/basti/org/Figs/statusAndProgress/CAST_2013/2013_CAST_gae_paper_data.pdf", width = 800, height = 480)
Figure 240: Data of the 2004 data taking period at CAST with the pn-CCD detector. \SI{1890}{\hour} of background data and \SI{197}{\hour} of tracking data. The background data is scaled to the tracking time. The data was extracted by hand from the paper. Table 23: Data of the 2004 data taking period at CAST with the pn-CCD detector. \SI{1890}{\hour} of background data and \SI{197}{\hour} of tracking data. The background data is scaled to the tracking time. The data was extracted by hand from the paper. Energy Energy, binCenter Candidates Background 0.7999 0.94287 1 2.27 1.0857 1.22857 3 1.58 1.3714 1.51428 1 2.4 1.6571 1.8 1 1.58 1.9428 2.08571 1 2.6 2.2285 2.37142 2 1.05 2.5142 2.65714 1 0.75 2.7999 2.94285 2 1.58 3.0857 3.22857 0 1.3 3.3714 3.51428 2 1.5 3.6571 3.79999 0 1.9 3.9428 4.08571 1 1.85 4.2285 4.37142 0 1.67 4.5142 4.65714 2 1.3 4.7999 4.94285 2 1.15 5.0857 5.22857 0 1.67 5.3714 5.51428 2 1.3 5.6571 5.8 1 1.3 5.9428 6.08571 2 2.27 6.2285 6.37142 2 1.3 Since no excess in the tracking is found in the data, a Poissonian binned likelihood is defined and used in a maximum likelihood estimation (MLE) to compute the best fit value for \(g^2_{ae}g^2_{aγ}\). This is done using the typical expression of MLE in form of a \(\chi^2\)
\[ \chi^2 = -2 \ln\mathcal{L} \]
The likelihood function used is: \[ \mathcal{L} = \prod_j^n \frac{e^{-\lambda_j} \lambda_j^{t_j}}{t_j!} \] where \(n = 20\) the number of "spectral bins", \(t_j\) the number of observed counts in tracking and \(\lambda_j\) the Poisson mean in bin \(j\).
They fit: \[ \lambda_j = \sigma_j + b_j \] where \(b_j\) is the background in bin \(j\) and \(\sigma_j \propto g^2_{ae}g^2_{aγ}\).
The paper only mentions the proportionality of \(sigma\) to the coupling constant. It is left as a (very questionable!) exercise to the reader to figure out the direct relation.
In theory it is obvious that \(\sigma\) is proportional to \(g^2_{ae}g^2_{aγ}\). The conversion probabilities of axions depend on the coupling constant squared. In the simplest case the creation in the Sun using the axion-electron coupling and then reconversion in the magnet using the axion-photon coupling. This is where the first major roadblock appears due to them giving no details about the proportionality. Depending on the ratio between assumed \(g_{ae}\) and \(g_{aγ}\) values, the solar axion production changes. If \(g_{ae}\) is "large" (whatever large means exactly) compared to \(g_{aγ}\), the axion flux has its peak near \SI{1}{\kilo\electronvolt}$, but the smaller it becomes compared to \(g_{aγ}\) the peak will shift towards the Primakoff peak near \SI{3}{\kilo\electronvolt}. The paper mentions:
For very small values of gae <= 10e−12 , the BCA flux is negligible and the CAST bound smoothly becomes gaγ < 0.88×10−10 GeV−1 as found in our previous study [5] where only Primakoff emission was assumed. However, for larger values of gae the BCA flux becomes dominant and we recover equation 1.1.
From this we can deduce that they assume \(g_{ae}\) to be large compared to \(g_{aγ}\) and thus \(g_{ae} \geq 1e-12\). In this regime we can assume that
- \(g_{aγ}\) is only relevant for conversion from axions to photons in the magnet and it is independent on the axion energy, i.e. a constant suppression \(\propto g^2_{aγ}\).
- \(g_{ae}\) is the only relevant contribution to axion production, it yields a non constant (i.e. energy dependent) flux \(\propto g^2_{ae}\) as long as \(g_{ae} < 1e-12\). This means in any product of \(g_{ae} g_{aγ}\) the \(g_{ae}\) contribution needs to be larger than \(1e-12\) if \(g_{aγ} \sim \mathcal{O}(\SI{1e-10}{\per\giga\electronvolt})\).
This is important, because when varying \(g^2_{ae}g^2_{aγ}\), we need to split up the \(g_{ae}\) and \(g_{aγ}\) contributions. The former varies the incoming flux and the latter the amount of reconverted photons. If we pick \(g_{aγ}\) "too large" varying the product to small values, pushes \(g_{ae}\) into a range, where it is not the dominant production mechanism anymore, and thus pushing the flux peak towards \SI{3}{\kilo\electronvolt}. A proper analysis should scan the phase space individually, keeping one of the coupling constants fixed while varying the other. This is why it is questionable to only talk about a proportionality of \(\sigma\) to the product of coupling constants.
For the sake of reproducing the results, we will pick a constant \(g_{aγ}\) to compute a conversion probability, which we will keep constant the whole time. The value will be chosen small enough, \(g_{aγ} = \SI{1e-12}{\per\giga\electronvolt}\) so that we can vary \(g_{ae}\) to values small enough without worrying about disturbing the shape of the differential flux. Also this is the value used for the provided differential solar axion flux computed by J. Redondo.
The paper gives the standard conversion probability for axions in a magnetic field via \(g_{aγ}\) as:
\[ P_{a \rightarrow γ} = \left( \frac{g_{aγ} B L}{2} \right)^2 \frac{\sin^2\left(\frac{qL}{2}\right)}{\left(\frac{qL}{2}\right)^2} \] where \(B\) the magnetic field, \(L\) the length of the magnet, \(q\) the momentum transfer \(q = m²_a / 2\omega\). Since we consider only coherent conversions (masses smaller than \(\sim \SI{10}{\milli\electronvolt}\)) the probability reduces to the first term (\(\sin(x) \approx x\) for small \(x\)).
The axion production is the one from Redondo's accompanying 2013 theory paper, reproduced in fig. 241.
Figure 241: The expected solar axion production computed for \(g_{ae} = 1e-13\) and \(g_{aγ} = \SI{1e-12}{\per\giga\electronvolt}\). With the production and conversion mechanisms laid out, we can now consider a specific relation for \(\sigma \propto g^2_{ae}g^2_{aγ}\) to express the actual flux visible in the detector \(F\):
\begin{equation} \label{eq_2013_flux_on_detector} F(g_{ae}, g_{aγ} = \SI{1e-12}{\per\giga\electronvolt}) = \alpha \frac{\mathrm{d}\Phi}{\mathrm{d}\omega}(g_{ae}, g_{aγ}) P_{a \rightarrow γ}(g_{aγ}) \end{equation}here \(\alpha\) is a combined scaling factor that includes the conversion of the differential flux in units of \(\si{\per\kilo\electronvolt\per\year\per\meter\squared}\) to actual counts on the detector in the time frame of the experiment, namely \(\SI{197}{\hour}\) of tracking time and finally the chosen bin width of the data.
This conversion factor is thus:
import unchained defUnit(keV⁻¹•yr⁻¹•m⁻²) defUnit(keV⁻¹•h⁻¹•m⁻²) # convert to `h⁻¹` (`yr` treated as distinct otherwise) let input = 1.keV⁻¹•yr⁻¹•m⁻².to(keV⁻¹•h⁻¹•m⁻²) let binWidth = 0.2857.keV let areaChip = 9.3.mm² let areaBore = π * (2.15 * 2.15).cm² # area of bore in cm² let time = 197.h let α = input * binWidth * areaBore * time echo "The scaling factor is ", α
The scaling factor is 9.32399e-06 UnitLess
So the conversion from a single count in units of the differential flux to counts on the detector per bin in energy is \(\alpha = \num{9.33038e-06}\). This conversion ignores 2 very important aspects:
- the telescope has a finite efficiency, which depends on the incoming angle of the photon
- the detector has a finite quantum efficiency (even though it is extremely high for the used pn-CCD detector)
These two facts will decrease the number of expected counts further. Our assumption of taking these numbers as 1 means we should underestimate the expected limit (i.e. get a better limit than reported in the paper).
This allows us to compute the maximum likelihood by varying \(g_{ae}\) using the relation:
\begin{equation} \label{eq_flux_rescaling_2013} \frac{\mathrm{d}\Phi}{\mathrm{d}\omega}(g_{ae}, g_{aγ}) = \frac{\mathrm{d}\Phi}{\mathrm{d}\omega}(g'_{ae}, g_{aγ}) \cdot \left( \frac{g_{ae}}{g'_{ae}} \right)^2 \end{equation} - Computing a limit according to the paper
Now we will compute the limit using the data and methods described in the previous section. That is perform a maximum likelihood estimation based on the given likelihood function. We will do this in a literate programming session.
We start by importing all required modules as well as defining a (numerically stable) Poisson distribution, the likelihood function and a procedure to compute the \(χ²\):
import std / [math, sequtils, sugar] import ggplotnim, seqmath, nlopt, unchained proc poisson(k: int, λ: float): float = # use mass function to avoid overflows # direct impl: #result = exp(-λ) * λ^k / fac(k).float result = exp(k.float * ln(λ) - λ - lgamma((k + 1).float)) proc L2013*(s, b: seq[float], d: seq[int]): float = result = 1.0 doAssert s.len == b.len doAssert s.len == d.len for i in 0 ..< s.len: result *= poisson(d[i], s[i] + b[i]) proc χ²(s, b: seq[float], d: seq[int]): float = result = -2 * ln(L2013(s, b, d))
With these procedures we can compute \(χ²\) values as shown for the \(χ²\) distribution in the paper once we have defined our input data as well as the expected signal hypothesis.
So next we read the input data from a CSV file and store the candidates and background counts in individual variables.
let df = readCsvTyped("/home/basti/org/Misc/2013_pnCCD_data.csv") let cands = df["Candidates", int].toRawSeq let backs = df["Background", float].toRawSeq echo df
For the expected signal we first read a CSV file containing the differential flux as shown in fig. 241. Things are a bit more complicated, because we have to rebin the differential flux to the binning used for the data. Because numerical efficiency doesn't matter here, we do things in individual steps. First we read the data of the differential flux, convert the energies from
eVtokeVand remove everything outside the data range we consider, namely in \(0.7999 <= E <= 6.2285 + 0.2857\) (these values are the bin edges as determined by studying the data):proc readAxModel(): DataFrame = let binWidth = 0.2857 let upperBin = 6.2285 + binWidth result = readCsvTyped("/home/basti/CastData/ExternCode/AxionElectronLimit/axion_diff_flux_gae_1e-13_gagamma_1e-12.csv") .mutate(f{"Energy / keV" ~ c"Energy / eV" / 1000.0}) .filter(f{float: c"Energy / keV" >= 0.7999 and c"Energy / keV" <= upperBin})
Now rebin everything to the bins as used in the paper and keep only those binned flux values and create a plot to see whether our rebinning actually works. So first define the binning itself:
let binWidth = 0.2857142857142858 let bins = @[0.7999,1.0857,1.3714,1.6571,1.9428,2.2285, 2.5142,2.7999,3.0857,3.3714,3.6571,3.9428, 4.2285,4.5142,4.7999,5.0857,5.3714,5.6571, 5.9428,6.2285, 6.2285 + binWidth]
and now rebin the solar flux according to that binning and plot in:
defUnit(keV⁻¹•yr⁻¹•m⁻²) proc rebinDf(df: DataFrame): seq[keV⁻¹•yr⁻¹•m⁻²] = let energy = df["Energy / keV", float].toRawSeq flux = df["Flux / keV⁻¹ m⁻² yr⁻¹", float].toRawSeq var count = 0 var binIdx = -1 var sumFlux = 0.0.keV⁻¹•yr⁻¹•m⁻² for i, el in flux: let E = energy[i] if binIdx < 0 and E.float > bins[0]: binIdx = 1 elif binIdx >= 0 and E.float > bins[binIdx]: result.add (sumFlux / count) count = 0 sumFlux = 0.0.keV⁻¹•yr⁻¹•m⁻² inc binIdx sumFlux = sumFlux + el.keV⁻¹•yr⁻¹•m⁻² inc count if binIdx > bins.high: break # add current sum as final entry result.add sumFlux / count echo result echo bins.mapIt(it + 0.5 * binWidth) let dfPlot = toDf({ "E" : bins[0 ..< ^1].mapIt(it + 0.5 * binWidth), "F" : result.mapIt(it.float) }) echo dfPlot.pretty(-1) ggplot(dfPlot, aes("E", "F")) + geom_point() + xlab("Energy [keV]") + ylab("Flux [keV⁻¹•yr⁻¹•m⁻²]") + ggtitle("Flux using data binning, g_ae = 1e-13, g_aγ = 1e-12 GeV⁻¹") + ggsave("/home/basti/org/Figs/statusAndProgress/CAST_2013/rebinned_df_flux_2013.pdf") discard readAxModel().rebinDf()
The rebinned flux then looks like shown in fig. 242, showing that indeed the rebinning works as expected.
Figure 242: Differential flux for \(g_{ae} = 1e-13\) and \(g_{aγ} = \SI{1e-12}{\per\giga\electronvolt}\) according to the binning used for the data in the paper. Further, we need to compute an actual number of counts expected on the detector. This means incorporating equation \eqref{eq_2013_flux_on_detector} into our code. Starting with the conversion probability, which we simplify to only consider the \(g_{aγ} B L\) term. Let's also compute the value for the numbers we will use, namely \(g_{aγ} = \SI{1e-12}{\per\GeV}, B = \SI{9}{\tesla}, L = \SI{9.26}{\m}\):
import unchained, math defUnit(GeV⁻¹) func conversionProb(B: Tesla, L: Meter, g_aγ: GeV⁻¹): UnitLess = ## simplified vacuum conversion prob. for small masses result = pow( (g_aγ * B.toNaturalUnit * L.toNaturalUnit / 2.0), 2.0 ) let B = 9.0.T let L = 9.26.m let g_aγ = 1e-12.GeV⁻¹ echo "Conversion probability is ", conversionProb(B, L, g_aγ)
Conversion probability is 1.70182e-21 UnitLess
Then we need to include the computation of the factor \(\alpha\) into the result:
defUnit(keV⁻¹•yr⁻¹•m⁻²) defUnit(keV⁻¹•h⁻¹•m⁻²) proc scaleToTracking(x: keV⁻¹•yr⁻¹•m⁻²): UnitLess = ## Convert the given flux in `keV⁻¹•yr⁻¹•m⁻²` to raw counts registered ## on the chip (assuming a perfect telescope!) during the full tracking period ## within a single energy bin. # convert to `h⁻¹` (`yr` treated as distinct otherwise) let input = x.to(keV⁻¹•h⁻¹•m⁻²) let binWidth = 0.2857.keV let areaChip = 9.3.mm² # not required! let areaBore = π * (2.15 * 2.15).cm² # area of bore in cm² let time = 197.h # factor 2.64 is a crude estimate of telescop efficiency of ~ 5.5 cm^2 result = input * binWidth * areaBore * time # / 2.64
and finally combine all this using eq. \eqref{eq_2013_flux_on_detector}.
let flux = readAxModel().rebinDf().mapIt((scaleToTracking(it) * conversionProb(B, L, g_aγ)).float)
It's a good idea to create a plot of the actual flux we expect given our initial coupling constants on the chip during their full tracking time in comparison to fig. 242.
let fluxInTracking = toDf({ "E" : bins, "F" : flux }) ggplot(fluxInTracking, aes("E", "F")) + geom_point() + xlab("Energy [keV]") + ylab("Counts") + margin(top = 2.0) + ggtitle("Expected X-ray flux for g_ae = 1e-13, g_aγ = 1e-12 during 197h of tracking assuming perfect X-ray optics") + ggsave("/home/basti/org/Figs/statusAndProgress/CAST_2013/flux_tracking_gae_1e-13_gag_1e-12.pdf")
which yields fig. 243:
Figure 243: The expected X-ray count during the 2004 data taking period of \SI{197}{\hour} of tracking time, using the initial coupling constants of \(g_{ae} = \num{1e-13}\) and \(g_{aγ} = \SI{1e-12}{\per\GeV}\). From this figure we can see that the flux needs to be significantly higher to be visible even theoretically. A number of counts of order
1e-6will of course not show up. For that we will scale upg_aeonly so as not to get into the troubles mentioned before.For testing purposes we can now compute the \(χ²\) value for the data and the expected flux using the initial coupling constants:
echo "The initial χ² value is: ", χ²(flux, backs, cands)
For the time being we will ignore that the this value is more than a factor of 2 larger than the values given in the paper.
The final step remaining is now the definition of rescaling the flux for a changed coupling constant, which will be done according to eq. \eqref{eq_flux_rescaling_2013}:
proc rescale(s: seq[float], new: float): seq[float] = let old = 1e-13 # initial value is always 1e-13 result = newSeq[float](s.len) for i, el in s: result[i] = el * pow(new / old, 2.0)
We will use it such that the input flux remains the previously computed
fluxwitholdremaining as1e-13from the initialg_ae. That way we don't have to carry around the last coupling constant.This allows us to define a non-linear optimizer that will scan the
g_aerange for the minimal \(χ²\) value (and thus doing MLE). First we need an object to store our signal, background and candidates information:type # need an object to store input flux as well as background and # candidates ScaleFn = proc(s: seq[float], new: float): seq[float] FitObject = object flux: seq[float] cands: seq[int] backs: seq[float] rescaleFn: ScaleFn var fitObject = FitObject( cands: cands, backs: backs, flux: flux )
Our procedure performing the optimization will simply receive the current parameter (our new coupling constant
g_aeto try), rescale the input flux according to it and compute the \(χ²\), which we return:proc optimize(p: seq[float], data: FitObject): float = # compute new "signals" using the new `g_ae` (parameter 0) let sigs = data.rescaleFn(data.flux, p[0]) result = χ²(sigs, data.backs, data.cands) echo "Current χ² ", result, " of g_ae = ", p[0]
and finally all that remains is some boilerplate to define the optimizer, by choosing the parameter:
template optMe(fn, rescaleProc, startParams, bound, algo: untyped): untyped = var opt = newNloptOpt[FitObject](algo, startParams.len, bounds = bound) # assign the user supplied rescaling function fitObject.rescaleFn = rescaleProc let varStruct = newVarStruct(fn, fitObject) opt.setFunction(varStruct) # set relative and absolute tolerance very small opt.xtol_rel = 1e-14 opt.ftol_rel = 1e-14 # start actual optimization let nloptRes = opt.optimize(startParams) echo nloptRes if opt.status < NLOPT_SUCCESS: echo opt.status echo "nlopt failed!" else: echo "Nlopt successfully exited with ", opt.status # clean up optimizer nlopt_destroy(opt.optimizer)
All that is left is to call the
optMetemplate by providing the function to optimize (theoptimizeprocedure), our starting parameter \(g_{ae} = \num{1e-13}\), the rescaling procedure we definedrescale, the minimization algorithm to use ((we will use a local, non gradient based, simplex routine here) and some reasonable bounds, which we will set very large from \(\num{1e-22} \leq g_{ae} \leq \num{1e-9}\):optMe(optimize, rescale, @[1e-13], @[(l: 1e-22, u: 1e-9)], LN_SBPLX)
As we can see, the lowest value is at the smallest possible coupling constant we allowed, \(g_{ae} = \num{1e-22}\). In particular the \(χ²\) value barely varies between different coupling constants. This makes sense, as the initial flux is already of the order of \(\sim\num{1e-6}\)! So the best fit indeed seems to be one with the smallest possible signal, essentially 0. Given the candidates are in most bins even lower than the expected background, this is "reasonable".
In the paper however, also negative values are allowed. While this does not make any physical sense at all we can try to see what happens in that case. For that we need to change our rescaling procedure to allow for negative values. In the existing
rescaleprocedure the input parameter is squared. To support negative parameters, we need to work with squares the whole way. Let's define a new rescaling procedure:proc rescaleSquares(s: seq[float], new: float): seq[float] = ## rescaling version, which takes a `new` squared coupling constant ## to allow for negative squares let old = 1e-13 # initial value is always 1e-13 result = newSeq[float](s.len) for i, el in s: result[i] = el * new / (old * old)
With this we can run a new optimization, using different bounds, let's say \(\num{-1e-18} \leq g^2_{ae} \leq \num{1e18}\). We of course have to change our starting parameter to start at \(g^2_{ae} = (\num{1e-13})^2\) too. For this we will change to a different optimization algorithm, one based on M. J. D. Powell's COBYLA (Constrained Optimization BY Linear Approximations):
optMe(optimize, rescaleSquares, @[1e-13 * 1e-13], @[(l: -1e-18, u: 1e-18)], LN_COBYLA)
where a few lines were removed in the output. The final best fit is for a coupling constant of \(g^2_{ae} = \num{-4.721e-22}\). With our fixed \(g_{aγ} = \SI{1e-12}{\per\GeV}\) this yields:
\[ g^2_{ae}g^2_{aγ}|_\text{bestfit} = \num{-4.721e-22} \cdot \left(\SI{1e-12}{\per\GeV}\right)^2 = \SI{-4.721e-46}{\per\GeV^2} \]
While not exactly the same as received in the paper (which was \(g^2_{ae}g^2_{aγ} = \SI{-1.136e-45}{\per\GeV^2}\)) it is at least in a similar ballpark. However, given the unphysicality of the number (a negative coupling constant squared requires a complex coupling constant!), the numbers are questionable.
Instead of relying on an optimization strategy, we can use a brute-force method of simply scanning the parameter space between
\[ \SI{-6e-45}{\per\GeV^2} \leq g^2_{ae}g^2_{aγ} \leq \SI{6e-45}{\per\GeV^2} \]
to see if we can at least recover the same behavior as shown in fig. 6 of the paper (aside from the glaring problem of a factor of 58 / 20.5 between the \(χ²\) values!).
import sugar let g_aγ² = 1e-12 * 1e-12 let couplings = linspace(-6e-45 / g_aγ², 6e-45 / g_aγ², 5000) let χ²s = block: var res = newSeq[float](couplings.len) for i, el in couplings: let newFlux = flux.rescaleSquares(el) res[i] = χ²(newFlux, backs, cands) res let dfχ² = toDf({"Couplings" : couplings, "χ²" : χ²s}) .mutate(f{"Couplings" ~ `Couplings` * 1e-12 * 1e-12}) .filter(f{`χ²` <= 100.0}) echo dfχ² ggplot(dfχ², aes("Couplings", "χ²")) + geom_line() + ylim(58, 63) + xlim(-6e-45, 6e-45) + xlab("g²_ae g²_aγ") + ggtitle("Scan of g²_ae g²_aγ for g_aγ = 1e-12 GeV⁻¹") + ggsave("/home/basti/org/Figs/statusAndProgress/CAST_2013/brute_force_chi2_scan.pdf")
Figure 244: Scan of the \(g²_{ae} g²_{aγ} / χ²\) phase space with yielding a behavior comparable to fig. 6 of the paper, but with \(χ²\) values that are too large. Also our curve is more asymmetric than the one in the paper. Having computed a minimum of the \(χ²\) distribution it is now up to us to compute an actual limit on the coupling constant (and not the square).
This is where things become the most problematic. One would normally expect to take the square root of the squared coupling constants and thus determine the limit. However, given that this square is negative, we can't do that.
Thus, it is curious that their given limit is \(g_{ae} g_{aγ} \leq \SI{8.1e-23}{\per\GeV}\). Let's compute the squared value for their limit and see if we can make sense of it:
\begin{align} g_{ae} g_{aγ} &= \SI{8.1e-23}{\per\GeV} \\ g²_{ae} g²_{aγ} &= \left(\SI{8.1e-23}{\per\GeV}\right)² \\ g²_{ae} g²_{aγ} &= \SI{6.561e-45}{\per\GeV^2} \\ \end{align}which curiously is even outside the shown plot fig. 6 in the paper. Of course the paper also gives statistical and systematic uncertainties. One might think that taking their squared value of \(g²_{ae} g²_{aγ} = \SI{-1.136e-45}{\per\GeV^2}\) and adding the
- statistical uncertainty of \(Δ\left(g²_{ae} g²_{aγ}\right)_{\text{stat.}} = \SI{3.09e-45}{\per\GeV^2}\)
- and the systematic uncertainty of \(Δ\left(g²_{ae} g²_{aγ}\right)_{\text{syst.}} = \SI{2.20e-45}{\per\GeV^2}\)
yields \(g²_{ae} g²_{aγ} = \SI{4.154e-45}{\per\GeV^2}\)! So even using that rather crude method yields numbers outside the
1 σrange.Did they really treat the sigmas of their statistical and systematic uncertainty as a form of confidence level? Using \(σ = 0.68\) and computing the \(σ_{95} = σ \cdot \frac{0.95}{0.68}\) and using these values, yields:
- \(σ_{\text{stat., } 95} = 3.09 \cdot \frac{0.95}{0.68} = 4.317\)
- \(σ_{\text{syst., } 95} = 2.20 \cdot \frac{0.95}{0.68} = 3.073\)
⇒ yielding a "limit" of \(-1.136 + 4.317 + 3.073 \approx 6.25\). So scarily that is sort of close…
Aside from the way these numbers are finally computed, the whole computation is based on the fact that the limit is allowed to be negative! All values between \(0 \leq g_{ae} g_{aγ} < x\) are completely ignored, in the sense that some arbitrarily small number may result from this if the input was different. The fact that \(\mathcal{O}(\num{1e-45})\) numbers appear are more or less "by accident".
Indeed, if their data had a larger background and candidates of the same ratio as in their real data, their limit would improve (by keeping the same amount of tracking time). This can be easily seen by increasing the background by a factor, e.g. 10 and rerunning this same analysis. The \(χ²\) scan for just this analysis yields fig. 245, which has the absolute minimum even further in negative values. Thus by using some arbitrary rule to determine a positive coupling limit (e.g. our attempted "shift by \(σ\)" rule, yields a lower number in the non-squared limit!
Figure 245: The \(χ²\) scan using artificially increased larger background and candidates of same ratio as now. This improves the limit by shifting it to even larger negative values. This is one of the biggest reasons I consider the analysis of the paper either flawed, or I completely misunderstand what is being done in this paper.
To summarize the main points:
- The limit calculation allows for negative squared values, which are unphysical.
- The \(χ²\) values as presented in the paper are not reproducible.
- The derivation of the actual limit of \(g_{ae} g_{aγ} = \SI{8.1}{\per\GeV}\) from the squared (negative) values is unclear and not reproducible.
- The papers approach would yield a better limit for a detector with larger background.
Finally, keep in mind that this retracing does not actually take into account any inaccuracies in the X-ray telescope or the efficiency of the detector. So the final limit should get worse including these.
- Note about likelihood function
One possible reason for the too large values of \(χ²\) would be a wrong likelihood function in the paper. In the calculations we do using
mclimit, we use a similar, yet different likelihood function, namely:\[ \mathcal{L} = \prod_j^m \sum_{d' = 0}^{d_\text{observed}} \frac{e^{-(s + b)} (s + b)^{d'}}{d'!} \]
where in addition to the product over the likelihood functions for each channel (i.e. bin), we also sum over all poisson contributions for all values larger than the number of observed candidates.
However, while using this approach yields lower \(χ²\) values, the behavior of the curve does not match the one of the paper anymore. Fig. 246 shows the same figure as fig. 244, but using this different likelihood function (and changing the phase space scanning range). We can see that the computation starts to break down below a certain value of around \(\SI{3e-45}{\per\GeV^2}\). (This was computed by temporarily changing the implementation of the likelihood function above).
Figure 246: Scan of the \(g²_{ae} g²_{aγ} / χ²\) phase space using a different likelihood function (same as for mclimit). Yields lower \(χ²\) values, but curve behavior is wrong and breaks below \(\SI{3e-45}{\per\GeV^2}\).
- UPDATE Notes by Klaus on the above section
Klaus wrote me a mail with the PDF of the above including comments. They are available here:
~/org/Mails/KlausUpdates/klaus_update_04_06_21/klaus_update_04_06_21_comments.pdfThe main takeaway is related to the determination of the actual limit from the best fit result. As he describes the limit is not the best fit of the MLE, but rather it assumes a more "normal" confidence limit calculation. I don't fully understand Klaus' logic in the written comments on pages 12-14, but it seems like essentially one takes some distribution as the basis and considers the 95% one sided range of that distribution. The limit then is the value of that 95% quantile essentially.
My main misunderstanding is what distribution this is supposed to be based on. The \(χ²\) distribution used is only a fit result. Surely we don't use that distribution (or the underlying \(\ln\mathcal{L}\) distribution) as the one to compute values from? In a sense of course this might make sense, but it still seems more than a bit bizarre.
Let's take a look at the actual likelihood distribution we compute during the \(χ²\) scan. For that add a few lines to compute it:
let Ls = block: var res = newSeq[float](couplings.len) for i, el in couplings: let newFlux = flux.rescaleSquares(el) res[i] = L2013(newFlux, backs, cands) res let dfLs = toDf({"Couplings" : couplings, "Ls" : Ls}) .mutate(f{"Couplings" ~ `Couplings` * 1e-12 * 1e-12}) .filter(f{float: `Ls` > 0.0}) echo dfLs ggplot(dfLs, aes("Couplings", "Ls")) + geom_line() + xlim(-6e-45, 6e-45) + xlab("g²_ae g²_aγ") + ylab("Likelihood") + ggtitle("Likelihood values for scan of g²_ae g²_aγ for g_aγ = 1e-12 GeV⁻¹") + ggsave("/home/basti/org/Figs/statusAndProgress/CAST_2013/likelihood_phase_space.pdf") echo dfLs echo dfLs.tail(20) let LsCumSum = dfLs["Ls", float].cumSum(axis = 0) let LsNorm = (LsCumSum /. LsCumSum.sum).toRawSeq var idx = 0 for el in LsNorm: if el >= 0.95: break inc idx echo "Quantile at ", couplings[idx] * 1e-12 * 1e-12, " for index ", idx
The resulting figure of those likelihood values is shown in fig. 247.
One of my assumptions would have been to compute some sort of quantile from this distribution and use the 95% quantile as the value for the real limit. But 1. Klaus talks about the \(\ln\mathcal{L}\) instead of \(\mathcal{L}\) (but that's negative so how does one compute a quantile properly?) and 2. the cut value deduced from this distribution would be too small (as seen from the code above at
3.6e-45instead of the required~6.5e-45). Also because the \(χ²\) curve in the paper is even more narrow than ours, implies that their \(\mathcal{L}\) distribution should be even more narrow as well.Figure 247: Likelihood values \(\mathcal{L}\) of the scanned phase space corresponding to to the \(χ²\) values shown in fig. 244. While the \(\ln\mathcal{L}\) would yield a wider distribution (in pure numbers), computing a quantile seems impossible.
TODO: continue this line of thought after discussing with Klaus.
- Trying to fix χ² values
Starting again from the code mentioned in the previous section to compute the \(χ²\) values:
import std / [math, sequtils, sugar] import ggplotnim, seqmath, nlopt, unchained proc poisson(k: int, λ: float): float = # use mass function to avoid overflows # direct impl: #result = exp(-λ) * λ^k / fac(k).float result = exp(k.float * ln(λ) - λ - lgamma((k + 1).float)) proc L2013*(s, b: seq[float], d: seq[int]): float = result = 1.0 doAssert s.len == b.len doAssert s.len == d.len for i in 0 ..< s.len: result *= poisson(d[i], s[i] + b[i]) proc χ²(s, b: seq[float], d: seq[int]): float = result = -2 * ln(L2013(s, b, d))
where we compute the poisson via the mass function instead of the mathematical definition.
If we now add the background and data into our code (as sequences for to make sure we have the correct data and to reproduce easier) and compute the \(χ²\) value for the case of no signal:
let cands = @[1, 3, 1, 1, 1, 2, 1, 2, 0, 2, 0, 1, 0, 2, 2, 0, 2, 1, 2, 2] let backs = @[2.27, 1.58, 2.40, 1.58, 2.6, 1.05, 0.75, 1.58, 1.3, 1.5, 1.90, 1.85, 1.67, 1.3, 1.15, 1.67, 1.3, 1.3, 2.27, 1.3] let sigs = newSeq[float](backs.len) # empty data for signal echo χ²(sigs, backs, cands)
Now let's change the implementation to use compute the logarithm of the poisson values first before multiplying them (in case we are worried about numerical problems due to too small numbers):
proc lnL2013*(s, b: seq[float], d: seq[int]): float = result = 0.0 doAssert s.len == b.len doAssert s.len == d.len for i in 0 ..< s.len: result += ln(poisson(d[i], s[i] + b[i])) proc χ²Alt(s, b: seq[float], d: seq[int]): float = result = -2 * lnL2013(s, b, d) echo χ²Alt(sigs, backs, cands)
So far this is almost exactly the same.
Now let's go one step further and directly compute the logarithm of the poisson distribution. This yields directly the argument of the mass function based Poisson implementation that we used in the original
poissonprocedure:Starting from the Poisson definition:
\[ P(k; λ) = \frac{e^{-λ} λ^k}{k!} \]
the mass function implementation instead is:
\[ P(k; λ) = \exp\left(k \ln(λ) - λ - \ln(Γ(k + 1))\right) \]
which is simply:
\begin{align} \mathcal{L} &= \prod_i Pois(k_i; λ_i) \\ \ln \mathcal{L} &= \ln \prod_i Pois(k_i; λ_i) \\ \ln \mathcal{L} &= \sum_i \ln Pois(k_i; λ_i) \\ \ln \mathcal{L} &= \sum_i \ln \frac{e^{-λ} λ^k}{k!} \\ \ln \mathcal{L} &= \sum_i \left[ -λ + k \ln(λ) - \ln(k!) \right] \\ \text{using } \ln(k!) &= ln(Γ(k + 1)) \\ \ln \mathcal{L} &= \sum_i \left[ -λ + k \ln(λ) - \ln(Γ(k+1)) \right] \\ \end{align}which, if one only considers the argument of the sum is exactly the log of the mass function.
proc lnPoisson(k: int, λ: float): float = result = (-λ + k * ln(λ))# - lgamma((k+1).float)) proc lnL2013Alt*(s, b: seq[float], d: seq[int]): float = result = 0.0 doAssert s.len == b.len doAssert s.len == d.len for i in 0 ..< s.len: result += lnPoisson(d[i], s[i] + b[i]) proc χ²Alt2(s, b: seq[float], d: seq[int]): float = result = -2 * lnL2013Alt(s, b, d) echo χ²Alt2(sigs, backs, cands)
So also this approach yields exactly the same numbers.
- (Re-)computing a 2013 limit using data extracted from the paper
After the fix of the photon conversion as discussed in the next section and in section 11.2, let's compute a new limit based on the 2013 data.
We will further use the ray tracing result from fig. 80, that is a ray tracing simulation without a detector window or gas absorption. The quantum efficiency of the pn-CCD detector is ignored (it is very high).
In addition we won't consider any systematic uncertainties.
We run the limit calculation at commit ADD COMMIT using:
./limitCalculation --axionModel ../../../AxionElectronLimit/axion_gae_1e13_gagamma_1e-12_no_window_no_gas_after_photon_abs_fix_flux_after_exp_N_10000000.csv \ --optimizeBy "<CLs>" \ --outfile /tmp/data_2013_limit.csv \ --eff 0.8 \ --limit2013
that is we first optimize by expected
CLs.This yields:
CLb = 0.35313 CLs = 0.03952134605418112 CLsb = 0.013956172932112978 <CLs> = 0.05102099877889168 <CLb> = 0.50001 NLOPT_MAXTIME_REACHED (p: @[1.2913472935557367e-10], f: 0.05102099638392309)
which is a limit of \(g_{ae} = 1.29e-10\) assuming a \(g_{aγ} = \SI{1e-12}{\per\GeV}\). The final limit plot is shown in fig. 248.
Figure 248: Final limit optimizing for the \(⟨CL_s⟩\) (expected \(CL_s\), hence the "candidates" shown can be fully ignored) of the 2004 pn-CCD data used in the 2013 paper. It yields a limit of \(g_{ae}g_{aγ} = \SI{1.29e-22}{\per\GeV}\) after the photon conversion fix has been applied. This is after ray tracing without a detector window or gas absorption. An (arbitrary) software efficiency of \(ε = \SI{80}{\percent}\) has been used due to no data about the applied cuts to reach the shown background levels. The resulting data of this limit calculation is:
axion signal Energy background exp. cand. 2.348 0.8 2.27 1 2.412 1.071 1.58 3 2.174 1.343 2.4 1 1.982 1.614 1.58 1 1.779 1.886 2.6 1 1.522 2.157 1.05 2 1.264 2.429 0.75 1 0.9741 2.7 1.58 2 0.7227 2.971 1.3 0 0.5349 3.243 1.5 2 0.3957 3.514 1.9 0 0.3069 3.786 1.85 1 0.2346 4.057 1.67 0 0.1933 4.329 1.3 2 0.1696 4.6 1.15 2 0.1419 4.871 1.67 0 0.1231 5.143 1.3 2 0.1042 5.414 1.3 1 0.0813 5.686 2.27 2 0.0587 5.957 1.3 2 0 6.228 0 0 Now we will re-run the limit using the background only data of 2017/18. For this we will use a ray tracing output that does include both the detector window as well as gas absorption of course. And again we optimize for the expected
CLs.The command we run:
./limitCalculation -b ../../Tools/backgroundRateDifferentEffs/out/lhood_2017_eff_0.8.h5 \ -b ../../Tools/backgroundRateDifferentEffs/out/lhood_2018_eff_0.8.h5 \ --axionModel ../../../AxionElectronLimit/axion_gae_1e13_gagamma_1e-12_flux_after_exp_N_10000000.csv \ --optimizeBy "<CLs>" \ --outfile /tmp/limit_2018_exp_cls.csv \ --eff 0.8
This yields:
CLb = 0.42891 CLs = 0.047280835600692024 CLsb = 0.020279223197492816 <CLs> = 0.05020267428478678 <CLb> = 0.50001 NLOPT_MAXTIME_REACHED (p: @[1.1058593749999999e-10], f: 0.050516099563680784)
which is a limit of \(g_{ae} = 1.11e-10\) assuming a \(g_{aγ} = \SI{1e-12}{\per\GeV}\).
It yields the following final plot as shown in fig.
Figure 249: Final limit optimizing for the \(⟨CL_s⟩\) (expected \(CL_s\), hence the "candidates" shown can be fully ignored) for the 2017/18 dataset. It yields a limit of \(g_{ae}g_{aγ} = \SI{1.29e-22}{\per\GeV}\) after the photon conversion fix has been applied. This is after ray tracing with a detector window and gas absorption. Our software efficiency of \(ε = \SI{80}{\percent}\) is used. The resulting data of this limit calculation (there are more bins in our energy range, which is also wider):
axion signal Energy background exp. cand. 0.00168 0 0.1022 0 0.07747 0.2 0.1534 0 0.1854 0.4 0.4601 0 0.4536 0.6 1.125 1 0.648 0.8 1.431 2 0.7907 1 1.636 2 0.7782 1.2 0.6135 1 0.7595 1.4 0.4601 0 0.7102 1.6 0.3067 1 0.5978 1.8 0.7157 0 0.5019 2 0.3067 0 0.4248 2.2 0.3579 1 0.3584 2.4 0.1534 0 0.2772 2.6 0.409 0 0.1956 2.8 0.6646 0 0.1521 3 1.278 2 0.2107 3.2 1.176 0 0.1686 3.4 1.176 1 0.143 3.6 0.7669 0 0.1139 3.8 0.5112 0 0.09143 4 0.1534 0 0.07709 4.2 0.1022 0 0.06915 4.4 0.1022 0 0.05822 4.6 0.1022 0 0.05121 4.8 0.1022 0 0.04503 5 0.2556 1 0.03912 5.2 0.3579 0 0.03487 5.4 0.5112 1 0.02772 5.6 0.4601 1 0.02147 5.8 0.7157 2 0.01606 6 0.2045 0 0.01105 6.2 0.5624 0 0.006703 6.4 0.3579 0 0.001406 6.6 0.3579 0 0 6.8 0.3579 0 0 7 0.2556 0 0 7.2 0.3067 0 0 7.4 0.2045 0 0 7.6 0.2045 1 0 7.8 0.3579 1 0 8 0.5112 2 0 8.2 0.7669 1 0 8.4 0.9202 2 0 8.6 0.7669 0 0 8.8 1.227 1 0 9 1.227 3 0 9.2 1.585 2 0 9.4 1.125 1 0 9.6 0.9202 1 0 9.8 0.9202 0 0 10 0 0
17.5.2. Photon conversion woes
During the study and retracing of the computations of the 2013 \(g_{ae}\) paper, I realized that the conversion from axions via the inverse Primakoff effect in the magnet is a bit more problematic than I initially realized (at least while writing the first notes about the 2013 paper).
The conversion obviously happens via:
\[ P_{a↦γ, \text{vacuum}} = \left(\frac{g_{aγ} B L}{2} \right)^2 \left(\frac{\sin\left(\delta\right)}{\delta}\right)^2 \]
where essentially for small masses the \(\sin(x)/x\) term becomes 1.
The tricky part now is the units of the \(g_{aγ} B L\) product. They need to become 1 of course.
Units of each part:
\begin{align} [g_{aγ}] &= \si{\per\GeV} \\ [B] &= \si{\tesla} \\ [L] &= \si{\m} \end{align}In terms of mass scales this works out, because \(B\) has mass scale 2 and \(L\) -1. So all in all it's of power 0 in mass. Now there's two possibilities:
- convert the coupling constant to \(J^{-1}\). This however begs the question, whether the coupling constant really only has units of inverse energy or it's actually more complicated than that? One might look at the lagrangian density (itself units of energy / volume \(\si{\joule\m^{-3}}\)) and look at the related terms to check whether something is missing or simply look at the units of the conversion probability to check for the missing units.
- convert \(B, L\) to natural units.
The latter is a bit simpler, as long as one converts both correctly. The problem now boils down to convention of natural units. Typically (at least in particle physics) people simply assume \(c = \hbar = 1\). This however leaves unspecified the correct way to convert amperes to natural units (and thus tesla). If one then is not careful, one might easily use the wrong conversion factor!
Things are more murky, because we might not know for certain (!) which notation is being used by the people having written the papers about the conversion probabilities in the first place!
Different sources all write different things about it. Wikipedia lists many different conventions: https://en.wikipedia.org/wiki/Natural_units but confusingly their definition for "particle and atomic physics" goes one step further and even sets \(m_e\) to 1!
The following lists more:
Confusingly one of the most prominent results from a quick search links to the following PDF: http://ilan.schnell-web.net/physics/natural.pdf This however sets \(4πε_0 = 1\) instead of the (in my experience) more common \(ε_0 = 1\).
In my understanding this is related to Lorentz-Heaviside units vs. Gauss units:
- https://en.wikipedia.org/wiki/Lorentz%E2%80%93Heaviside_units
- https://en.wikipedia.org/wiki/Gaussian_units
If one starts with the former and wishes to get to natural units, it makes sense to set \(c = \hbar = μ_0 = ε_0 = k_b = 1\) whereas starting from the latter it may be more convenient to set \(4πε_0 = 1\).
One comparison of conversions can be made with GNU units
(https://www.gnu.org/software/units/) using this natural_units file:
https://github.com/misho104/natural_units,
which explicitly mentions to base their units on Lorentz-Heaviside
units (i.e. \(ε_0 = μ_0 = 1\)).
- Comparison of previous computations
In any case though, let's look at the different implementations of axion to photon conversions in our code of the past.
- Code of my master thesis
First the code in my master thesis, in file ./../../CastData/AxionElectronAnalysis/src/signalGenerator.cc:
// in order to get the photon flux from the axion flux, we need the conversion // probability factor Double_t conversionProb; Double_t g_agamma = SIGNALGENERATOR_G_A_GAMMA; Double_t B = SIGNALGENERATOR_B_FIELD; Double_t L = SIGNALGENERATOR_B_LENGTH; Double_t m_a = SIGNALGENERATOR_AXION_MASS; // q is the momentum transfer of the axion to photon. below mass of axion of 10meV // given by q = m_a^2 / (2w) Double_t q; // delta is the argument of the sin Double_t delta; // B and L given in T and m. convert both to ~keV // 1 T = (eV^2 / (1.44 * 10^(-3))) = 10^(-6) keV^2 / (1.44 * 10^(-3)) B = B / 6.94e-4; // L conversion to 1/eV and then 1/keV L = L / (1.97227e-7 * 1e-3); // with these variables, we can calculate the conversion probability when we // run over the file for each energy while(axionSpecIfStream.good() && !axionSpecIfStream.eof()){ // if not a comment, line should be added // before we can add intensity, we need to include the conversion // probability from axions to photons iss_line >> _energy >> _axionIntensity; // calculate q using energy (m_a and _energy given in keV) q = m_a * m_a / (2.0 * _energy); // calculate argument of sin delta = q * L / 2.0; conversionProb = TMath::Power( ( (g_agamma * B * L / 2.0) * TMath::Sin(delta) / delta ) , 2 ); // now we just need to multiply our intensity with this factor // need to also multiply by 10**-19, since flux in file given in 1 / (10**19 keV etc...) _axionIntensity = _axionIntensity * conversionProb * 1.e-19; }
where I have removed most of the parsing related code as it's not part of the actual computations. From this code we can see that \(B\) is converted to \(\si{\eV^2}\) by division with \(\num{6.94e-4}\) and the length to \(\si{\eV^{-1}}\) by division with \(\num{1.973e-7}\). Note that there is a comment about the conversion of \(B\) that isn't actually used confusingly.
In nim:
import math func conversionProb(B, g_aγ, L: float): float = let B = B / 6.94e-4 let L = L / (1.97227e-7 * 1e-3) let g_aγ = g_aγ * 1e-6 # from GeV⁻¹ to keV⁻¹ result = pow( (g_aγ * B * L / 2.0), 2.0 ) echo conversionProb(1.0, 1.0, 1.0)
13344070575464.35
No matter what I do I cannot get reasonable numbers from this. In the other examples I use \(\SI{1}{\per\GeV}\), which should be \(1.0 * 1e-6\), because we deal with \(\si{\per\keV}\) here.
I'm confused.
- Code of current ray tracer by Johanna
The code doing the current ray tracing also contains the axion to photon conversion. I refactored the code somewhat a few months ago, but didn't check the implementation of the conversion.
The code is in ./../../CastData/ExternCode/AxionElectronLimit/raytracer2018.nim:
func conversionProb*(B, g_agamma, length: float): float {.inline.} = result = 0.025 * B * B * g_agamma * g_agamma * (1 / (1.44 * 1.2398)) * (1 / (1.44 * 1.2398)) * (length * 1e-3) * (length * 1e-3) #g_agamma= 1e-12 echo conversionProb(1.0, 1.0, 1e3)
0.00784353358442401
Here we see some other confusing things. Instead of a factor of
1/4we have1/40. Further the conversion of \(B\) and \(L\) to appropriate \(\si{\eV}\) units is done via factors of \(1/(1.44 \cdot 1.2398)\) instead. The \(\num{1.44}\) appears if one uses \(4πε_0 = 1\) for the conversion from tesla to electronvolt, but is missing a factor of1e-3in addition. The1.2398I can't make sense of. The length conversion with1e-3is simply because the lengths are given in \(\si{\mm}\) in the code.I suppose the
1/40and missing factors forg_agamma(it is really given inGeV⁻¹) are related to some contraction of all powers of 10 into one factor? - Code computing probability using
unchained
Spurred by this, I finally implemented a basic natural unit conversion proc into
unchained. With it we can write:import unchained, math defUnit(GeV⁻¹) func conversionProb(B: Tesla, L: Meter, g_aγ: GeV⁻¹): UnitLess = result = pow( (g_aγ * B.toNaturalUnit * L.toNaturalUnit / 2.0), 2.0 ) echo 1e-12.GeV⁻¹ * 9.T.toNaturalUnit() * 9.26.m.toNaturalUnit() echo 1.eV⁻¹ * 1.T.toNaturalUnit() * 9.0.m.toNaturalUnit() echo conversionProb(1.T, 1.m, 1.GeV⁻¹)
8.25062e-11 UnitLess 8.90996e+09 UnitLess 0.245023 UnitLess where we left out the
sin(x)/xterm of the probability.The
unchainedconvention uses \(c = \hbar = ε_0 = 1\), which (I hope) is the correct convention to use. - Cross checking with GNU units
Using the
natural_unitsfile mentioned above, we can compute the correct conversion factors for the equation (at least with the used conventions).units -f ~/src/natural_units/natural.units > You have: (1 GeV^-1 * 1 T * 1 m)^2 / 4 > You want: 1 > * 0.24502264 > / 4.0812555
So the absolute multiplier using \(g_{aγ} = \SI{1}{\per\GeV}, B = \SI{1}{\tesla}, L = \SI{1}{\m}\) is \(\sim\num{0.245}\).
This is precisely the result we also get in 17.5.2.1.3.
- Code of my master thesis
17.6. Applying the likelihood method (c/f 2013 paper) to 2017/18 data
With the method of the 2013 paper now understood (see section 17.5.1), it's a good idea to apply the same method to our own data as a comparison and cross check.
We will start from the code written in the previously mentioned section and add the required changes for our data.
First the required code to get started, including the telescope
efficiency conversion (factor 2.64 in scaleToTracking).
import std / [math, sequtils, sugar, strformat] import ggplotnim, seqmath, nlopt import unchained, math defUnit(GeV⁻¹) defUnit(keV⁻¹•yr⁻¹•m⁻²) defUnit(keV⁻¹•h⁻¹•m⁻²) proc poissonMF(k: int, λ: float): float = # use mass function to avoid overflows # direct impl: #result = exp(-λ) * λ^k / fac(k).float result = exp(k.float * ln(λ) - λ - lgamma((k + 1).float)) proc L2013*(s, b: seq[float], d: seq[int]): float = result = 1.0 doAssert s.len == b.len doAssert s.len == d.len for i in 0 ..< s.len: result *= poissonMF(d[i], s[i] + b[i]) proc χ²(s, b: seq[float], d: seq[int]): float = result = -2 * ln(L2013(s, b, d)) proc readAxModel(): DataFrame = let upperBin = 10.0 result = readCsv("/home/basti/CastData/ExternCode/AxionElectronLimit/axion_diff_flux_gae_1e-13_gagamma_1e-12.csv") .mutate(f{"Energy / keV" ~ c"Energy / eV" / 1000.0}) .filter(f{float: c"Energy / keV" <= upperBin}) proc rebin(bins, counts, toBins: seq[float], areRate = false): seq[float] = ## `toBins` must be less than `bins` doAssert toBins.len < bins.len, "Rebinning to more bins not supported" var count = 0 var binIdx = -1 var sumBin = 0.0 for i, el in counts: let bin = bins[i] if binIdx < 0 and bin.float > toBins[0]: binIdx = 1 elif binIdx >= 0 and bin.float > toBins[binIdx]: if areRate: result.add (sumBin / count) else: result.add sumBin count = 0 sumBin = 0.0 inc binIdx sumBin = sumBin + el inc count if binIdx > toBins.high: break # add current sum as final entry if areRate: result.add sumBin / count else: result.add sumBin proc rebinDf(df: DataFrame, binWidth: float, bins: seq[float]): seq[keV⁻¹•yr⁻¹•m⁻²] = let energy = df["Energy / keV", float].toRawSeq flux = df["Flux / keV⁻¹ m⁻² yr⁻¹", float].toRawSeq var count = 0 var binIdx = -1 result = rebin(energy, flux, bins, areRate = true).mapIt(it.keV⁻¹•yr⁻¹•m⁻²) echo bins.mapIt(it + 0.5 * binWidth) let dfPlot = toDf({ "E" : bins[0 ..< ^1].mapIt(it + 0.5 * binWidth), "F" : result.mapIt(it.float) }) echo dfPlot.pretty(-1) ggplot(dfPlot, aes("E", "F")) + geom_point() + xlab("Energy [keV]") + ylab("Flux [keV⁻¹•yr⁻¹•m⁻²]") + ggtitle("Flux using data binning, g_ae = 1e-13, g_aγ = 1e-12 GeV⁻¹") + ggsave("/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/rebinned_df_flux_2018.pdf") func conversionProb(): UnitLess = ## simplified vacuum conversion prob. for small masses let B = 9.0.T let L = 9.26.m let g_aγ = 1e-12.GeV⁻¹ result = pow( (g_aγ * B.toNaturalUnit * L.toNaturalUnit / 2.0), 2.0 ) proc scaleToTracking(x: keV⁻¹•yr⁻¹•m⁻², binWidth: keV, trackingTime: Hour, useConstTelEff = true): UnitLess = ## Convert the given flux in `keV⁻¹•yr⁻¹•m⁻²` to raw counts registered ## on the chip (assuming a perfect telescope!) during the full tracking period ## within a single energy bin. # convert to `h⁻¹` (`yr` treated as distinct otherwise) let input = x.to(keV⁻¹•h⁻¹•m⁻²) let areaBore = π * (2.15 * 2.15).cm² # area of bore in cm² if useConstTelEff: # factor 2.64 is a crude estimate of telescop efficiency of ~ 5.5 cm^2 result = input * binWidth * areaBore * trackingTime / 2.64 else: result = input * binWidth * areaBore * trackingTime proc rescale(s: seq[float], new: float): seq[float] = ## rescaling version, which takes a `new` squared coupling constant ## to allow for negative squares let old = 1e-13 # initial value is always 1e-13 result = newSeq[float](s.len) for i, el in s: result[i] = el * new / (old * old) template linearScan(fn: untyped, flux, backs, cands: untyped): DataFrame {.dirty.} = block: let vals = block: var res = newSeq[float](couplings.len) for i, el in couplings: let newFlux = flux.rescale(el) res[i] = fn(newFlux, backs, cands) res toDf({"CouplingsRaw" : couplings, astToStr(fn) : vals}) .mutate(f{"Couplings" ~ `CouplingsRaw` * 1e-12 * 1e-12}) type Limit = object coupling: float χ²: float proc computeχ²(flux, backs: seq[float], cands: seq[int], coupling: float): float = let newFlux = flux.rescale(coupling) result = χ²(newFlux, backs, cands) proc performScans(flux, backs: seq[float], cands: seq[int], range = (-6e-45, 6e-45), couplings: seq[float] = @[], toPlot = true, verbose = true, suffix = ""): Limit = # due to `linearScan` being `dirty` template can define couplings, gaγ here let g_aγ² = 1e-12 * 1e-12 let couplings = if couplings.len > 0: couplings else: linspace(range[0] / g_aγ², range[1] / g_aγ², 5000) let couplingStep = couplings[1] - couplings[0] let dfχ² = linearScan(χ², flux, backs, cands) .filter(f{ `χ²` <= 150.0 }) let χ²vals = dfχ²["χ²", float] let χ²argmin = χ²vals.toRawSeq.argmin let χ²min = χ²vals[χ²argmin] # need to read the minimum coupling from DF, because input coupling contains χ² equivalent # NaN values (`χ²vals.len != couplings.len` is the problem) let couplingsRaw = dfχ²["CouplingsRaw", float] proc findLimit(vals: seq[float], start: int, couplingStart: float): float = ## walk to right until start + 5.5 and print value. We step through the ## couplings using the step size in couplings defined by the input ## `couplings` sequence. ## Returns the coupling constant of the limit (χ² + 5.5) let startVal = vals[start] #echo "Min χ² ", startVal, " at index ", start var curCoupling = couplingStart var curχ² = startVal while curχ² < startVal + 5.5: #echo "Current χ² ", curχ², " at coupling ", curCoupling, " still smaller ", startVal + 5.5 # compute next coupling step and χ² value curCoupling += couplingStep curχ² = computeχ²(flux, backs, cands, curCoupling) result = curCoupling let couplingLimit = findLimit(χ²vals.toRawSeq, χ²argmin, couplingsRaw[χ²argmin]) if verbose: echo "Limit of χ² (+ 5.5) is = ", couplingLimit result = Limit(coupling: couplingLimit, χ²: computeχ²(flux, backs, cands, couplingLimit)) if toPlot: echo dfχ² ggplot(dfχ², aes("Couplings", "χ²")) + geom_line() + #ylim(50, 100) + xlim(-6e-45, 6e-45) + xlab("g²_ae g²_aγ") + ggtitle("Scan of g²_ae g²_aγ for g_aγ = 1e-12 GeV⁻¹") + ggsave(&"/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/brute_force_chi2_scan_{result.χ²}{suffix}.pdf") let dfLs = linearScan(L2013, flux, backs, cands).filter(f{`L2013` > 0.0}) echo dfLs ggplot(dfLs, aes("Couplings", "L2013")) + geom_line() + xlim(-6e-45, 6e-45) + xlab("g²_ae g²_aγ") + ylab("Likelihood") + ggtitle("Likelihood values for scan of g²_ae g²_aγ for g_aγ = 1e-12 GeV⁻¹") + ggsave(&"/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/likelihood_phase_space_{result.χ²}{suffix}.pdf") echo dfLs echo dfLs.tail(20) let LsCumSum = dfLs["L2013", float].cumSum(axis = 0) let LsNorm = (LsCumSum /. LsCumSum.sum).toRawSeq var idx = 0 for el in LsNorm: if el >= 0.95: break inc idx echo "Quantile at ", couplings[idx] * 1e-12 * 1e-12, " for index ", idx proc readFluxAndPlot(binWidth: float, bins: seq[float], trackingTime: Hour, useConstTelEff = true, telEff: seq[float] = @[], title = "", suffix = "", toPlot = true, telEffIsNormalized = false): seq[float] = # never use constant effective area if specific value sgiven let useConstTelEff = if telEff.len > 0: false else: useConstTelEff result = readAxModel().rebinDf(binWidth, bins) .mapIt((scaleToTracking(it, binWidth.keV, trackingTime, useConstTelEff = useConstTelEff) * conversionProb()).float) if telEff.len > 0: doAssert telEff.len == bins.len doAssert telEff.len == result.len, " tel eff " & $telEFf.len & " " & $result.len for i in 0 ..< result.len: if not telEffIsNormalized: result[i] *= telEff[i] / (π * (2.15 * 2.15)) # compute efficiency from effective area else: result[i] *= telEff[i] # input is already a pure efficiency (possibly including SB + ε) let fluxInTracking = toDf({ "E" : bins, "F" : result }) let titlePrefix = &"Expected X-ray flux for g_ae = 1e-13, g_aγ = 1e-12 during {trackingTime} of tracking" let title = if title.len == 0 and not useConstTelEff: titlePrefix & " assuming perfect X-ray optics" elif title.len == 0 and useConstTelEff: titlePrefix & " assuming constant telescope eff area ~5.5 cm² (factor 2.64)" else: titlePrefix & title if toPlot: ggplot(fluxInTracking, aes("E", "F")) + geom_point() + xlab("Energy [keV]") + ylab("Counts") + margin(top = 2.0) + ggtitle(title) + ggsave(&"/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/flux_tracking_gae_1e-13_gag_1e-12_useConstTelEff_{useConstTelEff}{suffix}.pdf") if result.len == bins.len: # remove the last bin (it's technically just the bin of the right most edge!) result = result[0 ..< ^1]
With this in place we just need a bit of code to read our actual data and feed into our:
- bin width
- bin edges
- tracking time
We can take that code straight from the proper limit calculation code in ./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/limitCalculation.nim.
import nimhdf5, os import ingrid / tos_helpers proc readDsets(h5f: H5FileObj, names: varargs[string]): DataFrame = ## reads all likelihood data in the given `h5f` file as well as the ## corresponding energies. Flattened to a 1D seq. ## This proc is for TPA generated H5 files! (i.e. containing run_* groups, ...) # iterate over all groups, read all likelihood and energy dsets result = newDataFrame() for name in names: var data = newSeq[float]() for run, grp in runs(h5f, likelihoodBase()): let group = h5f[grp.grp_str] let centerChip = if "centerChip" in group.attrs: "chip_" & $group.attrs["centerChip", int] else: "chip_3" if grp / centerChip in h5f: doAssert grp / centerChip / name in h5f[(group.name / centerChip).grp_str] data.add h5f[grp / centerChip / name, float64] else: echo &"INFO: Run {run} does not have any candidates" result[name] = toColumn data proc flatten(dfs: seq[DataFrame]): DataFrame = ## flatten a seq of DFs, which are identical by stacking them for df in dfs: result.add df.clone proc readFiles(s: seq[H5File]): DataFrame = result = s.mapIt( it.readDsets(likelihoodBase(), some((chip: 3, dsets: @["energyFromCharge"]))) .rename(f{"Energy" <- "energyFromCharge"})).flatten import random / mersenne import alea / [core, rng, gauss, poisson] proc drawExpCand(h: seq[float], rnd: var Random): seq[int] = ## given a histogram as input, draws a new histogram using Poisson ## statistics var pois: Poisson result = newSeq[int](h.len) for i in 0 ..< h.len: let cnt = h[i] pois = poisson(cnt) let cntDraw = rnd.sample(pois) result[i] = cntDraw.int proc toHisto(df: DataFrame, nBins = 50): (seq[float], seq[float]) = let energy = df["Energy", float].toRawSeq let (histo, bins) = histogram(energy, range = (0.0, 10.0), bins = nBins) # rescale to tracking time let trackToBackRatio = 19.56 # this is the ratio of background to tracking result[0] = histo.mapIt(it.float / trackToBackRatio) result[1] = bins
With all required procedures in place, we can now insert our data files (background, ε = 80%), draw a set of expected candidates from a Poisson and compute a toy limit:
let path = "/home/basti/CastData/ExternCode/TimepixAnalysis/Tools/backgroundRateDifferentEffs/out/" let backFiles = @["lhood_2017_eff_0.8.h5", "lhood_2018_eff_0.8.h5"] var df2018 = readFiles(backFiles.mapIt(H5file(path / it, "r"))) let (backs, bins) = df2018.toHisto() # a random number generator var rnd = wrap(initMersenneTwister(299792458)) let cands = drawExpCand(backs, rnd) echo backs echo bins echo cands ## using constant telescope efficiency of effective area ~5.5 cm² let flux = readFluxAndPlot(bins[1] - bins[0], bins, 180.Hour) echo "The initial χ² value is: ", χ²(flux, backs, cands) # now scan discard performScans(flux, backs, cands)
Limit of χ² (+ 5.5) is = 4.778155631125587e-21
So we get a (single toy) limit of \(g²_{ae} = 4.78-21\) if we take the similar Δχ² of about 5.5.
Now let's write a limit calculation procedure, which does a Monte Carlo of the scans, each time with a different set of candidates.
proc plotScatter(bins: seq[float], flux, backs: seq[float], cands: seq[int], limit: float, nBins = 50, suffix = "") = ## creates a plot of the flux, background and candidates let flux = flux.rescale(limit) let df = toDf(flux, backs, cands, bins) .gather(@["flux", "backs", "cands"], key = "Type", value = "Counts") #echo flux #echo df.pretty(-1) ggplot(df, aes("bins", "Counts", color = "Type")) + geom_point() + margin(top = 2) + ggtitle(&"Flux, background and toy candidates at a limit of g²_ae = {limit}") + ggsave(&"/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/scatter_flux_backs_cands_bins_{nBins}{suffix}.pdf") proc monteCarloLimit(bins: seq[float], flux, backs: seq[float], rnd: var Random, nBins = 50, suffix = "", verbose = true, fn = performScans): DataFrame = ## Returns the DataFrame of the coupling constants and χ² values at the determined limit. const nmc = 5000 var limits = newSeq[Limit](nmc) var cands: seq[int] let g_aγ² = 1e-12 * 1e-12 let couplings = linspace(-6e-45 / g_aγ², 6e-45 / g_aγ², 5000) for i in 0 ..< limits.len: # draw a set of candidates if i mod 500 == 0: echo "Iteration ", i cands = drawExpCand(backs, rnd) #echo "New candidates ", cands limits[i] = fn(flux, backs, cands, couplings = couplings, toPlot = false, verbose = verbose) if i == 0: plotScatter(bins, flux, backs, cands, limits[i].coupling, nBins = nBins, suffix = suffix) if verbose: echo limits # plot the different limits as scatter plot let df = toDf({ "χ²" : limits.mapIt(it.χ²), "Coupling" : limits.mapIt(it.coupling)}) ggplot(df, aes("Coupling", "χ²")) + geom_point() + xlab("Coupling limit g²_ae and g_aγ = 1e-12") + ggtitle("χ² values at coupling limit") + ggsave(&"/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/mc_likelihood_bins_{nBins}{suffix}.pdf") ggplot(df, aes("Coupling")) + geom_histogram(bins = 100, hdKind = hdOutline) + ggtitle("Limits on coupling constant g²_ae for g_aγ = 1e-12 of toy experiments") + ggsave(&"/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/mc_likelihood_2018_histogram_bins_{nBins}{suffix}.pdf") echo "Mean coupling constant of MC toy experiments (for g_aγ = 1e-12 GeV⁻¹): ", df["Coupling", float].mean result = df #discard monteCarloLimit(bins, flux, backs, rnd)
Mean coupling constant of MC toy experiments (for g_aγ = 1e-12 GeV⁻¹): 6.4455161432276914e-21
We created two plots from the sets of toy experiments. Fig. 250 shows a scatter plot of the χ² values at each coupling limit for each toy experiment. It's more of a cross check to see that each limit is sensible and different.
Then fig. 251 is the histogram of all toy experiments in the coupling limits only. The mean of it is our expected coupling limit.
The limit for \(g²_{ae}\) we get from the MC is \(g²_{ae} = \num{6.44e-21}\), which reduces to \(g_{ae} = \num{8.03e-11}\) or \(g_{ae} g_{aγ} = \SI{8.03e-23}{\per\GeV}\).
The individual \(χ²\) distributions look very different each time. In some cases like fig. 252 the χ² distribution jumps straight from decreasing to ∞. In other cases the shape is much better defined and has a steady increase like fig. 253. If one were to zoom in much more, an increase could certainly be found in the first example, but on a much smaller scale in coupling constant change.
If we look at an example of the counts for signal, background and flux at a coupling limit, see fig. 254, we can see that many of our candidate bins are zero. For that reason it may be a good idea to perform another MC limit calculation by using a smaller binning to get less
From here a few more things are interesting to test.
- computing the limit with fewer bins, e.g. 20
- computing the limit with a signal efficiency of ε = 60%
- including the correct telescope efficiency for each detector (the LLNL values are in ./../Code/CAST/XrayFinderCalc/llnl_telescope_trans_cropped.txt)
- utilize our ray tracing result for both datasets
17.6.1. Limit with 20 bins
So let's change the toHisto parameter for the number of bins
used. Let's try 20 instead of 50 and see what the limit histogram and
such a scatter plot of the counts look like.
block: let nBins = 20 let (backs, bins) = df2018.toHisto(nBins = nBins) let flux = readFluxAndPlot(bins[1] - bins[0], bins, 180.Hour, title = " using 20 bins, tel eff. area ~5.5cm²", suffix = "_bins_20") #discard monteCarloLimit(bins, flux, backs, rnd, nBins = nBins)
Mean coupling constant of MC toy experiments (for g_aγ = 1e-12 GeV⁻¹): 6.6883919183826745e-21
In this case the plot of a single example of counts including candidates is shown in fig. 255. We see that now we have counts in more bins. The histogram of the limit computation is fig. 256. This yields a limit of \(g²_{ae} = \num{6.69e-21}\) or \(g_{ae} = \num{8.18e-11}\).
\clearpage
17.6.2. Limit with ε = 60%
Number 2 is pretty simple. All we need to do is read the different input files (ε = 60% instead of 80%). For the following computations we will again use 50 bins.
block: let backFiles = @["lhood_2017_eff_0.6.h5", "lhood_2018_eff_0.6.h5"] var df2018_60 = readFiles(backFiles.mapIt(H5file(path / it, "r"))) let (backs, bins) = df2018_60.toHisto() let flux = readFluxAndPlot(bins[1] - bins[0], bins, 180.Hour, title = " using ε = 60%, tel eff. area ~5.5cm²", suffix = "_eff_0.6") #discard monteCarloLimit(bins, flux, backs, rnd, suffix = "_eff_60")
Mean coupling constant of MC toy experiments (for g_aγ = 1e-12 GeV⁻¹): 4.467202240447578e-21
In this case we get a limit of \(g²_{ae} = \num{4.47e-21}\) or \(g_{ae} = \num{6.69e-11}\).
And again the two plots, example counts and histogram in figs. 257 and 258.
\clearpage
17.6.3. TODO Limit using exact telescope efficiencies
[ ]UPDATE THE LLNL EFFICIENCY WITH CORRECT BEHAVIOR! See journal [BROKEN LINK: sec:journal:2023_07_13]!!
First we need to source the telescope efficiencies.
For the LLNL telescope we already have the data file in:
./../Code/CAST/XrayFinderCalc/llnl_telescope_trans_cropped.txt
extracted from the plot in fig. 259,
which is found in the paper arxiv:1509.06190.
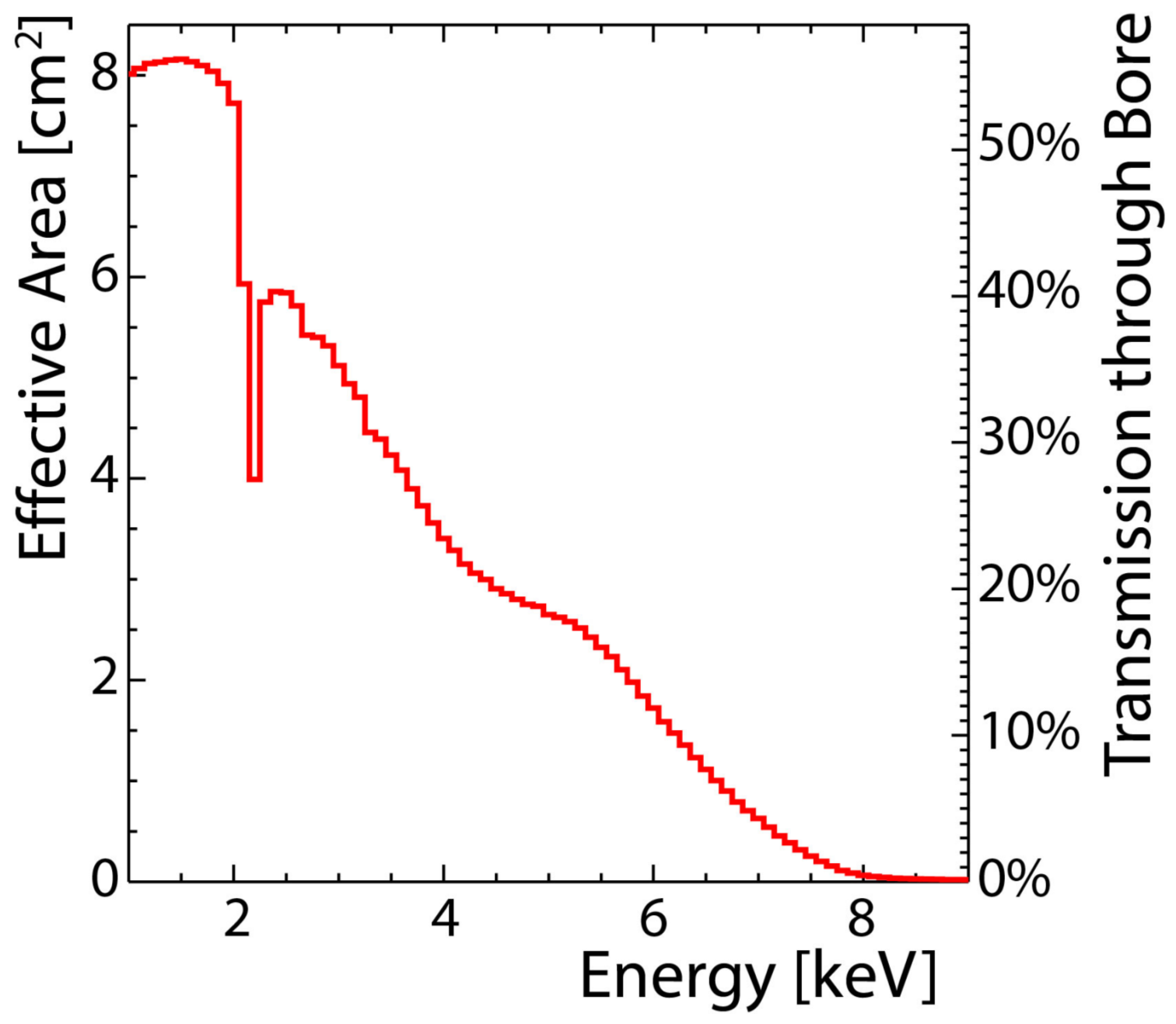
The data is extracted using ./../Misc/extractDataFromPlot.nim. The resulting extracted data is stored in ./../resources/llnl_xray_telescope_cast_effective_area.csv.
For the MPE telescope we get the corresponding plot in the paper
https://iopscience.iop.org/article/10.1088/1367-2630/9/6/169/pdf
or ~/org/Papers/the_xray_telescope_of_cast_mpe_10.1088.pdf.
The figure we need to extract data from is fig. 260.
Its data is stored accordingly in ./../resources/mpe_xray_telescope_cast_effective_area.csv.
Let's plot both together.
import ggplotnim, os, strutils, sequtils, unchained proc readFiles(fs: seq[string], names: seq[string]): DataFrame = result = newDataFrame() for i, f in fs: var df = readCsv(f) df["File"] = constantColumn(names[i], df.len) result.add df let boreArea = π * (2.15 * 2.15).cm² result = result.mutate(f{"Transmission" ~ idx("EffectiveArea[cm²]") / boreArea.float}) let path = "/home/basti/org/resources/" let df = readFiles(@["llnl_xray_telescope_cast_effective_area.csv", "mpe_xray_telescope_cast_effective_area.csv"].mapIt(path / it), names = @["LLNL", "MPE"]) df.writeCsv("/tmp/llnl_mpe_effective_areas.csv") ggplot(df, aes("Energy[keV]", "EffectiveArea[cm²]", color = "File")) + geom_line() + ggtitle("Comparison of the effective area of the LLNL and MPE telescopes") + ggsave("/home/basti/org/Figs/statusAndProgress/llnl_mpe_effective_area_comparison.pdf") ggplot(df, aes("Energy[keV]", "Transmission", color = "File")) + geom_line() + ggtitle("Comparison of the transmission of the LLNL and MPE telescopes") + ggsave("/home/basti/org/Figs/statusAndProgress/llnl_mpe_transmission_comparison.pdf")
This yields fig. 261.
To apply these telescope efficiencies to a limit calculation we need to do 1 1/2 things:
- for the LLNL telescope we need to extend the data range to cover the full 0 - 10 keV
- we need to rebin the efficiency data to match our counts to actually apply it to the expected flux
The former we will do by computing the ratio between LLNL and MPE at the boundaries of the LLNL and then extending the efficiency by:
\[ ε_{\text{LLNL,extension}}(E) = \frac{ε_{\text{LLNL,@bound}}}{ε_{\text{MPE,@bound}}} \cdot ε_{\text{MPE}}(E) \]
Extension can be done independent of the limit code above. For the
rebinning we will use the rebin procedure we defined in the limit
code.
First let's extend the effective area and store it in a new CSV file.
var dfLLNL = df.filter(f{`File` == "LLNL"}) .arrange("Energy[keV]") let dfMPE = df.filter(f{`File` == "MPE"}) .arrange("Energy[keV]") let boundsLLNL = (dfLLNL["EffectiveArea[cm²]", float][0], dfLLNL["EffectiveArea[cm²]", float][dfLLNL.high]) let dfMpeLess = dfMPE.filter(f{ idx("Energy[keV]") <= 1.0 }) let dfMpeMore = dfMPE.filter(f{ idx("Energy[keV]") >= 9.0 }) let mpeAtBounds = (dfMpeLess["EffectiveArea[cm²]", float][dfMpeLess.high], dfMpeMore["EffectiveArea[cm²]", float][0]) echo boundsLLNL echo mpeAtBounds echo dfMpeLess.pretty(-1) let ratios = (boundsLLNL[0] / mpeAtBounds[0], boundsLLNL[1] / mpeAtBounds[1]) echo ratios let Eless = dfMpeLess["Energy[keV]", float] let εless = dfMpeLess["EffectiveArea[cm²]", float] let εLlnlLess = εless.map_inline(x * ratios[0]) let Emore = dfMpeMore["Energy[keV]", float] let εmore = dfMpeMore["EffectiveArea[cm²]", float] let εLlnlMore = εmore.map_inline(x * ratios[1]) var dfExt = toDf({ "Energy[keV]" : concat(Eless, dfLLNL["Energy[keV]", float], Emore, axis = 0), "EffectiveArea[cm²]" : concat(εLlnlLess, dfLLNL["EffectiveArea[cm²]", float], εLlnlMore, axis = 0) }) dfExt["File"] = constantColumn("LLNL_extended", dfExt.len) dfExt.add dfMpe.select(@["Energy[keV]", "EffectiveArea[cm²]", "File"]) dfExt.add dfLLNL.select(@["Energy[keV]", "EffectiveArea[cm²]", "File"]) ggplot(dfExt, aes("Energy[keV]", "EffectiveArea[cm²]", color = "File")) + geom_line() + ggtitle("Comparison of the effective area of the LLNL and MPE telescopes") + ggsave("/home/basti/org/Figs/statusAndProgress/llnl_mpe_effective_area_comparison_extended.pdf") # write away extended LLNL as CSV to load into limit code echo dfExt dfExt.filter(f{`File` == "LLNL_extended"}).drop("File").writeCsv("/home/basti/org/resources/llnl_xray_telescope_cast_effective_area_extended.csv")
Fig. 262 shows the effective area of two telescope with the added extension of the LLNL telescope (by using the ratio of LLNL / MPE at the LLNL boundary). We can see that the extension looks very reasonable. Reality might differ somewhat, but as for a start it should be enough.
For testing let's try to load the extended efficiencies and rebin it to the background binning:
let dfLLNL = readCsv("/home/basti/org/resources/llnl_xray_telescope_cast_effective_area_extended.csv") echo dfLLNL let llnlEffs = rebin(dfLLNL["Energy[keV]", float].toRawSeq, dfLLNL["EffectiveArea[cm²]", float].toRawSeq, bins, areRate = true) let effDf = toDf({ "Energy[keV]" : bins, "EffectiveArea[cm²]" : llnlEffs }) ggplot(effDf, aes("Energy[keV]", "EffectiveArea[cm²]")) + geom_point() + ggtitle("Extended LLNL effective area in 2017/18 data binning") + ggsave("/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/llnl_eff_area_2018_binning.pdf")
This yields fig. 263.
The same can be done for the 2013 binning and the MPE telescope effective areas:
let dfMPE = readCsv("/home/basti/org/resources/mpe_xray_telescope_cast_effective_area.csv") echo dfMPE ## binning for 2013 data let bins2013 = @[0.7999,1.0857,1.3714,1.6571,1.9428,2.2285, 2.5142,2.7999,3.0857,3.3714,3.6571,3.9428, 4.2285,4.5142,4.7999,5.0857,5.3714,5.6571, 5.9428,6.2285, 6.2285 + 0.2857142857142858] let mpeEffs = rebin(dfMPE["Energy[keV]", float].toRawSeq, dfMPE["EffectiveArea[cm²]", float].toRawSeq, bins2013, areRate = true) let effDfMPE = toDf({ "Energy[keV]" : bins2013, "EffectiveArea[cm²]" : mpeEffs }) ggplot(effDfMPE, aes("Energy[keV]", "EffectiveArea[cm²]")) + geom_point() + ggtitle("MPE effective area in 2013 data binning") + ggsave("/home/basti/org/Figs/statusAndProgress/CAST_2013/mpe_eff_area_2013_binning.pdf")
which in turn yields fig. 264.
With this we may now apply this as an additional scaling factor for the expected flux. Let's plot a comparison (in the 2018 binning) of 3 cases:
- perfect optics
- constant efficiency corresponding to ~5.5 cm²
- real LLNL effective area (2017/18 binning + 2017/18 tracking time)
- real MPE effective area (2013 binning + assuming same 2017/18 tracking time of 180 h)
proc plotEfficiencyComparison(dfs: seq[DataFrame] = @[], suffix = "") = ## plots the LLNL, perfect, constant, MPE telescope setups and any added ## ones from the arguments. The input ones need the columns ## `Energy[keV]`, `Setup` and `Flux` let fluxLLNL = readFluxAndPlot(bins[1] - bins[0], bins, 180.Hour, telEff = llnlEffs, title = " extended LLNL eff. area", suffix = "_llnl_extended_eff_area", toPlot = false) let fluxPerfect = readFluxAndPlot(bins[1] - bins[0], bins, 180.Hour, useConstTelEff = false, title = " assuming perfect X-ray optics", toPlot = false) let fluxConst = readFluxAndPlot(bins[1] - bins[0], bins, 180.Hour, useConstTelEff = true, title = " assuming constant tel. eff. ~5.5", toPlot = false) var fluxDf = toDf({ "Energy[keV]" : bins, "perfect" : fluxPerfect, "5.5cm²" : fluxConst, "LLNL" : fluxLLNL }) .gather(@["perfect", "5.5cm²", "LLNL"], key = "Setup", value = "Flux") # and add flux for 2013 data (different binning, so stack let fluxMpe2013 = readFluxAndPlot(bins2013[1] - bins2013[0], bins2013, 180.Hour, telEff = mpeEffs, toPlot = false) let mpe2013Df = toDf({"Energy[keV]" : bins2013, "Flux" : fluxMpe2013, "Setup" : "MPE"}) fluxDf.add mpe2013Df for df in dfs: fluxDf.add df.select(["Energy[keV]", "Setup", "Flux"]) ggplot(fluxDf, aes("Energy[keV]", "Flux", color = "Setup")) + geom_point() + ggtitle("Comparison of expected fluxes for g_ae = 1e-13, g_aγ = 1e-12 in 180 h of tracking " & "assuming different 'telescope setups' (MPE uses 2017/18 tracking time)") + margin(top = 2.5) + ggsave(&"/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/flux_comparison_telescope_setups{suffix}.pdf") plotEfficiencyComparison()
Which yields fig. 265. When looking at the comparison between the MPE and LLNL cases, keep in mind that these are not rates anymore, but total expected counts. That means while the numbers seem higher between \SIrange{1}{2}{\keV}, the absolute flux is lower because there are only <4 bins within that range and 5 for the LLNL case!
With these in place, we can now compute the next step. Namely, an actual limit for the 2017/18 data with the LLNL efficiencies and for the 2013 data with the MPE efficiencies. For the 2013 data we use their real tracking time now of course (in contrast to the comparison of fig. 265.
let fluxLLNL = readFluxAndPlot(bins[1] - bins[0], bins, 180.Hour, telEff = llnlEffs, title = " extended LLNL eff. area", suffix = "_llnl_extended_eff_area", toPlot = false) # `bins` and `backs` are the 2017/18 ε = 0.8 variables from before echo "Monte Carlo experiments for 2017/18 + LLNL effective area:\n\n" #discard monteCarloLimit(bins, fluxLLNL, backs, rnd, suffix = "llnl_extended_eff_area", verbose = false) let cands2013 = @[1, 3, 1, 1, 1, 2, 1, 2, 0, 2, 0, 1, 0, 2, 2, 0, 2, 1, 2, 2] let backs2013 = @[2.27, 1.58, 2.40, 1.58, 2.6, 1.05, 0.75, 1.58, 1.3, 1.5, 1.90, 1.85, 1.67, 1.3, 1.15, 1.67, 1.3, 1.3, 2.27, 1.3] let fluxMpe2013 = readFluxAndPlot(bins2013[1] - bins2013[0], bins2013, 197.0.Hour, telEff = mpeEffs, toPlot = false) # actual cnadidates echo "Limit scan for 2013 + MPE effective area:\n\n" echo performScans(flux, backs, cands) # monte carlo toys echo "Monte Carlo experiments for 2013 + MPE effective area:\n\n" #discard monteCarloLimit(bins2013, fluxMPE2013, backs2013, rnd, suffix = "mpe_eff_area_2013_toy_exps", verbose = false)
Monte Carlo experiments for 2017/18 + LLNL effective area: Mean coupling constant of MC toy experiments (for g_aγ = 1e-12 GeV⁻¹): 5.354519543908154e-21 Limit scan for 2013 + MPE effective area: Limit of χ² (+ 5.5) is = 5.411882376474744e-21 Monte Carlo experiments for 2013 + MPE effective area: Mean coupling constant of MC toy experiments (for g_aγ = 1e-12 GeV⁻¹): 9.864804640926203e-21
Summarizing these outputs in a table, tab 24:
| Setup | Toy (y/n) | g²ae | gae |
|---|---|---|---|
| 2017/18 + LLNL | y | 5.35e-21 | 7.31e-11 |
| 2013 + MPE | n | 5.41e-21 | 7.36e-11 |
| 2013 + MPE | y | 9.86e-21 | 9.93e-11 |
We can see the effect of "being lucky" in the 2013 case very well. The observed limit is much better than the expected one (from MC).
The distributions of the MC limits are further shown in fig. 266 for the 2017/18 case and in fig. 267.
17.6.4. Limit with detector window & gas absorption
UPDATE : The below code to compute the
efficiency now lives in
./../../CastData/ExternCode/TimepixAnalysis/Tools/septemboardDetectionEff/septemboardDetectionEff.nim
using xrayAttenuation and allowing to read different effective area
files for the LLNL telescope.
The next step of the limit calculation is now to include the detector window and argon gas. Both of these cause further losses of the expected flux, so they need to be applied to the flux normalization.
For the \SI{300}{\nm} SiN window there are two different things to consider:
- the actual window that's \SI{300}{\nm} thick
- the strongback support structure that is for all intents and purposes opaque for X-rays
For reference again the SiN occlusion figure from chapter 2 in fig. 268.

Let's now extend fig. 4 to also include the LLNL telescope efficiency for our detector:
import ggplotnim, numericalnim, math proc interpLLNL(energies: Tensor[float]): DataFrame = ## interpolates the LLNL telescope efficiencies to the given energies let llnl = readCsv("/home/basti/org/resources/llnl_xray_telescope_cast_effective_area_extended.csv") .mutate(f{"Efficiency" ~ idx("EffectiveArea[cm²]") / (PI * 2.15 * 2.15)}) let interp = newHermiteSpline(llnl["Energy[keV]", float].toRawSeq, llnl["Efficiency", float].toRawSeq) var effs = newSeq[float](energies.size) let eMin = llnl["Energy[keV]", float].min let eMax = llnl["Energy[keV]", float].max for idx in 0 ..< effs.len: effs[idx] = if energies[idx] < eMin: 0.0 elif energies[idx] > eMax: 0.0 else: interp.eval(energies[idx]) result = toDf({"Energy [keV]" : energies, "LLNL" : effs}) let al = readCsv("/home/basti/org/resources/Al_20nm_transmission_10keV.txt", sep = ' ', header = "#") let siN = readCsv("/home/basti/org/resources/Si3N4_density_3.44_thickness_0.3microns.txt", sep = ' ') let si = readCsv("/home/basti/org/resources/Si_density_2.33_thickness_200microns.txt", sep = ' ') let argon = readCsv("/home/basti/org/resources/transmission-argon-30mm-1050mbar-295K.dat", sep = ' ') let llnl = interpLLNL( siN.mutate( f{"Energy [keV]" ~ idx("PhotonEnergy(eV)") / 1000.0})["Energy [keV]", float] ) var df = newDataFrame() df["300nm SiN"] = siN["Transmission", float] df["200μm Si"] = si["Transmission", float] df["30mm Ar"] = argon["Transmission", float][0 .. argon.high - 1] df["20nm Al"] = al["Transmission", float] df["LLNL"] = llnl["LLNL", float] df["Energy [eV]"] = siN["PhotonEnergy(eV)", float] df["SB"] = constantColumn(1.0 - 0.222, df.len) df["ε"] = constantColumn(0.8, df.len) df = df.mutate(f{"Energy [keV]" ~ idx("Energy [eV]") / 1000.0}, f{"30mm Ar Abs." ~ 1.0 - idx("30mm Ar")}, f{"Efficiency" ~ idx("30mm Ar Abs.") * idx("300nm SiN") * idx("20nm Al")}, f{"Eff • SB • ε" ~ `Efficiency` * `SB` * `ε`}, f{"Eff • ε" ~ `Efficiency` * `ε`}, f{"Eff • ε • LLNL" ~ `Efficiency` * `ε` * `LLNL`}, f{"full Eff." ~ idx("Eff • SB • ε") * `LLNL`}) # strongback occlusion of 22% and ε = 80% .drop(["Energy [eV]", "Ar"]) df.writeCsv("/home/basti/org/resources/combined_detector_efficiencies.csv") showBrowser(df, "df_initial.html") block: let df = df .gather(["300nm SiN", "Efficiency", "full Eff.", "Eff • SB • ε", "30mm Ar Abs.", "200μm Si", "20nm Al", "LLNL", "Eff • ε", "Eff • ε • LLNL"], key = "Type", value = "Efficiency") ggplot(df, aes("Energy [keV]", "Efficiency", color = "Type")) + geom_line() + ggtitle("Full detector efficiencies, including window, gas, ε, window SB, telescope") + margin(top = 1.5) + ggsave("/home/basti/org/Figs/statusAndProgress/detector/window_plus_argon_efficiency.pdf", width = 800, height = 600) block: echo df let df = df.drop(["200μm Si", "SB", "Efficiency", "Eff • ε", "Eff • SB • ε", "full Eff."]) .rename(f{"full eff." <- "Eff • ε • LLNL"}) .gather(["300nm SiN", "30mm Ar Abs.", "20nm Al", "LLNL", "full eff."], key = "Type", value = "Efficiency") echo df ggplot(df, aes("Energy [keV]", "Efficiency", color = "Type")) + geom_line() + ggtitle("Detection efficiencies of window, software eff., LLNL efficiency and Argon absorption") + margin(top = 1.5) + ggsave("/home/basti/org/Figs/statusAndProgress/detector/detection_efficiency.pdf", width = 800, height = 600)
This yields us the rather depressing fig. 269 of our detection efficiency in the gold region assuming a software efficiency of ε = 80%. We see that our peak efficiency is a total of ~30% at around 1.5 keV. I suppose it could be worse?
Let's apply this to the limit calculation. If we use the full Eff.
field of the above as the input for the "telescope efficiency"
argument to the readFluxAndPlot proc, we can get the flux for the
full detector efficiency:
let combEffDf = readCsv("/home/basti/org/resources/combined_detector_efficiencies.csv") let combEff = rebin(combEffDf["Energy [keV]", float].toRawSeq, combEffDf["full Eff.", float].toRawSeq, bins, areRate = true) let combinedEffFlux = readFluxAndPlot(bins[1] - bins[0], bins, 180.Hour, telEff = combEff, title = ", total combined detector efficiency", suffix = "_total_combined_det_eff", toPlot = false, telEffIsNormalized = true) let combEffFluxDf = toDf({ "Energy[keV]" : bins, "Flux" : combinedEffFlux, "Setup" : "full Eff." }) echo combEffFluxDf plotEfficiencyComparison(@[combEffFluxDf], suffix = "with_total_combined_det_eff")
The flux we now expect at the detector is shown in fig. 270.
Now we can finally compute the limit using this efficiency:
#discard monteCarloLimit(bins, combinedEffFlux, backs, rnd, suffix = "full_detector_efficiency", verbose = false)
#+RESULTS
Mean coupling constant of MC toy experiments (for g_aγ = 1e-12 GeV⁻¹): 1.2019404040805464e-20
So, a limit of \(g²_{ae} = \num{1.202e-20}\) or \(g_{ae} = \num{1.096e-10}\), which is quite a bit worse than before. But it makes sense, because the window + gas absorb quite a bit. Also now we actually take the software efficiency into account, which we did not do above yet (not even in the ε = 60% case).
However, this represents a worst case scenario, as the window strongback occlusion is included, which on paper occludes 22.2% of the sensitive area. The actual flux isn't homogeneous in reality though. This means the real losses will be a bit smaller (we can compute that from the raytracer).
The "χ²" distribution of the toys is found in fig. 271.
17.6.5. Limit with artificially reduced background
To get a better understanding of what the impact of lower background would be, we will also now compute the limit using an artificially reduced background. We will scale it down by a factor of 5.
let backsReduced = backs.mapIt(it / 5.0) #discard monteCarloLimit(bins, combinedEffFlux, backsReduced, rnd, suffix = "full_det_eff_background_reduced_5", verbose = false)
Mean coupling constant of MC toy experiments (for g_aγ = 1e-12 GeV⁻¹): 7.266601800358901e-21
The limit does improve quite a bit over the value in the previous section. We now get \(g²_{ae} = \num{7.267e-21}\) or \(g_{ae} = \num{8.52e-11}\). And again, this is with the full detector efficiency and the slight overestimation of the strongback occlusion.
After a 5 times reduction using our binning there are barely any expected candidates in each bin for the tracking time we took. An example plot after one toy experiment is shown in fig. 272.
17.6.6. Limit using correct unphysical χ² rescaling
For the final step we will now compute the \(χ²\) limit value based on the correct MLE approach.
This means we need to:
- scan the computed \(χ²\) phase space and determine the range of \(g²_{ae}\) covered by \(Δχ² = 1\)
- this distance in \(g²_{ae}\) is the 1σ of a gaussian distribution that we imagine centered at \(g²_{ae,\text{limit}}\).
- cut this gaussian distribution at the physical (\(g²_{ae} = 0\)) range. Compute the 95% point of the CDF. That point is our limit.
Let's implement this (we need to make some small changes in the code above too, to make it a bit more generic):
proc findConstantCutLimit(flux, backs: seq[float], cands: seq[int], vals: Tensor[float], start: int, couplingStart: float, couplingStep: float, cutValue = 1.0, searchLeft = false): float = ## Finds the the coupling constant such that `χ² = start + cutValue`. Our scan starts ## at `couplingStart` let startVal = vals[start] var curCoupling = couplingStart var curχ² = startVal while curχ² < startVal + cutValue and classify(curχ²) == fcNormal: # compute next coupling step and χ² value if searchLeft: curCoupling -= couplingStep else: curCoupling += couplingStep curχ² = computeχ²(flux, backs, cands, curCoupling) result = curCoupling proc computeSigma(flux, backs: seq[float], cands: seq[int], vals: Tensor[float], start: int, couplingStart, couplingStep: float, verbose = false): float = ## Computes the 1σ value of the current `χ²` "distribution" let limitLeft = findConstantCutLimit(flux, backs, cands, vals, start, couplingStart, couplingStep, searchLeft = true) let limitRight = findConstantCutLimit(flux, backs, cands, vals, start, couplingStart, couplingStep, searchLeft = false) let gaussSigma = limitRight - limitLeft if verbose: echo "Min at ", start echo "g²_ae,min = ", couplingStart echo "left = ", limitLeft echo "right = ", limitRight echo "1σ of gauss = ", gaussSigma echo "Naive + Δχ² = 4.0 limit ", findConstantCutLimit(flux, backs, cands, vals, start, couplingStart, couplingStep, 4.0, searchLeft = false) echo "Naive + Δχ² = 5.5 limit ", findConstantCutLimit(flux, backs, cands, vals, start, couplingStart, couplingStep, 5.5, searchLeft = false) result = gaussSigma proc findχ²Minimum(flux, backs: seq[float], cands: seq[int], range = (-6e-45, 6e-45)): tuple[vals: Tensor[float], idx: int, minVal: float] = let g_aγ² = 1e-12 * 1e-12 let couplings = linspace(range[0] / g_aγ², range[1] / g_aγ², 5000) let couplingStep = couplings[1] - couplings[0] let dfχ² = linearScan(χ², flux, backs, cands) .filter(f{ `χ²` <= 200.0 and classify(`χ²`) == fcNormal }) let χ²vals = dfχ²["χ²", float] let χ²argmin = χ²vals.toRawSeq.argmin let χ²min = χ²vals[χ²argmin] let couplingsRaw = dfχ²["CouplingsRaw", float] result = (vals: χ²vals, idx: χ²argmin, minVal: couplingsRaw[χ²argmin]) ## Compute the integral of normal distibution using CDF written using error function proc cdf(x: float, μ = 0.0, σ = 1.0): float = 0.5 * (1.0 + erf((x - μ) / (σ * sqrt(2.0)))) proc calcSigmaLimit(μ, σ: float, ignoreUnphysical = false): tuple[limit, cdf: float] = ## Computes the limit based on a 1 σ gaussian distrubition around the computed χ² ## results. The 1 σ range is determined based on the coupling range covered by ## χ²_min + 1. ## The limit is then the gaussian CDF at a value of 0.95. Either in the full ## data range (`ignoreUnphysical = false`) or the CDF@0.95 only in the physical ## range (at x = 0). var x = μ var offset = 0.0 if ignoreUnphysical: x = 0.0 offset = x.cdf(μ, σ) while x.cdf(μ, σ) < (1.0 - (1.0 - offset) * 0.05): x += (σ / 1000.0) result = (limit: x, cdf: x.cdf(μ, σ)) proc performScansCorrect(flux, backs: seq[float], cands: seq[int], range = (-6e-45, 6e-45), couplings: seq[float] = @[], verbose = false, toPlot = true, suffix = ""): Limit = # due to `linearScan` being `dirty` template can define couplings, gaγ here # need to read the minimum coupling from DF, because input coupling contains χ² equivalent # NaN values (`χ²vals.len != couplings.len` is the problem) let (χ²vals, χ²argmin, couplingMinVal) = findχ²Minimum(flux, backs, cands, range) let σ = computeSigma(flux, backs, cands, χ²vals, χ²argmin, couplingMinVal, (range[1] - range[0] / 5000 / (1e-12 * 1e-12)), verbose = verbose) # compute the real limit based on the sigma let limit = calcSigmaLimit(couplingMinVal, σ, ignoreUnphysical = false) let limitPhys = calcSigmaLimit(couplingMinVal, σ, ignoreUnphysical = true) result = Limit(coupling: limitPhys[0], χ²: computeχ²(flux, backs, cands, limitPhys[0])) if toPlot: const g²_aγ = 1e-12 * 1e-12 let μ = couplingMinVal let xs = linspace(μ - 3 * σ, μ + 3 * σ, 2000) let df = toDf(xs) .mutate(f{float: "gauss" ~ gauss(`xs`, μ, σ)}, f{"xs" ~ `xs` * g²_aγ}) let couplings = linspace(range[0] / g²_aγ, range[1] / g²_aγ, 5000) let dfχ² = linearScan(χ², flux, backs, cands) .filter(f{ `χ²` <= 200.0 and classify(`χ²`) == fcNormal }) let lim = limit.limit let limPhys = limitPhys.limit let xLimLow = (μ - 3 * σ) * g²_aγ let xLimHigh = (μ + 3 * σ) * g²_aγ let χ²minVal = χ²vals[χ²argmin] ggmulti([ggplot(df, aes("xs", "gauss")) + geom_line() + geom_linerange(aes(x = lim * g²_aγ, yMin = 0.0, yMax = 1.0)) + geom_linerange(aes(x = limPhys * g²_aγ, yMin = 0.0, yMax = 1.0)) + xlim(xlimLow, max(xlimHigh, dfχ²["Couplings", float].max)) + xlab("g²_ae g²_aγ") + annotate(&"Limit at: {limit.limit:.2e}\nCorresponds to CDF cut @{limit.cdf:.2f}", x = lim * g²_aγ, bottom = 0.5) + annotate(&"Physical limit at: {limitPhys.limit:.2e}\nCorresponds to CDF cut @{limitPhys.cdf:.2f}", x = limPhys * g²_aγ, bottom = 0.3), ggplot(dfχ², aes("Couplings", "χ²")) + geom_line() + geom_linerange(aes(y = χ²minVal + 1.0, xMin = xlimLow, xMax = xLimHigh)) + geom_linerange(aes(y = χ²minVal + 4.0, xMin = xlimLow, xMax = xLimHigh)) + geom_linerange(aes(x = lim * g²_aγ, yMin = χ²minVal, yMax = χ²minVal + 4.0)) + geom_linerange(aes(x = limPhys * g²_aγ, yMin = χ²minVal, yMax = χ²minVal + 4.0)) + annotate("χ²_min + 1", y = χ²minVal + 1.1, left = 0.2) + annotate("χ²_min + 4", y = χ²minVal + 4.1, left = 0.2) + xlim(xlimLow, max(xlimHigh, dfχ²["Couplings", float].max)) + xlab("g²_ae g²_aγ") + ggtitle("Scan of g²_ae g²_aγ for g_aγ = 1e-12 GeV⁻¹")], &"/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/chi_square_gauss_limit{suffix}.pdf", 1200, 500)
Let's run this performScansCorrect procedure on a single toy
experiment and look at the resulting plot. Namely the comparison of
the χ² "distribution", it's 1σ environment and the generated Gaussian
plus its 95% value taken from the Gaussian CDF both for the full range
as well as the only physical case (cut off at \(g²_{ae} = 0\)).
block: let cand2 = drawExpCand(backs, rnd) echo performScansCorrect(combinedEffFlux, backs, cand2, range = (-1e-44, 1e-44), suffix = "_initial_example")
This yields fig. 274. The plot shows a subplot of a single χ² phase scan compared to the Gaussian distribution that is created at the χ² minimum using a 1σ range determined from the χ² values by checking for the width in coupling space at \(χ²_{\text{min}} + 1\). From this Gaussian we compute the CDF value at 0.95 for the full and only physical range (\(g²_{ae} \geq 0\)).
We can see that the initial assumption of \(\sim χ²_{\text{min}} + 4\) does not really match the CDF 0.95 value for the Gaussian in the full range. The value is closer to ~5.
To get a better understanding of the behavior for multiple of these
plots (for multiple toys), let's compute 10 different ones and move
them to
~/org/Figs/statusAndProgress/CAST_2018_Likelihood/chi_sq_gauss_comparison/:
import strutils block: let path = "/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/chi_square_gauss_limit$#.pdf" let toPath = "/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/chi_sq_gauss_comparison/chi_square_gauss_limit$#.pdf" for i in 0 ..< 10: let cand2 = drawExpCand(backs, rnd) echo performScansCorrect(combinedEffFlux, backs, cand2, range = (-1e-44, 1e-44), suffix = $i) moveFile(path % $i, toPath % $i)
With this let's now compute the full Monte Carlo toy experiments and make a plot comparing the resulting distributions of the obtained limits from the scan that only looks at the physical part of the Gaussian with our initial assumption of using a χ² offset of 5.5 all the time. We'll also include the 2013 data into this approach using the MPE telescope efficiencies but not any other (no software efficiency etc.).
block: let dfPhys = monteCarloLimit(bins, combinedEffFlux, backs, rnd, suffix = "_correct_phys_limit", verbose = false, fn = performScansCorrect) let dfConst = monteCarloLimit(bins, combinedEffFlux, backs, rnd, suffix = "_constant_plus_5.5", verbose = false) let df2013 = monteCarloLimit(bins2013, fluxMPE2013, backs2013, rnd, suffix = "_mpe_eff_area_2013_correct_physical_limit", verbose = false, fn = performScansCorrect) let dfComb = bind_rows([("2018_1σ", dfPhys), ("2018Const+5.5", dfConst), ("2013_1σ", df2013)], "From") ggplot(dfComb, aes("Coupling", color = "From", fill = "From")) + geom_histogram(bins = 100, position = "identity", alpha = some(0.5), hdKind = hdOutline) + ggtitle("Comparison of gaussian (physical only) CDF@95% vs. χ²+5.5") + ggsave("/home/basti/org/Figs/statusAndProgress/CAST_2018_Likelihood/comparison_coupling_dist_physical_vs_constant.pdf")
This finally yields the following plot in fig. 275.
Most strikingly is the large peak of the 2017/18 data 2018_1σ close
to very small values. It then has another "main" peak at larger
values.
The resulting limits for each of these cases (mean of the distribution, which is somewhat questionable for the 2018 + Gaussian case) is shown in tab. 25.
| Input | Method | \(g²_{ae}\) | \(g_{ae}\) |
|---|---|---|---|
| 2017/18 | \(χ²_{\text{min}} + 5.5\) | 1.213e-20 | 1.10e-10 |
| 2017/18 | physical CDF@0.95 | 1.358e-20 | 1.16e-10 |
| 2013 | physical CDF@0.95 | 1.346e-20 | 1.16e-10 |
We can see that barring the weird behavior in the 2017/18 data the results aren't that far off. If one were to look at only the wider peak in the Gaussian 2017/18 distribution, the numbers would probably become a bit worse. But it's still uplifting to know that this somewhat pessimistic approach (overestimation of window occlusion, not taking into account flux distribution on chip) is pretty close to the 2013 limit on a fair basis.
Maybe will need to turn monteCarloLimit into a template to hand this
performScansCorrect procedure instead of the original one. Other
than that we should be able to compute the correct limit just fine
then. Add a couple of plots (compare limit histogram in particular!).
Create a histogram of perform "normal" (+5.5) and perform correct in
one plot.
import math import seqmath, ggplotnim, strformat ## Compute the integral of normal distibution proc cdf(x: float, μ = 0.0, σ = 1.0): float = 0.5 * (1.0 + erf((x - μ) / (σ * sqrt(2.0)))) proc calcLimit(μ, σ: float, ignoreUnphysical = false): tuple[limit, cdf: float] = var x = μ var offset = 0.0 if ignoreUnphysical: x = 0.0 offset = x.cdf(μ, σ) while x.cdf(μ, σ) < (1.0 - (1.0 - offset) * 0.05): x += (σ / 1000.0) echo x echo x.cdf(μ, σ) echo "At 0 it is ", cdf(0.0, μ, σ) result = (limit: x, cdf: x.cdf(μ, σ)) discard calcLimit(0.0, 1.0) #let μ = -1.05e-21 #let σ = 7.44e-21 let μ = -1.55e-21 let σ = 5.71e-21 let limit = calcLimit(μ, σ) echo limit let limit2 = calcLimit(μ, σ, ignoreUnphysical = true) echo limit2 const g²_aγ = 1e-12 * 1e-12 let xs = linspace(μ - 3 * σ, μ + 3 * σ, 2000) let df = toDf(xs) .mutate(f{float: "gauss" ~ gauss(`xs`, μ, σ)}) .mutate(f{"xs" ~ `xs` * g²_aγ}) echo df let lim = limit.limit let limPhys = limit2.limit ggplot(df, aes("xs", "gauss")) + geom_line() + geom_linerange(aes(x = lim * g²_aγ, yMin = 0.0, yMax = 1.0)) + geom_linerange(aes(x = limPhys * g²_aγ, yMin = 0.0, yMax = 1.0)) + #xlim(-4e-21, 2e-20) + annotate(&"Limit at: {limit.limit:.2e}\nCorresponds to CDF cut @{limit.cdf:.2f}", left = 0.7, bottom = 0.5) + annotate(&"Physical limit at: {limit2.limit:.2e}\nCorresponds to CDF cut @{limit2.cdf:.2f}", left = 0.7, bottom = 0.3) + ggsave("/tmp/plot.pdf")
18. Signal vs. background classification
This should be the main chapter for the logL method used currently for limits etc.
18.1. TODO Neural network based separation
Two types mainly:
- taking geometrical objects after clustering as inputs
- pros: small networks, fast training
- cons: bias due to clustering etc.
- purely image based sig / back using CNNs
- pros: no bias
- cons: longer training, huge networks
Should start as such:
- take Flambeau, build ultra simple MLP
- read CDL based data, apply CDL cut that we use to determine reference dataset
- take a background run
- play around with this setup, mainly to test out:
- Flambeau
- 3090
If that works, we can extend Flambeau for more complex layouts etc.
19. Gas gains vs. energy calibration factor fits
See the previous discussions about this topic at random places throughout the document and in particular in section 15.1.5 for the latest plots up to this point ().
With the lessons learned in 15.1.7, the 90 minute intervals and the fix of the last gas gain slice possibly being too short (c/f: https://github.com/Vindaar/TimepixAnalysis/issues/50) the gas gain intervals as well as the gas gain vs. energy calibration factors had to be recomputed.
A small script was used to do this in parallel ./../../CastData/ExternCode/TimepixAnalysis/Tools/recomputeGasGainSlices.nim.
The resulting plots for 2017 and 2018 are shown in fig. 276 and 277.
gcIndividual). The last slice is possibly merged into the second last, if it is shorter than 25 min. Each slice is ~90 min. These plots do not have enlarged error bars anymore.gcIndividual). The last slice is possibly merged into the second last, if it is shorter than 25 min. Each slice is ~90 min. These plots do not have enlarged error bars anymore.19.1. Gas gain slice fits
Generated with ./../../CastData/ExternCode/TimepixAnalysis/Plotting/plotGasGainFitParameters/plotGasGainFitParameters.nim
19.2. Gas gain vs. energy calibration fit, improved χ²
:DATE:
I wondered why the χ²/dof was so horribly large for the gas gain vs. energy calib fit. Turns out the Fe charge spectrum fits used "errors" of 1 for every single bin!
Changing this to sqrt(counts) for all bins with bin content > 0, results in much better fits, e.g. fig. 281 and 282.
And the energy calibration:
20. Background rate
While the background rate was already mentioned in previous chapters, maybe it's a good idea to have its own chapter.
The plots shown before were all done using the Python plotting script:
In order to have a unified style of the plots (and to have it ready
come my thesis), I rewrote a script to plot it using ggplotnim:
Using the background files created for the discussions in sec. 17.2, we can generate the background rate plot shown in fig. 285.
The plotting script of course works on tho likelihood output H5 files
generated by the likelikood program, using the following commands:
/likelihood /mnt/1TB/CAST/2017/DataRuns2017_Reco.h5 --h5out /tmp/lhood_2017.h5 --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 --cdlYear=2018 /likelihood /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 --h5out /tmp/lhood_2018.h5 --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 --cdlYear=2018
where me make sure to use the 2019 CDL data and do not use the
--tracking flag.
These files are now found in:
- ./../../CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_no_tracking.h5
- ./../../CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2018_no_tracking.h5
The plot is then generated using:
./plotBackgroundRate ../../resources/LikelihoodFiles/lhood_2017_no_tracking.h5 ../../resources/LikelihoodFiles/lhood_2018_no_tracking.h5 --show2014
Note that there is a major difference between this program and the old Python srcipt: Namely, this one handles adding data of one kind properly. That is both 2017 and 2018 (technically Run 2 & 3) are added together.
These are the baseline background rate "benchmark" plots as of ~ .
20.1. STARTED Investigate total duration
:
We've probably done this before. But I want to investigate the
totalDuration value in the output of the likelihood data again.
We get about 3300h for 2017/18 data in background.
Is that due to solar tracking being taken out?
Compare event duration & event numbers to total number of events. What events are missing? If an event is missing, doesn't that still imply the detector was active during that time?
I don't think tracking accounts for the difference (even though it
sort of matches the discrepancy) as I don't see anything of the like
in the code (the total duration is computed from the full
eventDuration dataset, instead of the filtered numbers.
: I've started looking into this by extending the ./../../CastData/ExternCode/TimepixAnalysis/Tools/printXyDataset.nim script to allow to combine all the runs in a single file.
Looking at the sum of all event durations for the background data, it seems like we end up at the 3318h number for background data (contrasting with the ~3500h number). The difference between 3300 and 3500h are then likely just from the total event numbers * 2.4s vs. actual event duration times.
This implies the following: The actual background time without solar tracking is actually 3300-170h (180h in total by frame numbers multiplied by 3318/3526) hours. This unfortunately further reduces our background time (as well as obviously our solar tracking time!).
In principle the "background time" is even longer than the 3526h though, as the readout time is again very significant! But that does not seem very helpful unfortunately.
20.2. TODO investigate the shape
There is still some weirdness going on in this background rate.
To me it really seems like the energy calibration (more than the likelihood method for instance) is to blame for the shape. I'm under the impression the energies are simply not reconstructed properly. The 2018 part has a shift to too large energies. The 2017 is slightly better.
One thing to look at is the ridgeline plots of the background and signal plots for this data and see what the distributions look like maybe?
Certainly need to check the gas gain fit again, as well as the fit to all gas gains vs. calibration factors. We probably have to take into account the movement of the peak position over time in a much better way, also check out the sec. 14.5 for something to keep in mind.
20.3. Recompute background rate with new energy calibration
After the fixes done due to section 15.1, it is time to recompute the energy calibration.
:Date:
The gas gain intervals were recomputed for each file, as well as the gas gain vs. energy calibration factor fit, sec. 19 and the energy itself using the following config:
# this TOML document is used to configure the raw data manipulation, reconstruction and likelihood # tools [Calibration] showPlots = false plotDirectory = "out" # minutes for binning of gas gain gasGainInterval = 90 # minutes the gas gain interval has to be long at least. This comes into play # at the end of a run `(tRun mod gasGainInterval) = lastSliceLength`. If the # remaining time `lastSliceLength` is less than `minimumGasGainInterval`, the # slice will be absorbed into the second to last, making that longer. minimumGasGainInterval = 25 # decides if `charge` dataset is deleted when running `only_charge`. # by default this is disabled, because the attributes of the dataset # contain things beyond the actual charge calibration! deleteChargeDset = true # the gas gain vs energy calibration factor computation to use # - "": the default of just one gas gain per calibration run based on # one gas gain interval for each run # - "Mean": use the mean of all gas gain time slices # - "Individual": use the factors obtained from one Fe fit per gas gain slice gasGainEnergyKind = "Individual" [RawData] plotDirectory = "out"
The new computation was done using ./../../CastData/ExternCode/TimepixAnalysis/Tools/recomputeGasGainSlices.nim, a simple helper script.
The computations were done at commit 01584d2.
The files resulting from likelihoon.nim are:
- ./../../CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_gasgain_slices_no_tracking.h5
- ./../../CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2018_gasgain_slices_no_tracking.h5
Let's compare the background rate we can achieve by
- 2017/2018 vs. 20182 dataset
- 2017/2018 last baseline vs. current
- 20182 last baseline vs. current
- 2017/20182 vs. 2014
First up comparison of the Sep 2020 background rate with the new rate, Jan 2021 for the 2017/18 dataset in fig. 286.
2017/18_0) and Jan 2021 (2017/18_2).Secondly the same, but for the end of 2018 dataset in fig. 287. In particular the 20182 (Run 3) data has moved more towards an energy range that we would expect. The shift to too high energies has been corrected somewhat. However, not to the extent that we would have liked.
2017/18_0) and Jan 2021 (2017/18_2).And finally a comparison of the new background rate Jan 2021 for both datasets combined vs. the 2014 background rate in fig. 288. As mentioned about the 20182 dataset, the full dataset still does not show a nice peak at \(\SI{8}{\kilo\electronvolt}\) as we might expect from theory.
2017/18_2) compared to the 2014 background rate.20.4. Note on comparison between 2014/15 and 2017/18 at very low energies
A possibly important distinction between the 2014/15 and 2017/18
detectors is the window. The old detector used a Mylar window. Mylar
is stretched PET, i.e. (C10H8O4)n.
That implies there are a lot more C and O atoms present, which both have fluorescence lines below 1 keV (ref. tab 4 / https://xdb.lbl.gov/Section1/Table_1-2.pdf). This could in theory explain the large discrepency.
On the other hand it is important to take into account that the C Kα line is explicitly part of the CDL dataset. So it seems not particularly likely that that would not correctly classify such C Kα X-rays (unless their properties are so vague that it is indeed possible).
\clearpage
20.5. STARTED Evaluate 8 keV peak (TODO: fix computation)
See the study under 23.
20.6. Background rates for different signal efficiencies
Comparison of the effect of the signal efficiency (the percentage of desired X-rays of the reference distributions to be recovered, by default 80%) on the background rate.
The following efficiencies were considered:
const effs = [0.5, 0.6, 0.7, 0.8, 0.85, 0.9, 0.95, 0.975, 0.9875, 0.99, 0.995]
First the background rate plots with the 2017 and 2018 data combined and secondly the end of 2017 / beginning of 2018 dataset separate from the end of 2018 dataset.
20.6.1. 2017/18 combined
\clearpage
20.6.2. 2017/18 separate
20.6.3. Summary & thoughts
From 26.18: #+beginquote
- background rates for different logL efficiencies:
- curious that 8-10 keV peak barely changes for increasing efficiency. Implies that the clusters in that peak are so photonic that they are not removed even for very sharp cuts. In theory "more photonic" than real photons: check: what is the difference in number of clusters at 0.995 vs. 0.5? Should in theory be almost a factor of 2. Is certainly less, but how much less?
- 6 keV peak smaller at 50% eff. than 8-10 keV, but larger at 99.5% peak -> more / less photonic in former / latter
- barely any background at 5 keV even for 99.5% efficiency
- largest background rate at lowest energy somewhat makes sense, due to worst separation of signal / background at those energies
- interval boundaries of the different CDL distributions becomes more and more prevalent the higher the efficiency is.
- pixel density of orthogonal muon tracks should have a different drop off than X-ray, due to convolution of many different gaussians. An X-ray has one point of origin from which all electrons drift according to diffusion from height h, ⇒ gaussian profile. In muon each pixel has diffusion d(hi) where each electron has its own height. Could probably compute the expected distribution based on: mean distance between interactions = detector height / num electrons and combine D = Σ d(hi) or something like this? Ref my plot from my bachelor thesis… :) Is this described by kurtosis? Also in theory (but maybe not in practice due to integration time) the FADC signal should also have encoded that information (see section on FADC veto / SiPM)
20.6.4. Background rates (and muons)
- combine background rate plots for different logL signal efficiencies
into a single plot. Possibly just use
geom_line(aes = aes(color = Efficiency))or histogram with outlines and no fill. - make histogram of length of all remaining clusters. Cut at median and produce a background rate for left and right side. Or possibly do the same with some other measure on the size of each cluster or rather a likely conversion origin in the detector (near the cathode vs. near anode)
- compute other geometrical properties of the remaining clusters (c/f last week eccentricity, but also other observables)
- compute "signal" / √background of each bin. Since we don't have signal, use ε = efficiency. Plot all these ratios of all signal efficiencies in one plot. In theory we want the efficiency that produces the largest ε / √B. Is 80% actually a good value in that regard?
- orthogonal muons: try to find other ways to investigate shape of muons vs. x-rays. Possibly plot kurtosis of 8-10 keV events?
#+endquote
20.7. Implementation of: Analysis of geometric properties of logL results
- Reuse some logic from
plotBackgroundRate(or add to that script?) for reading the logL cluster information. Pretty trivial. - After reading clusters into DF, cut on energy.
3a. for single signal efficiency: just generate plots for each
property (we could do a facet plot with free scales after gathering
all properties into a pair of key: Property, value: Value columns.
3b: combine DFs of all signal efficiencies and run same plots as 3a.
20.8. Effects of Morphing on background rate
See 22
20.9. STARTED Weirdness of larger background rate if considering all clusters
Hard to explain: There may be an issue that means we have more background in the gold region iff we actually compute clusters after LogL on the whole chip.
Our background rate plotting script as well as the background rate interpolation (can) work with lhood h5 files that contain all clusters. If we filter a full chip file to the gold region we see more background in the gold region than if we never included it in the likelihood program.
relevant comparison plots:
likelihood only gold region using default clustering, 65 pixels septem veto. Generated from the following H5 files: In dir:
~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugBelow2keVFromAllChip/lhood_2017_septemveto.h5lhood_2018_septemveto.h5
Which themselves were generated using:
./likelihood /mnt/1TB/CAST/201{7,8_2}/DataRuns201{7,8}_Reco.h5 --h5out /tmp/lhood_201{7,8}_septemveto.h5 \ --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crGold --septemvetousing the following settings in the ./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/config.toml:
[Reconstruction] # the search radius for the cluster finding algorithm in pixel searchRadius = 65 # for default clustering algorithm # clustering algorithm to use clusterAlgo = "default" # choose from {"default", "dbscan"} epsilon = 65 # for DBSCAN algorithm
likelihood only gold region using dbscan clustering, 65 pixels septem veto
Generated from the following H5 files: In dir:
~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugBelow2keVFromAllChip/lhood_2017_septemveto_dbscan.h5lhood_2018_septemveto_dbscan.h5
Which themselves were generated using:
./likelihood /mnt/1TB/CAST/201{7,8_2}/DataRuns201{7,8}_Reco.h5 --h5out /tmp/lhood_201{7,8}_septemveto.h5 \ --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crGold --septemvetousing the following settings in the ./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/config.toml:
[Reconstruction] # the search radius for the cluster finding algorithm in pixel searchRadius = 65 # for default clustering algorithm # clustering algorithm to use clusterAlgo = "dbscan" # choose from {"default", "dbscan"} epsilon = 65 # for DBSCAN algorithm
Generated from the following H5 files: In dir:
~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugBelow2keVFromAllChip/lhood_2017_septemveto_all_chip_dbscan.h5lhood_2018_septemveto_all_chip_dbscan.h5
Which themselves were generated using:
./likelihood /mnt/1TB/CAST/201{7,8_2}/DataRuns201{7,8}_Reco.h5 \ --h5out /tmp/lhood_201{7,8}_septemveto_all_chip_dbscan.h5 \ --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crAll --septemvetousing the following settings in the ./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/config.toml:
[Reconstruction] # the search radius for the cluster finding algorithm in pixel searchRadius = 65 # for default clustering algorithm # clustering algorithm to use clusterAlgo = "dbscan" # choose from {"default", "dbscan"} epsilon = 65 # for DBSCAN algorithm
These must be compared with the same plot as the last (generated from full chip data) if we use the following (already created in the past) likelihood files:
/home/basti/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_all_chip_septem_dbscan.h5/home/basti/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2018_all_chip_septem_dbscan.h5
which are the files we mostly used to develop the background interpolation (during which at some point we discovered the weirdness in the background interpolation, i.e. a peak behavior at around 1 keV that shouldn't be there according to the most recent background, i.e. the dbscan gold region one above for example or the one we sent do Esther in Dec 2021 by mail).
So, generating the background rate plot for these two files:
./plotBackgroundRate ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_all_chip_septem_dbscan.h5 \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2018_all_chip_septem_dbscan.h5 \ --combName bla \ --combYear 2018 \ --title "GridPix background rate based on CAST data in 2017/18" \ --region crGold
yields the following plot:
which clearly also shows much more background than expected below 2 keV!
It is likely that file was not properly utilizing the septem veto resulting in more background in the area where the background suppression of that is most likely?
Try to find out where the file is from / if it's referenced in these notes here.
UPDATE: the only reference in this file is in this section, the
background interpolation / limit code & the notes talking about the
input files for the background interpolation (stating they are created
using septem veto, but maybe the likelihood binary was old?).
Nothing of particular interest in the HDF5 file attributes either. We
should add the latest commit hash as an attribute to know which file
generated it.
Let's compare the clusters & total number for these files:
- the old files
LikelihoodFiles/lhood_201{7,8}_all_chip_septem_dbscan.h5generate exactly the same plot as the clustering shows in 414 (which references said file as well), also stored in:i.e. 9939 clusters.
- the files:
LikelihoodFiles/debugBelow2keVFromAllChip/lhood_201{7,8}_septemveto.h5:492 clusters only in gold.
- the files:
LikelihoodFiles/debugBelow2keVFromAllChip/lhood_201{7,8}_septemveto_dbscan.h5:505 clusters only in gold.
- the files:
LikelihoodFiles/debugBelow2keVFromAllChip/lhood_201{7,8}_septemveto_all_chip_dbscan.h5: ./../Figs/statusAndProgress/backgroundRates/debugBelow2keVFromAllChip/background_clusters_all_chip_dbscan.pdf 677 clusters over the whole chip!!!
IMPORTANT turns out the issue is extremely glaring. We simply remove most of the clusters that exist. Why? Didn't I see this before at some point? Let's check the H5 files of these:
Ahh, it seems like for some reason we are removing almost everything on the center chip (aside from gold region!), but keep everything on the other chips, as would be desired….
UPDATE: The important message above this was simply due to the way we the septem veto was implemented. Essentially, it included the 'line veto' by default, which added an additional check on the cluster being in the gold region. It's more complicated than that though. There could somehow be clusters that were initially outside the gold region that still pass it. I'm still not entirely sure about why that part is the way it is.
Since the above, we have now pushed our reconstructed data again through the likelihood tool. But this time we disabled the 'line veto' fully, so that we only make use of the regular septem veto.
Filtering with likelihood.nim using the whole chip on both 2017 and
2018 data files (see paths in below) and then plotting the result with the background rate
script:
./plotBackgroundRate ../../resources/LikelihoodFiles/debugBelow2keVFromAllChip/lhood_2017_septemveto_all_dbscan_no_lineveto.h5 \ ../../resources/LikelihoodFiles/debugBelow2keVFromAllChip/lhood_2018_septemveto_all_dbscan_no_lineveto.h5 \ --combName bla \ --combYear 2018 \ --title "septem veto, all chip, no lineveto" \ --region crGold
yields the following plot, fig. 312.
Now look at no septem veto at all, but only the gold region
./plotBackgroundRate ../../resources/LikelihoodFiles/debugBelow2keVFromAllChip/lhood_2017_septemveto_gold_dbscan_no_septemveto.h5 \ ../../resources/LikelihoodFiles/debugBelow2keVFromAllChip/lhood_2018_septemveto_gold_dbscan_no_septemveto.h5 \ --combName bla \ --combYear 2018 \ --title "no septem veto, gold, no lineveto" \ --region crGold
yields the following plot, fig. 313
So we can see that not using the septem veto yields much more background below 2 keV.
Next we look at using the septem veto, but still no line veto and only the gold region
./plotBackgroundRate ../../resources/LikelihoodFiles/debugBelow2keVFromAllChip/lhood_2017_septemveto_gold_dbscan_no_lineveto.h5 \ ../../resources/LikelihoodFiles/debugBelow2keVFromAllChip/lhood_2018_septemveto_gold_dbscan_no_lineveto.h5 \ --combName bla \ --combYear 2018 \ --title "septem veto, gold, no lineveto" \ --region crGold
yields the following plot, fig. 314.
So, surprise, we can see that the last plot looks pretty much identical to the no lineveto plot from the whole chip, fig. 314. That means the explanation is that the plot we sent to Esther of course included the line veto, but it doesn't work for the whole chip. The files we used were generated on my desktop where we modified the following line:
let inGoldRegion = inRegion(cl.centerX - 14.0, cl.centerY - 14.0, crGold) # to let inGoldRegion = inRegion(cl.centerX - 14.0, cl.centerY - 14.0, crAll)
This effectively disabled the line veto. I have no idea what the code like this would really do to be honest (it could have been worse). We need to think about whether there is a way to apply it on the whole chip?
20.9.1. TODO Can we apply line veto to whole chip?
See the discussion above. Simple question.
20.10. TODO Compute background rates for IAXO TDR and IAXO CM talk
As the mess in 6.3.5 was just that, a mess, let's take notes about the background rates we compute.
Files are computed now:
/home/basti/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/
Just quickly plotted the comparison of the scinti veto files with the "regular" files (no vetoes). They are exactly the same…. So time to debug the scinti veto. :( UPDATE: Uhh, well. Apparently I did something dumb. They do differ after all!
All the files are in the mentioned place.
Now generate the correct plots. Start with "no veto" background rate:
cd ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/ plotBackgroundRate lhood_2017_crGold.h5 lhood_2018_crGold.h5 --combName 2017/18 --combYear 2018 \ --suffix "no vetoes" \ --title "Background rate CAST GridPix 2017/18, no vetoes, $ε = \SI{80}{\percent}$" \ --useTeX --genTikZ
Scinti veto (only 2018 comparison):
cd ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/ plotBackgroundRate lhood_2018_crGold.h5 lhood_2018_crGold_scintiveto.h5 -n "no vetoes" -n "scinti veto" \ --suffix "scinti veto" \ --hideErrors \ --title "Background rate CAST GridPix 2018, scinti veto, $ε = \SI{80}{\percent}$" \ --useTeX --genTikZ
Septem veto:
cd ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/ plotBackgroundRate lhood_2017_crGold.h5 lhood_2018_crGold.h5 \ lhood_2017_crGold_scintiveto_septemveto.h5 lhood_2018_crGold_scintiveto_septemveto.h5 \ -n "no vetoes" -n "no vetoes" -n "septem veto" -n "septem veto" \ --suffix "septem veto" \ --hideErrors \ --title "Background rate CAST GridPix 2017/18, scinti \& septem veto, $ε = \SI{80}{\percent}$" \ --useTeX
Septem veto + scinti veto:
cd ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/ plotBackgroundRate lhood_2017_crGold.h5 lhood_2018_crGold.h5 \ lhood_2017_crGold_scintiveto.h5 lhood_2018_crGold_scintiveto.h5 \ lhood_2017_crGold_scintiveto_septemveto.h5 lhood_2018_crGold_scintiveto_septemveto.h5 \ -n "no vetoes" -n "no vetoes" -n "scinti veto" -n "scinti veto" -n "septem veto" -n "septem veto" \ --suffix "scinti veto septem veto" \ --hideErrors \ --title "Background rate CAST GridPix 2017/18, scinti \& septem veto, $ε = \SI{80}{\percent}$" \ --useTeX --genTikZ
Line veto:
cd ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/ plotBackgroundRate lhood_2017_crGold.h5 lhood_2018_crGold.h5 \ lhood_2017_crGold_scintiveto_septemveto_lineveto.h5 lhood_2018_crGold_scintiveto_septemveto_lineveto.h5 \ -n "no vetoes" -n "no vetoes" -n "line veto" -n "line veto" \ --suffix "line veto" \ --hideErrors \ --title "Background rate CAST GridPix 2017/18, scinti \& septem \& line veto, $ε = \SI{80}{\percent}$" \ --useTeX
Line veto + Septem veto + scinti veto:
cd ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/ plotBackgroundRate lhood_2017_crGold.h5 lhood_2018_crGold.h5 \ lhood_2017_crGold_scintiveto.h5 lhood_2018_crGold_scintiveto.h5 \ lhood_2017_crGold_scintiveto_septemveto.h5 lhood_2018_crGold_scintiveto_septemveto.h5 \ lhood_2017_crGold_scintiveto_septemveto_lineveto.h5 lhood_2018_crGold_scintiveto_septemveto_lineveto.h5 \ -n "no vetoes" -n "no vetoes" -n "scinti veto" -n "scinti veto" \ -n "septem veto" -n "septem veto" -n "line veto" -n "line veto" \ --suffix "scinti veto septem veto line veto" \ --hidePoints --hideErrors \ --title "Background rate CAST GridPix 2017/18, scinti \& septem \& line veto, $ε = \SI{80}{\percent}$" \ --useTeX --genTikZ
20.10.1. Generate event displays side by side
The event displays currently used based on:
/home/basti/org/Figs/statusAndProgress/exampleEvents/background_event_run267_chip3_event1456_region_crAll_hits_200.0_250.0_centerX_4.5_9.5_centerY_4.5_9.5_applyAll_true_numIdxs_100.pdf/home/basti/org/Figs/statusAndProgress/exampleEvents/calibration_event_run266_chip3_event5791_region_crAll_hits_200.0_250.0_centerX_4.5_9.5_centerY_4.5_9.5_applyAll_true_numIdxs_100.pdf
are too ugly for the TDR.
Added to the ingridEventIter in plotData:
for (tup, subDf) in groups(group_by(events, "Index")): let event = tup[0][1].toInt if run == 266 and event == 5791 or run == 267 and event == 1456: subDf.writeCsv(&"/tmp/run_{run}_event_{event}.csv")
Now regenerate the plots:
./plotData --h5file /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 \ --runType=rtBackground --cuts '("hits", 200, 250)' \ --cuts '("centerX", 4.5, 9.5)' --cuts '("centerY", 4.5, 9.5)' \ --applyAllCuts --eventDisplay 267 --head 100 --chips 3 ./plotData --h5file /mnt/1TB/CAST/2018_2/CalibrationRuns2018_Reco.h5 \ --runType=rtCalibration --cuts '("hits", 200, 250)' \ --cuts '("centerX", 4.5, 9.5)' --cuts '("centerY", 4.5, 9.5)' \ --applyAllCuts --eventDisplay 266 --head 100 --chips 3
which generated the corresponding CSV files, which now live in:
/home/basti/org/resources/exampleEvents/run_266_event_5791.csv/home/basti/org/resources/exampleEvents/run_267_event_1456.csv
Now we generate a pretty TeX plot from it:
import ggplotnim let dfCalib = readCsv("/home/basti/org/resources/exampleEvents/run_266_event_5791.csv") let dfBack = readCsv("/home/basti/org/resources/exampleEvents/run_267_event_1456.csv") let df = bind_rows([(r"$ ^{55}\text{Fe}$ Calibration", dfCalib), ("Background", dfBack)], id = "Type") .rename(f{"ToT" <- "ch"}) ggplot(df, aes("x", "y", color = "ToT")) + facet_wrap("Type") + geom_point() + ylab(margin = 2.0, tickMargin = -0.5) + xlab(tickMargin = 2.5) + xlim(0, 256) + ylim(0, 256) + ggsave("/home/basti/org/Figs/statusAndProgress/exampleEvents/calibration_background_comparison.tex", width = 900, height = 480, useTeX = true, onlyTikZ = true)
And now also generate the background cluster plot as a facet plot.
./plotBackgroundClusters \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2017_crAll_new_septemveto_lineveto_fixed_inRegion.h5 \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/debugSeptemVeto/lhood_2018_crAll_new_septemveto_lineveto_fixed_inRegion.h5 \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_all_chip_no_septemveto_dbscan.h5 \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2018_all_chip_no_septemveto_dbscan.h5 \ -n "all vetoes" -n "all vetoes" -n "no vetoes" -n "no vetoes" \ --suffix "_veto_comparison"
which generates the plot:
~/org/Figs/statusAndProgress/IAXO_TDR/background_cluster_centers_veto_comparison.pdf
Next, let's compute a comparison of the effect of the vetoes on the center region alone. We do this by generating the background plot comparison, same as above here, only for the gold data. Then we can read off the number of clusters in each.
./plotBackgroundClusters \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/lhood_2017_crGold.h5 \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/lhood_2018_crGold.h5 \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/lhood_2017_crGold_scintiveto_septemveto_lineveto.h5 \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/lhood_2018_crGold_scintiveto_septemveto_lineveto.h5 \ -n "all vetoes" -n "all vetoes" -n "no vetoes" -n "no vetoes" \ --suffix "_crGold_veto_comparison"
So from 960 clusters we go down to 534, which is a reduction to 55% of the original.
And finally the integrated background rates while excluding the Ar peak.
plotBackgroundRate \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/lhood_2017_crGold_scintiveto_septemveto_lineveto.h5 \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/lhood_2018_crGold_scintiveto_septemveto_lineveto.h5 \ --combName "vetoes" --combYear 2018
which yields:
Dataset: vetoes
Integrated background rate in range: 0.0 .. 12.0: 1.5637e-04 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.0 .. 12.0: 1.3031e-05 keV⁻¹ cm⁻² s⁻¹
Dataset: vetoes
Integrated background rate in range: 0.5 .. 2.5: 1.4565e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.5 .. 2.5: 7.2827e-06 keV⁻¹ cm⁻² s⁻¹
Dataset: vetoes
Integrated background rate in range: 0.5 .. 5.0: 5.9433e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.5 .. 5.0: 1.3207e-05 keV⁻¹ cm⁻² s⁻¹
Dataset: vetoes
Integrated background rate in range: 0.0 .. 2.5: 1.9086e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.0 .. 2.5: 7.6342e-06 keV⁻¹ cm⁻² s⁻¹
Dataset: vetoes
Integrated background rate in range: 4.0 .. 8.0: 2.4945e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 4.0 .. 8.0: 6.2363e-06 keV⁻¹ cm⁻² s⁻¹
Dataset: vetoes
Integrated background rate in range: 0.0 .. 8.0: 8.6053e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.0 .. 8.0: 1.0757e-05 keV⁻¹ cm⁻² s⁻¹
So, let's compute the background rate in the region 0-2.5, 4 - 8 keV:
echo ((7.6342e-06 * 2.5) + (6.2363e-06 * 4.0)) / (2.5 + 4.0)
Meaning we get a background rate of 6.77e-06 keV⁻¹ cm⁻² s⁻¹ in that region.
And with the aggressive veto:
plotBackgroundRate \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/lhood_2017_aggressive.h5 \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_TDR/lhood_2018_aggressive.h5 \ --combName "vetoes" --combYear 2018
yields:
Dataset: vetoes
Integrated background rate in range: 0.0 .. 12.0: 5.6755e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.0 .. 12.0: 4.7295e-06 keV⁻¹ cm⁻² s⁻¹
Dataset: vetoes
Integrated background rate in range: 0.5 .. 2.5: 4.0180e-06 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.5 .. 2.5: 2.0090e-06 keV⁻¹ cm⁻² s⁻¹
Dataset: vetoes
Integrated background rate in range: 0.5 .. 5.0: 1.4063e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.5 .. 5.0: 3.1251e-06 keV⁻¹ cm⁻² s⁻¹
Dataset: vetoes
Integrated background rate in range: 0.0 .. 2.5: 5.8596e-06 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.0 .. 2.5: 2.3438e-06 keV⁻¹ cm⁻² s⁻¹
Dataset: vetoes
Integrated background rate in range: 4.0 .. 8.0: 1.2221e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 4.0 .. 8.0: 3.0554e-06 keV⁻¹ cm⁻² s⁻¹
Dataset: vetoes
Integrated background rate in range: 0.0 .. 8.0: 2.7289e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.0 .. 8.0: 3.4111e-06 keV⁻¹ cm⁻² s⁻¹
which itself results in a background rate in the same 0 - 2.5, 4 - 8 keV region:
echo ((2.3438e-06 * 2.5) + (3.0554e-06 * 4.0)) / (2.5 + 4.0)
So a background rate of 2.78e-06 keV⁻¹ cm⁻² s⁻¹
20.11. Background rate after LogL mapping bug fixed
I noticed a HUGE bug in the likelihood code (see sec. 14.7). The mapping of the data from the CDL target / filter combination to the determination of the cut value was wrong. That caused the cut values to be associated with the wrong energies.
Let's find out what this means for the background rate:
likelihood /mnt/1TB/CAST/201{7,8_2}/DataRuns201{7,8}_Reco.h5 --h5out ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_201{7,8}_gold_cdl_mapping_fixed.h5 \
--altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \
--altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \
--cdlYear=2018 --region=crGold
and the background rate:
plotBackgroundRate \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_gold_cdl_mapping_fixed.h5 \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2018_gold_cdl_mapping_fixed.h5 \ --combName "cdl_fixed" --combYear 2018
which results in this background rate:
And the relevant background rate output:
Dataset: No vetoes
Integrated background rate/keV in range: 0.0 .. 12.0: 1.7160e-05 keV⁻¹ cm⁻² s⁻¹
Dataset: No vetoes
Integrated background rate/keV in range: 0.5 .. 2.5: 2.6619e-05 keV⁻¹ cm⁻² s⁻¹
Dataset: No vetoes
Integrated background rate/keV in range: 0.5 .. 5.0: 2.1988e-05 keV⁻¹ cm⁻² s⁻¹
Dataset: No vetoes
Integrated background rate/keV in range: 0.0 .. 2.5: 3.0202e-05 keV⁻¹ cm⁻² s⁻¹
Dataset: No vetoes
Integrated background rate/keV in range: 0.0 .. 8.0: 1.7725e-05 keV⁻¹ cm⁻² s⁻¹
and now including all vetoes…
likelihood /mnt/1TB/CAST/201{7,8_2}/DataRuns201{7,8}_Reco.h5 --h5out ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_201{7,8}_gold_vetoes_cdl_mapping_fixed.h5 \
--altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \
--altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \
--cdlYear=2018 --region=crGold --lineveto --septemveto --scintiveto
Background rates:
Dataset: Vetoes
Integrated background rate in range: 0.0 .. 12.0: 1.2339e-04 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.0 .. 12.0: 1.0282e-05 keV⁻¹ cm⁻² s⁻¹
Dataset: Vetoes
Integrated background rate in range: 0.5 .. 2.5: 1.5068e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.5 .. 2.5: 7.5338e-06 keV⁻¹ cm⁻² s⁻¹
Dataset: Vetoes
Integrated background rate in range: 0.5 .. 5.0: 4.7379e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.5 .. 5.0: 1.0529e-05 keV⁻¹ cm⁻² s⁻¹
Dataset: Vetoes
Integrated background rate in range: 0.0 .. 2.5: 2.1932e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.0 .. 2.5: 8.7727e-06 keV⁻¹ cm⁻² s⁻¹
Dataset: Vetoes
Integrated background rate in range: 4.0 .. 8.0: 1.8081e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 4.0 .. 8.0: 4.5203e-06 keV⁻¹ cm⁻² s⁻¹
Dataset: Vetoes
Integrated background rate in range: 0.0 .. 8.0: 6.9981e-05 cm⁻² s⁻¹
Integrated background rate/keV in range: 0.0 .. 8.0: 8.7476e-06 keV⁻¹ cm⁻² s⁻¹
And for the complete chip (for the limit calculation):
likelihood DataRuns2017_Reco.h5 \ --h5out ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_all_vetoes_dbscan_cdl_mapping_fixed.h5 \ --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crAll --scintiveto --septemveto --lineveto
likelihood DataRuns2018_Reco.h5 \ --h5out ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2018_all_vetoes_dbscan_cdl_mapping_fixed.h5 \ --altCdlFile /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 \ --altRefFile /mnt/1TB/CAST/CDL_2019/XrayReferenceFile2018.h5 \ --cdlYear=2018 --region=crAll --scintiveto --septemveto --lineveto
20.12. Compute background rate for IAXO CM Sep 2022 Zaragoza
We compute the background rate for the IAXO CM talk at Zaragoza in 2022 using the files mentioned in the 20.11 now.
In addition a few further likelihood files will be generated in the same way as shown above, but for the individual veto contributions.
The following plots will be generated:
- crGold no vetos
- crGold with scinti veto
- crGold with scinti + septem veto
- crGold with scinti + septem veto + line veto
- with 'aggressive' veto, but based on older file for simplicity.
20.12.1. Generate the additional likelihood files
The new files will be stored in ./../../CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_CM_2022/
- Script to generate all outputs
This just generates all lhood files for the different scinti setups based on the current H5
DataReco*files on my laptop on .import shell, os, strformat, strutils const outpath = "/home/basti/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_CM_2022" const datapath = "/mnt/1TB/CAST/" const cdlPath = dataPath / "CDL_2019/calibration-cdl-2018.h5" const filePrefix = "DataRuns$#_Reco.h5" const outPrefix = "lhood_$#_crGold_$#.h5" const scv = "--scintiveto" const svv = "--septemveto" const liv = "--lineveto" const ag const yearPaths = ["2017", "2018_2"] const yearNames = ["2017", "2018"] const vetoes = ["", scv, &"{scv} {svv}", &"{scv} {svv} {liv}"] const vetoNames = ["no_vetoes", "scinti", "scinti_septem", "scinti_septem_line"] for i, path in yearPaths: for j, veto in vetoes: if j == 0: continue # skip no vetoes let filePath = dataPath / path let fileName = filePath / (filePrefix % yearNames[i]) let outName = outpath / (outPrefix % [yearNames[i], vetoNames[j]]) shell: one: cd ($filePath) likelihood -f ($fileName) --h5out ($outName) --cdlFile ($cdlPath) --cdlYear 2018 --region crGold ($veto)
20.12.2. Generate background rate plots from likelihood files
crGoldwithout vetoes
DATA=~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_CM_2022/ plotBackgroundRate $DATA/lhood_2017_crGold_no_vetoes.h5 $DATA/lhood_2018_crGold_no_vetoes.h5 \ -n "No vetoes" -n "No vetoes" --combYear 2018
Dataset: No vetoes Integrated background rate in range: 0.0 .. 12.0: 2.1111e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.7593e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 2.5: 5.4746e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.7373e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 5.0: 1.0229e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.2732e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 2.5: 7.7180e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.0872e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 4.0 .. 8.0: 2.4945e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 6.2363e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 8.0: 1.4666e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.8332e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 2.0 .. 8.0: 7.4166e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.2361e-05 keV⁻¹·cm⁻²·s⁻¹
crGoldwith scinti veto
DATA=~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_CM_2022/ plotBackgroundRate $DATA/lhood_2017_crGold_no_vetoes.h5 $DATA/lhood_2018_crGold_no_vetoes.h5 \ $DATA/lhood_2017_crGold_scinti.h5 $DATA/lhood_2018_crGold_scinti.h5 \ -n "No vetoes" -n "No vetoes" -n "Scinti" -n "Scinti" --combYear 2018 --hidePoints --hideErrors
Dataset: No vetoes Integrated background rate in range: 0.0 .. 12.0: 2.1111e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.7593e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 12.0: 1.9303e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.6086e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 2.5: 5.4746e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.7373e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.5 .. 2.5: 5.3574e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.6787e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 5.0: 1.0229e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.2732e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.5 .. 5.0: 9.3921e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.0871e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 2.5: 7.7180e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.0872e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 2.5: 7.5673e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.0269e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 4.0 .. 8.0: 2.4945e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 6.2363e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 4.0 .. 8.0: 2.2936e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 5.7341e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 8.0: 1.4666e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.8332e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 8.0: 1.3594e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.6993e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 2.0 .. 8.0: 7.4166e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.2361e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 2.0 .. 8.0: 6.4623e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.0771e-05 keV⁻¹·cm⁻²·s⁻¹
crGoldwith scinti + septem veto
DATA=~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_CM_2022/ plotBackgroundRate $DATA/lhood_2017_crGold_no_vetoes.h5 $DATA/lhood_2018_crGold_no_vetoes.h5 \ $DATA/lhood_2017_crGold_scinti.h5 $DATA/lhood_2018_crGold_scinti.h5 \ $DATA/lhood_2017_crGold_scinti_septem.h5 $DATA/lhood_2018_crGold_scinti_septem.h5 \ -n "No vetoes" -n "No vetoes" -n "Scinti" -n "Scinti" -n "Septem" -n "Septem" --combYear 2018 --hidePoints --hideErrors
Dataset: No vetoes Integrated background rate in range: 0.0 .. 12.0: 2.1111e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.7593e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 12.0: 1.9303e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.6086e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.0 .. 12.0: 1.3661e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.1384e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 2.5: 5.4746e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.7373e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.5 .. 2.5: 5.3574e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.6787e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.5 .. 2.5: 1.9253e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 9.6265e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 5.0: 1.0229e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.2732e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.5 .. 5.0: 9.3921e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.0871e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.5 .. 5.0: 5.2402e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 1.1645e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 2.5: 7.7180e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.0872e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 2.5: 7.5673e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.0269e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.0 .. 2.5: 3.0805e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 1.2322e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 4.0 .. 8.0: 2.4945e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 6.2363e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 4.0 .. 8.0: 2.2936e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 5.7341e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 4.0 .. 8.0: 1.9755e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 4.9388e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 8.0: 1.4666e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.8332e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 8.0: 1.3594e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.6993e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.0 .. 8.0: 8.1198e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.0150e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 2.0 .. 8.0: 7.4166e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.2361e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 2.0 .. 8.0: 6.4623e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.0771e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 2.0 .. 8.0: 5.3741e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 8.9568e-06 keV⁻¹·cm⁻²·s⁻¹
crGoldwith scinti + septem + line veto
DATA=~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_CM_2022/ plotBackgroundRate $DATA/lhood_2017_crGold_no_vetoes.h5 $DATA/lhood_2018_crGold_no_vetoes.h5 \ $DATA/lhood_2017_crGold_scinti.h5 $DATA/lhood_2018_crGold_scinti.h5 \ $DATA/lhood_2017_crGold_scinti_septem.h5 $DATA/lhood_2018_crGold_scinti_septem.h5 \ $DATA/lhood_2017_crGold_scinti_septem_line.h5 $DATA/lhood_2018_crGold_scinti_septem_line.h5 \ -n "No vetoes" -n "No vetoes" -n "Scinti" -n "Scinti" -n "Septem" -n "Septem" -n "Line" -n "Line" \ --combYear 2018 --hidePoints --hideErrors
Dataset: Line Integrated background rate in range: 0.0 .. 12.0: 1.2138e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.0115e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 12.0: 2.1111e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.7593e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 12.0: 1.9303e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.6086e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.0 .. 12.0: 1.3661e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.1384e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 0.5 .. 2.5: 1.3226e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 6.6130e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 2.5: 5.4746e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.7373e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.5 .. 2.5: 5.3574e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.6787e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.5 .. 2.5: 1.9253e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 9.6265e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 0.5 .. 5.0: 4.4700e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 9.9334e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 5.0: 1.0229e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.2732e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.5 .. 5.0: 9.3921e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.0871e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.5 .. 5.0: 5.2402e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 1.1645e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 0.0 .. 2.5: 1.9923e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 7.9691e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 2.5: 7.7180e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.0872e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 2.5: 7.5673e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.0269e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.0 .. 2.5: 3.0805e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 1.2322e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 4.0 .. 8.0: 1.8751e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 4.6877e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 4.0 .. 8.0: 2.4945e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 6.2363e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 4.0 .. 8.0: 2.2936e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 5.7341e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 4.0 .. 8.0: 1.9755e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 4.9388e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 0.0 .. 8.0: 6.7972e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 8.4964e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 8.0: 1.4666e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.8332e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 8.0: 1.3594e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.6993e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.0 .. 8.0: 8.1198e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.0150e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 2.0 .. 8.0: 5.1062e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 8.5104e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 2.0 .. 8.0: 7.4166e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.2361e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 2.0 .. 8.0: 6.4623e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.0771e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 2.0 .. 8.0: 5.3741e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 8.9568e-06 keV⁻¹·cm⁻²·s⁻¹
20.12.3. Generate lhood files using aggresive veto
Can we regenerate the background rate using the aggressive veto?
Copied over
/home/basti/CastData/ExternCode/TimepixAnalysis/Tools/NN_playground/trained_model.pt
to /tmp (as in
~/CastData/ExternCode/TimepixAnalysis/Tools/NN_playground/predict_event.nim
we read the model from tmp).
Then recompiled likelihood:
nim cpp -d:danger -d:cuda likelihood.nim
after modifying the code to actual predict events if the
fkAggressive flag is set.
Then we run:
likelihood -f ~/CastData/data/DataRuns201{7,8}_Reco.h5 \
--h5out /tmp/lhood_201{7,8}_crGold_scinti_septem_line_aggressive.h5 \
--cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \
--cdlYear=2018 --region=crGold \
--scintiveto --septemveto --lineveto --aggressive
both of which we copied over to
~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_CM_2022
together with the other files to generate the background plots.
NOTE: as these likelihood files were not generated using the same
input DataRuns* files, there may be differences…
Generate the background rate:
DATA=~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_CM_2022/ plotBackgroundRate $DATA/lhood_2017_crGold_no_vetoes.h5 $DATA/lhood_2018_crGold_no_vetoes.h5 \ $DATA/lhood_2017_crGold_scinti.h5 $DATA/lhood_2018_crGold_scinti.h5 \ $DATA/lhood_2017_crGold_scinti_septem.h5 $DATA/lhood_2018_crGold_scinti_septem.h5 \ $DATA/lhood_2017_crGold_scinti_septem_line.h5 $DATA/lhood_2018_crGold_scinti_septem_line.h5 \ $DATA/lhood_2017_crGold_scinti_septem_line_aggressive.h5 $DATA/lhood_2018_crGold_scinti_septem_line_aggressive.h5 \ -n "No vetoes" -n "No vetoes" -n "Scinti" -n "Scinti" -n "Septem" -n "Septem" \ -n "Line" -n "Line" -n "MLP" -n "MLP" \ --combYear 2018 --hidePoints --hideErrors
Dataset: Line
[88/10104]Integrated background rate in range: 0.0 .. 12.0: 1.2138e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.0115e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 0.0 .. 12.0: 4.7212e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 3.9343e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 12.0: 2.1111e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.7593e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 12.0: 1.9303e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.6086e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.0 .. 12.0: 1.3661e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 1.1384e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 0.5 .. 2.5: 1.3226e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 6.6130e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 0.5 .. 2.5: 4.0180e-06 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.0090e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 2.5: 5.4746e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.7373e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.5 .. 2.5: 5.3574e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 2.6787e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.5 .. 2.5: 1.9253e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 9.6265e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 0.5 .. 5.0: 4.4700e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 9.9334e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 0.5 .. 5.0: 1.2389e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.7531e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.5 .. 5.0: 1.0229e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.2732e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.5 .. 5.0: 9.3921e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 2.0871e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.5 .. 5.0: 5.2402e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 1.1645e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 0.0 .. 2.5: 1.9923e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 7.9691e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 0.0 .. 2.5: 6.6967e-06 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 2.6787e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 2.5: 7.7180e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.0872e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 2.5: 7.5673e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.0269e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.0 .. 2.5: 3.0805e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 1.2322e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 4.0 .. 8.0: 1.8751e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 4.6877e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 4.0 .. 8.0: 9.8776e-06 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 2.4694e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 4.0 .. 8.0: 2.4945e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 6.2363e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 4.0 .. 8.0: 2.2936e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 5.7341e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 4.0 .. 8.0: 1.9755e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 4.9388e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 0.0 .. 8.0: 6.7972e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 8.4964e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 0.0 .. 8.0: 2.4276e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 3.0344e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 0.0 .. 8.0: 1.4666e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.8332e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 8.0: 1.3594e-04 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.6993e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.0 .. 8.0: 8.1198e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.0150e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 2.0 .. 8.0: 5.1062e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 8.5104e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 2.0 .. 8.0: 1.9253e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 3.2088e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: No vetoes Integrated background rate in range: 2.0 .. 8.0: 7.4166e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.2361e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 2.0 .. 8.0: 6.4623e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.0771e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 2.0 .. 8.0: 5.3741e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 8.9568e-06 keV⁻¹·cm⁻²·s⁻¹
i.e. a background rate of 3.0344e-06 keV⁻¹·cm⁻²·s⁻¹!
- Background rate with
ε = 50%and aggressive vetoε = 80%
DATA=~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/IAXO_CM_2022/ plotBackgroundRate #$DATA/lhood_2017_crGold_no_vetoes.h5 $DATA/lhood_2018_crGold_no_vetoes.h5 \ $DATA/lhood_2017_crGold_scinti_eff_50.h5 $DATA/lhood_2018_crGold_scinti_eff_50.h5 \ $DATA/lhood_2017_crGold_scinti_septem_eff_50.h5 $DATA/lhood_2018_crGold_scinti_septem_eff_50.h5 \ $DATA/lhood_2017_crGold_scinti_septem_line_eff_50.h5 $DATA/lhood_2018_crGold_scinti_septem_line_eff_50.h5 \ $DATA/lhood_2017_crGold_scinti_septem_line_aggressive_eff_50.h5 $DATA/lhood_2018_crGold_scinti_septem_line_aggressive_eff_50.h5 \ -n "Scinti" -n "Scinti" -n "Septem" -n "Septem" \ -n "Line" -n "Line" -n "MLP" -n "MLP" \ --combYear 2018 --hidePoints --hideErrors
Dataset: Line Integrated background rate in range: 0.0 .. 12.0: 7.0483e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 5.8736e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 0.0 .. 12.0: 2.8461e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 2.3717e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 12.0: 9.7437e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 8.1198e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.0 .. 12.0: 7.7180e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 12.0: 6.4316e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 0.5 .. 2.5: 7.1990e-06 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 3.5995e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 0.5 .. 2.5: 2.0090e-06 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 1.0045e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.5 .. 2.5: 2.1597e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 1.0798e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.5 .. 2.5: 9.5428e-06 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 2.5: 4.7714e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 0.5 .. 5.0: 2.7791e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 6.1758e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 0.5 .. 5.0: 7.0315e-06 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 1.5626e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.5 .. 5.0: 4.5705e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 1.0157e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.5 .. 5.0: 3.1140e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.5 .. 5.0: 6.9199e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 0.0 .. 2.5: 9.8776e-06 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 3.9511e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 0.0 .. 2.5: 3.0135e-06 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 1.2054e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 2.5: 3.0637e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 1.2255e-05 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.0 .. 2.5: 1.4230e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 2.5: 5.6922e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 4.0 .. 8.0: 1.1384e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 2.8461e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 4.0 .. 8.0: 5.1899e-06 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 1.2975e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 4.0 .. 8.0: 1.3226e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 3.3065e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 4.0 .. 8.0: 1.2389e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 4.0 .. 8.0: 3.0972e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 0.0 .. 8.0: 4.1017e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 5.1272e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 0.0 .. 8.0: 1.3226e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 1.6532e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 0.0 .. 8.0: 6.6800e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 8.3500e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 0.0 .. 8.0: 4.7044e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 0.0 .. 8.0: 5.8805e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Line Integrated background rate in range: 2.0 .. 8.0: 3.2479e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 5.4132e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: MLP Integrated background rate in range: 2.0 .. 8.0: 1.1217e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 1.8695e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Scinti Integrated background rate in range: 2.0 .. 8.0: 3.8673e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 6.4456e-06 keV⁻¹·cm⁻²·s⁻¹ Dataset: Septem Integrated background rate in range: 2.0 .. 8.0: 3.4488e-05 cm⁻² s⁻¹ Integrated background rate/keV in range: 2.0 .. 8.0: 5.7480e-06 keV⁻¹·cm⁻²·s⁻¹
Relevant background rates for 0-8 keV:
- Line veto: 5.1272e-06 keV⁻¹·cm⁻²·s⁻¹
- MLP veto: 1.6532e-06 keV⁻¹·cm⁻²·s⁻¹
21. Energy calibration of clusters
The energy calibration of individual clusters performed by Krieger and then also applied in TimepixAnalysis is reasonably complex.
21.1. TODO explain calculation of gas gain
21.2. TODO explain fit of Fe spectrum
21.3. TODO explain scatter plot of gas gain vs fit parameter
21.4. TODO reference problems due to detector instability
21.5. TODO explain new changes, not all runs anymore
22. STARTED Morphing of CDL reference spectra
One problem with the current approach of utilizing the CDL data is that the reference distributions for the different logL variables are non continuous between two energy bins. This means that if a cluster is moved from one bin to another it suddenly has a very different cut for each property.
It might be possible to morph CDL spectra between two energies. That is to allow interpolation between the shape of two neighboring reference datasets.
This is the likely cause for the sudden steps visible in the background rate. With a fully morphed function this should hopefully disappear.
22.1. References & ideas
Read up on morphing of different functions:
- in HEP: https://indico.cern.ch/event/507948/contributions/2028505/attachments/1262169/1866169/atlas-hcomb-morphwshop-intro-v1.pdf
- https://mathematica.stackexchange.com/questions/208990/morphing-between-two-functions
- https://mathematica.stackexchange.com/questions/209039/convert-symbolic-to-numeric-code-speed-up-morphing
Aside from morphing, the theory of optimal transport seems to be directly related to such problems:
- https://de.wikipedia.org/wiki/Optimaler_Transport (funny, this can be described by topology using GR lingo; there's no English article on this)
- https://en.wikipedia.org/wiki/Transportation_theory_(mathematics)
see in particular:

This seems to imply that given some functions \(f(x)\), \(g(x)\) we are looking for the transport function \(T\) which maps \(T: f(x) \rightarrow g(x)\) in the language of transportation theory.
See the linked article about the Wasserstein metric:
- https://en.wikipedia.org/wiki/Wasserstein_metric in particular the section about its connection to the optimal transport problem
It describes a distance metric between two probability distributions. In that sense the distance between two distributions to be transported is directly related to the Wasserstein distance.
One of the major constraints of transportation theory is that the transportation has to preserve the integral of the transported function. Technically, this is not the case for our application to the CDL data, due to different amount of data available for each target. However, of course we normalize the CDL data and assume that the given data actually is a PDF. At that point each distribution is normalized to 1 and thus each morphed function has to be normalized to 1 as well. This is a decent check for a morph result. If a morphing technique does not satisfy this property we need to renormalize the result.
On the other hand, considering the slides by Verkerke (indico.cern.ch link above), the morphing between two functions can also be interpreted as a simple interpolation problem.
In that sense there are multiple approaches to compute an intermediate step of the CDL distributions.
- visualize the CDL data as a 2D heatmap:
- value of the logL variable is x
- energy of each fluorescence line is y
- linear interpolation at a specific energy \(E\) based on the two neighboring CDL distributions (interpolation thus only along y axis)
- spline interpolation over all energy ranges
- KDE along the energy axis (only along y) Extend KDE to 2D?
- bicubic interpolation (-> problematic, because our energies/variables are not spread on a rectilinear grid (energy is not spread evenly)
- other distance based interpolations, i.e. KD-tree? Simply perform an interpolation based on all neighboring points in a certain distance?
Of these 2 and 4 seem to be the easiest implementations. KD-tree would also be easy, provided I finish the implementation finally.
We will investigate different ideas in ./../../CastData/ExternCode/TimepixAnalysis/Tools/cdlMorphing/cdlMorphing.nim.
22.1.1. DONE Visualize CDL data as a 2D heatmap
In each of the following plots each distribution (= target filter combination) is normalized to 1.
First let's visualize all of the CDL data as a scatter plot. That is pretty simple and gives an idea where the lines are and what the shape is roughly, fig. 317.
Now we can check out what the data looks like if we interpret the whole (value of each variable, Energy) phase space as a tile map. In this way, morphing can be interpreted as performing interpolation along the energy axis in the resulting tile map.
Each figure contains in addition as colored lines the start of each energy range as currently used. So clusters will be with the distribution defined by the line below.
In addition the energy of each fluorescence lines is plotted in red at the corresponding energy value. This also shows that the intervals and the energy of the lines are highly asymmetric.
22.1.2. DONE Morph by linear interpolation bin by bin
Based on the tile maps in the previous section it seems like a decent idea to perform a linear interpolation for any point in between two intervals:
where \(f_\text{low,high}\) are the distributions below / above the given energy \(E\). \(L_\text{low,high}\) corresponds to the energy of the fluorescence line corresponding to the distribution below / above \(E\). \(\Delta E\) is the difference in energy between the lower and higher fluorescence lines. \(x\) is the value of the given logL variable.
In code this is:
let E = ... # given as argument to function let lineEnergies = getXrayFluorescenceLines() let refLowT = df.filter(f{string -> bool: `Dset` == refLow})["Hist", float] let refHighT = df.filter(f{string -> bool: `Dset` == refHigh})["Hist", float] result = zeros[float](refLowT.size.int) let deltaE = abs(lineEnergies[idx] - lineEnergies[idx+offset]) # walk over each bin and compute linear interpolation between for i in 0 ..< refLowT.size: result[i] = refLowT[i] * (1 - (abs(lineEnergies[idx] - E)) / deltaE) + refHighT[i] * (1 - (abs(lineEnergies[idx+offset] - E)) / deltaE)
Since doing this for a point between two lines is not particularly helpful,
because we do not know what the distribution in between does actually look
like. Instead for validation we will now try to compute the Cu-EPIC-0.9kV
distribution (corresponding to the \(\text{O K}_{\alpha}\) line at
\(\SI{0.525}{\kilo \electronvolt}\)) based on the C-EPIC-0.6kV and Cu-EPIC-2kV
distributions.
That means the interpolate the second ridge from the first and third in the
CDL ridgeline plots.
This is shown in fig. 321, 322 and 323. The real data for each distribution is shown in red and the morphed linear bin-wise interpolation for the second ridge is shown in red.
Cu-EPIC-0.9kV distribution for the eccentricity logL variable using bin wise linear interpolation based on the C-EPIC-0.6kV and Cu-EPIC-2kV distributions. The real data is shown in the second ridge in red and the morphed interpolation is is shown in blue. The agreement is remarkable for the simplicity of the method.Cu-EPIC-0.9kV distribution for the length / transverse RMS logL variable using bin wise linear interpolation based on the C-EPIC-0.6kV and Cu-EPIC-2kV distributions. The real data is shown in the second ridge in red and the morphed interpolation is is shown in blue. The agreement is remarkable for the simplicity of the method.Cu-EPIC-0.9kV distribution for the fraction in transverse RMS logL variable using bin wise linear interpolation based on the C-EPIC-0.6kV and Cu-EPIC-2kV distributions. The real data is shown in the second ridge in red and the morphed interpolation is is shown in blue. This in particular is the problematic variable, due to the integer nature of the data at low energies. However, even here the interpolation works extremely well.22.1.3. DONE Compute all reference spectra from neighbors
Similar to the plots in the previous section we can now compute all reference spectra based on the next neighboring spectras.
This is done in fig. 324, 325, 326.
22.1.4. DONE Compute full linear interpolation between fluorescence lines
We can now apply the lessons from the last section to compute arbitrary reference spectra. We will use this to compute a heatmap of all possible energies in between the first and last fluorescence line.
For all three logL variables, these are shown in fig.
22.2. KDE approach
Using a KDE is problematic, because our data is already pre-binned of course. This leads to a very sparse phase space, which either makes the local prediction around a known distribution good, but fails miserably in between them (small bandwidth) or gives decent predictions in between, but pretty bad reconstruction of the known distributions (larger bandwidth).
There is also a strong conflict in bandwidth selection, due to the non-linear steps in energy between the different CDL distributions. This leads to a too large / too small bandwidth at either end of the energy range.
Fig. 330, 331, 332 show the default bandwidth (Silverman's rule of thumb). In comparison Fig. 333, 334, 335 show the same plot using a much smaller custom bandwidth of 0.3 keV. The agreement is much better, but the actual prediction between the different distributions becomes much worse. Compare fig. 336 (default bandwidth) to fig. 337. The latter has regions of almost no counts, which is obviously wrong.
Note that also the fig. 336 is problematic. An effect of a bad KDE input is visible, namely that the bandwidth vs. number of datapoints is such that the center region (in energy) has higher values than the edges, due to the fact that predictions near the boundaries see no signal (this boundary effect could be corrected for by assuming a suitable boundary conditions, e.g. just extending the first/last distributions in the respective ranges. It is not clear however in what spacing such a distribution should be placed etc.
\clearpage
\clearpage
22.3. Spline approach
Another idea is to use a spline interpolation. This has the advantage that the existing distributions will be correctly predicted (as for the linear interpolation), but possibly yields better results between distributions (or in the case of predicting a known distributions.
Fig. 338, 339, 340 show the prediction using a spline. Same as for the linear interpolation each morphed distribution was computed by excluding that distribution from the spline definition and then predicting the energy of the respective fluorescence line.
The result looks somewhat better in certain areas than the linear interpolation, but has unphysical artifacts in other areas (negative values) while also deviating quite a bit. For that reason it seems like simpler is better in case of CDL morphing (at least if it's done bin-wise).
\clearpage
22.4. Summary
For the time being we will use the linear interpolation method and see where this leads us. Should definitely be a big improvement over the current interval based option.
For the results of applying linear interpolation based morphing to the likelihood analysis see section 20.8.
22.5. Implementation in likelihood.nim
Thoughts on the implememntition in likelihood.nim or CDL morphing.
- Add interpolation code from
cdlMorphing.niminprivate/cdl_cuts.nim. - Add a field to
config.nimthat describes the morphing technique to be used. - Add an enum for the possible morphing techniques,
MorphingKindwith fieldsmkNone,mkLinear - In
calcCutValueTabwe currently return aTable[string, float]mapping target/filter combinations to cut values. This needs to be modified such that we have something that hides away input -> output and yields what we need. Define anCutValueInterpolatortype, which is returned instead. It will be a variant object with casekind: MorphingKind. This object will allow access to cut values based on:string: a target/filter combination.mkNone: access internalTable[string, float]as done currentlymkLinear: raise exception, since does not make sense
float: an energy in keV:mkNone: convert energy to a target/filter combination and access internalTablemkLinear: access the closest energy distribution and return its cut value
- in
filterClustersByLogLreplacecutTabname and access by energy of cluster instead of converted to target/filter dataset
With these steps we should have a working interpolation routine. The
code used in the cdlMorphing.nim test script needs to be added of
course to provide the linearly interpolated logic (see step 0).
22.5.1. Bizarre Al-Al 4kV behavior with mkLinear
After the first implementation we see some very bizarre behavior in the case of linear interpolation for the logL distributions.
This is both visible with the plotCdl.nim as well as plotting code
in likelihood.nim.
See fig. 341.
mkLinear. The Al-Al 4kV line is noweher near where we expect it. The code currently recomputes the logL values by default, in which the mkLinear plays a role. The bug has to be somewhere in that part of the interpolation.
UPDATE: The issue was a couple of bugs & design choices in the
implementation of the linear interpolation in
likelihood_utils.nim. In particular about the design of the DF
returned from getInterpolatedWideDf and a bug accessing not the sub
DF, but the actual DF in the loop.
The fixed result is shown in fig. 342 and in comparison the result using no interpolation (the reference in a way) in fig. 343.
mkLinear and after the above mentioned bug has been fixed. This is the same result as for mkNone, see fig. 343.mkLinear and after the above mentioned bug has been fixed. This is the same result as for mkNone, see fig. 343.23. TODO Muon spectra
Study muon behavior as it happens at CAST wrt. angles & energies to see what we can learn about 8-10 keV hump.
See 26.18 for more information.
Markus: Ratio of Cu fluorescence peaks is 170/17 roundabout Kα to Kβ Tobi: Take note of Cu pipe, which didn't exist in 2014/15 data! (look at paper he sent) Jochen:
- take a look at FADC data spectra for 8 keV peak / SiPM
- 5-8 keV peak in background rate could be Ar escape process of the 8-11 keV hump
According to theory the 8 keV peak is a mix of:
- Cu K alpha line
- orthogonal muons
Muons should deposit their energy according to the Bethe-Bloch equation. So let's compute the expected value according to that.
References:
- Paper about cosmic muons for simulations:
https://arxiv.org/pdf/1606.06907.pdf
~/org/Papers/energy_angular_muons_earth_1606.06907.pdf - Bethe formula: https://en.wikipedia.org/wiki/Bethe_formula
- Lecture about particle matter interactions: https://indico.cern.ch/event/145296/contributions/1381063/attachments/136866/194145/Particle-Interaction-Matter-upload.pdf
- Database of all sorts of element properties by the PDG: https://pdg.lbl.gov/2020/AtomicNuclearProperties/index.html
- PDG itself
- PDG part about muon stopping power: https://pdg.lbl.gov/2020/AtomicNuclearProperties/adndt.pdf (NOTE: includes table of mean energy loss!)
- NIST: X-ray database, contains mean exctitation potential of different elements (I(Z)): https://physics.nist.gov/PhysRefData/XrayMassCoef/tab1.html
Note: the computation is the mean energy loss. The Bethe-Bloch equation of course gives the mean value. For a thin absorber the data will be Landau distributed with a much lower most probable value! Is a 3 cm Ar detector a thin absorber?
| Element | Symbol | Z | A | State | ρ | ⟨−dE/dx⟩min | Eµc | ⟨−dE/dx⟩ | b | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| [g/cm3] | [MeV cm2/g] | [GeV] | & Range | |||||||
| Hydrogen gas | H | 1 | 1.00794 | D | 8.375e−5 | 4.103 | 3611. | I– 1 | VI– 1 | |
| Liquid hydrogen | H | 1 | 1.00794 | L | 7.080e−2 | 4.034 | 3102. | I– 2 | VI– 1 | 1 |
| Helium gas | He | 2 | 4.002602 | G | 1.663e−4 | 1.937 | 2351. | I– 3 | VI– 2 | |
| Liquid helium | He | 2 | 4.002602 | L | 0.125 | 1.936 | 2020. | I– 4 | VI– 2 | 2 |
| Lithium | Li | 3 | 6.941 | S | 0.534 | 1.639 | 1578. | I– 5 | VI– 3 | |
| Beryllium | Be | 4 | 9.012182 | S | 1.848 | 1.595 | 1328. | I– 6 | VI– 4 | |
| Boron | B | 5 | 10.811 | S | 2.370 | 1.623 | 1169. | I– 7 | VI– 5 | |
| Carbon (compact) | C | 6 | 12.0107 | S | 2.265 | 1.745 | 1056. | I– 8 | VI– 6 | |
| Carbon (graphite) | C | 6 | 12.0107 | S | 1.700 | 1.753 | 1065. | I– 9 | VI– 6 | |
| Nitrogen gas | N | 7 | 14.00674 | D | 1.165e−3 | 1.825 | 1153. | I–10 | VI– 7 | |
| Liquid nitrogen | N | 7 | 14.00674 | L | 0.807 | 1.813 | 982. | I–11 | VI– 7 | 2 |
| Oxygen gas | O | 8 | 15.9994 | D | 1.332e−3 | 1.801 | 1050. | I–12 | VI– 8 | |
| Liquid oxygen | O | 8 | 15.9994 | L | 1.141 | 1.788 | 890. | I–13 | VI– 8 | 2 |
| Fluorine gas | F | 9 | 18.9984032 | D | 1.580e−3 | 1.676 | 959. | I–14 | VI– 9 | |
| Liquid fluorine | F | 9 | 18.9984032 | L | 1.507 | 1.634 | 810. | I–15 | VI– 9 | 2 |
| Neon gas | Ne | 10 | 20.1797 | G | 8.385e−4 | 1.724 | 906. | I–16 | VI–10 | |
| Liquid neon | Ne | 10 | 20.1797 | L | 1.204 | 1.695 | 759. | I–17 | VI–10 | 2 |
| Sodium | Na | 11 | 22.989770 | S | 0.971 | 1.639 | 711. | I–18 | VI–11 | |
| Magnesium | Mg | 12 | 24.3050 | S | 1.740 | 1.674 | 658. | I–19 | VI–12 | |
| Aluminum | Al | 13 | 26.981538 | S | 2.699 | 1.615 | 612. | I–20 | VI–13 | |
| Silicon | Si | 14 | 28.0855 | S | 2.329 | 1.664 | 581. | I–21 | VI–14 | 1 |
| Phosphorus | P | 15 | 30.973761 | S | 2.200 | 1.613 | 551. | I–22 | VI–15 | |
| Sulfur | S | 16 | 32.066 | S | 2.000 | 1.652 | 526. | I–23 | VI–16 | |
| Chlorine gas | Cl | 17 | 35.4527 | D | 2.995e−3 | 1.630 | 591. | I–24 | VI–17 | |
| Liquid chlorine | Cl | 17 | 35.4527 | L | 1.574 | 1.608 | 504. | I–25 | VI–17 | 2 |
| Argon gas | Ar | 18 | 39.948 | G | 1.662e−3 | 1.519 | 571. | I–26 | VI–18 | |
| Liquid argon | Ar | 18 | 39.948 | L | 1.396 | 1.508 | 483. | I–27 | VI–18 | 2 |
| Potassium | K | 19 | 39.0983 | S | 0.862 | 1.623 | 470. | I–28 | VI–19 | |
| Calcium | Ca | 20 | 40.078 | S | 1.550 | 1.655 | 445. | I–29 | VI–20 | |
| Scandium | Sc | 21 | 44.955910 | S | 2.989 | 1.522 | 420. | I–30 | VI–21 | |
| Titanium | Ti | 22 | 47.867 | S | 4.540 | 1.477 | 401. | I–31 | VI–22 | |
| Vanadium | V | 23 | 50.9415 | S | 6.110 | 1.436 | 383. | I–32 | VI–23 | |
| Chromium | Cr | 24 | 51.9961 | S | 7.180 | 1.456 | 369. | I–33 | VI–24 | |
| Manganese | Mn | 25 | 54.938049 | S | 7.440 | 1.428 | 357. | I–34 | VI–25 | |
| Iron | Fe | 26 | 55.845 | S | 7.874 | 1.451 | 345. | I–35 | VI–26 | |
| Cobalt | Co | 27 | 58.933200 | S | 8.900 | 1.419 | 334. | I–36 | VI–27 | |
| Nickel | Ni | 28 | 58.6934 | S | 8.902 | 1.468 | 324. | I–37 | VI–28 | |
| Copper | Cu | 29 | 63.546 | S | 8.960 | 1.403 | 315. | I–38 | VI–29 | |
| Zinc | Zn | 30 | 65.39 | S | 7.133 | 1.411 | 308. | I–39 | VI–30 | |
| Gallium | Ga | 31 | 69.723 | S | 5.904 | 1.379 | 302. | I–40 | VI–31 | |
| Germanium | Ge | 32 | 72.61 | S | 5.323 | 1.370 | 295. | I–41 | VI–32 | |
| Element | Symbol | Z | A | State | ρ | ⟨−dE/dx⟩min | Eµc | ⟨−dE/dx⟩ | b | Notes |
| [g/cm3] | [MeV cm2/g] | [GeV] | & Range | |||||||
| Arsenic | As | 33 | 74.92160 | S | 5.730 | 1.370 | 287. | I–42 | VI–33 | |
| Selenium | Se | 34 | 78.96 | S | 4.500 | 1.343 | 282. | I–43 | VI–34 | |
| Bromine | Br | 35 | 79.904 | L | 3.103 | 1.385 | 278. | I–44 | VI–35 | 2 |
| Krypton gas | Kr | 36 | 83.80 | G | 3.478e−3 | 1.357 | 321. | I–45 | VI–36 | |
| Liquid krypton | Kr | 36 | 83.80 | L | 2.418 | 1.357 | 274. | I–46 | VI–36 | 2 |
| Rubidium | Rb | 37 | 85.4678 | S | 1.532 | 1.356 | 271. | I–47 | VI–37 | |
| Strontium | Sr | 38 | 87.62 | S | 2.540 | 1.353 | 262. | I–48 | VI–38 | |
| Zirconium | Zr | 40 | 91.224 | S | 6.506 | 1.349 | 244. | I–49 | VI–39 | |
| Niobium | Nb | 41 | 92.90638 | S | 8.570 | 1.343 | 237. | I–50 | VI–40 | |
| Molybdenum | Mo | 42 | 95.94 | S | 10.220 | 1.330 | 232. | I–51 | VI–41 | |
| Palladium | Pd | 46 | 106.42 | S | 12.020 | 1.289 | 214. | I–52 | VI–42 | |
| Silver | Ag | 47 | 107.8682 | S | 10.500 | 1.299 | 211. | I–53 | VI–43 | |
| Cadmium | Cd | 48 | 112.411 | S | 8.650 | 1.277 | 208. | I–54 | VI–44 | |
| Indium | In | 49 | 114.818 | S | 7.310 | 1.278 | 206. | I–55 | VI–45 | |
| Tin | Sn | 50 | 118.710 | S | 7.310 | 1.263 | 202. | I–56 | VI–46 | |
| Antimony | Sb | 51 | 121.760 | S | 6.691 | 1.259 | 200. | I–57 | VI–47 | |
| Iodine | I | 53 | 126.90447 | S | 4.930 | 1.263 | 195. | I–58 | VI–48 | |
| Xenon gas | Xe | 54 | 131.29 | G | 5.485e−3 | 1.255 | 226. | I–59 | VI–49 | |
| Liquid xenon | Xe | 54 | 131.29 | L | 2.953 | 1.255 | 195. | I–60 | VI–49 | 2 |
| Cesium | Cs | 55 | 132.90545 | S | 1.873 | 1.254 | 195. | I–61 | VI–50 | |
| Barium | Ba | 56 | 137.327 | S | 3.500 | 1.231 | 189. | I–62 | VI–51 | |
| Cerium | Ce | 58 | 140.116 | S | 6.657 | 1.234 | 180. | I–63 | VI–52 | |
| Dysprosium | Dy | 66 | 162.50 | S | 8.550 | 1.175 | 161. | I–64 | VI–53 | |
| Tantalum | Ta | 73 | 180.9479 | S | 16.654 | 1.149 | 145. | I–65 | VI–54 | |
| Tungsten | W | 74 | 183.84 | S | 19.300 | 1.145 | 143. | I–66 | VI–55 | |
| Platinum | Pt | 78 | 195.078 | S | 21.450 | 1.128 | 137. | I–67 | VI–56 | |
| Gold | Au | 79 | 196.96655 | S | 19.320 | 1.134 | 136. | I–68 | VI–57 | |
| Mercury | Hg | 80 | 200.59 | L | 13.546 | 1.130 | 136. | I–69 | VI–58 | |
| Lead | Pb | 82 | 207.2 | S | 11.350 | 1.122 | 134. | I–70 | VI–59 | |
| Bismuth | Bi | 83 | 208.98038 | S | 9.747 | 1.128 | 133. | I–71 | VI–60 | |
| Thorium | Th | 90 | 232.0381 | S | 11.720 | 1.098 | 124. | I–72 | VI–61 | |
| Uranium | U | 92 | 238.0289 | S | 18.950 | 1.081 | 120. | I–73 | VI–62 | |
| Plutonium | Pu | 94 | 244.064197 | S | 19.840 | 1.071 | 117. | I–74 | VI–63 |
And the table for common mixtures:
| Compound or mixture | Formula | ⟨Z/A⟩ | State | ρ | ⟨−dE/dx⟩min | Eµc | ⟨−dE/dx⟩ | b | Notes |
|---|---|---|---|---|---|---|---|---|---|
| [g/cm3] | [MeV cm2/g] | [GeV] | & Range | ||||||
| Acetone | (CH3CHCH3) | 0.55097 | L | 0.790 | 2.003 | 1160. | II– 1 | VII– 1 | |
| Acetylene | (C2H2) | 0.53768 | G | 1.097e−3 | 2.025 | 1400. | II– 2 | VII– 2 | |
| Aluminum oxide | (Al2O3) | 0.49038 | S | 3.970 | 1.647 | 705. | II– 3 | VII– 3 | |
| Barium fluoride | (BaF2) | 0.42207 | S | 4.890 | 1.303 | 227. | II– 4 | VII– 4 | |
| Beryllium oxide | (BeO) | 0.47979 | S | 3.010 | 1.665 | 975. | II– 5 | VII– 5 | |
| Bismuth germanate | (BGO, Bi4(GeO4)3) | 0.42065 | S | 7.130 | 1.251 | 176. | II– 6 | VII– 6 | |
| Butane | (C4H10) | 0.59497 | G | 2.493e−3 | 2.278 | 1557. | II– 7 | VII– 7 | |
| Calcium carbonate | (CaCO3) | 0.49955 | S | 2.800 | 1.686 | 630. | II– 8 | VII– 8 | |
| Calcium fluoride | (CaF2) | 0.49670 | S | 3.180 | 1.655 | 564. | II– 9 | VII– 9 | |
| Calcium oxide | (CaO) | 0.49929 | S | 3.300 | 1.650 | 506. | II–10 | VII–10 | |
| Carbon dioxide | (CO2) | 0.49989 | G | 1.842e−3 | 1.819 | 1094. | II–11 | VII–11 | |
| Solid carbon dioxide | (dry ice) | 0.49989 | S | 1.563 | 1.787 | 927. | II–12 | VII–11 | 2 |
| Cesium iodide | (CsI) | 0.41569 | S | 4.510 | 1.243 | 193. | II–13 | VII–12 | |
| Diethyl ether | ((CH3CH2)2O) | 0.56663 | L | 0.714 | 2.072 | 1220. | II–14 | VII–13 | |
| Ethane | (C2H6) | 0.59861 | G | 1.253e−3 | 2.304 | 1603. | II–15 | VII–14 | |
| Ethanol | (C2H5OH) | 0.56437 | L | 0.789 | 2.054 | 1178. | II–16 | VII–15 | |
| Lithium fluoride | (LiF) | 0.46262 | S | 2.635 | 1.614 | 903. | II–17 | VII–16 | |
| Lithium iodide | (LiI) | 0.41939 | S | 3.494 | 1.272 | 207. | II–18 | VII–17 | |
| Methane | (CH4) | 0.62334 | G | 6.672e−4 | 2.417 | 1715. | II–19 | VII–18 | |
| Octane | (C8H18) | 0.57778 | L | 0.703 | 2.123 | 1312. | II–20 | VII–19 | |
| Paraffin | (CH3(CH2)n≈23CH3) | 0.57275 | S | 0.930 | 2.088 | 1287. | II–21 | VII–20 | |
| Plutonium dioxide | (PuO2) | 0.40583 | S | 11.460 | 1.158 | 136. | II–22 | VII–21 | |
| Liquid propane | (C3H8) | 0.58962 | L | 0.493 | 2.198 | 1365. | II–23 | VII–22 | 1 |
| Silicon dioxide | (fused quartz, SiO2) | 0.49930 | S | 2.200 | 1.699 | 708. | II–24 | VII–23 | 1 |
| Sodium iodide | (NaI) | 0.42697 | S | 3.667 | 1.305 | 223. | II–25 | VII–24 | |
| Toluene | (C6H5CH3) | 0.54265 | L | 0.867 | 1.972 | 1203. | II–26 | VII–25 | |
| Trichloroethylene | (C2HCl3) | 0.48710 | L | 1.460 | 1.656 | 568. | II–27 | VII–26 | |
| Water (liquid) | (H2O) | 0.55509 | L | 1.000 | 1.992 | 1032. | II–28 | VII–27 | |
| Water (vapor) | (H2O) | 0.55509 | G | 7.562e−4 | 2.052 | 1231. | II–29 | VII–2 |
From table: Argon ⟨dE/dx⟩ should be ~ 1.519. Note that this is given
as a mass stopping power, i.e. divided by the material density. At
CAST using Ar at 1050 mbar at room temperature yields a density of
about 1.72 g/cm³.
1.519 * 1.72 = 2.614 keV / cm
or 7.84 keV in 3 cm of gas.
However, I'm apparently too dumb to compute these numbers using the equations below…
Problem was I given in eV instead of MeV…
import math, macros, unchained macro `^`(x: untyped, num: static int): untyped = result = nnkInfix.newTree(ident"*") proc addInfix(n, x: NimNode, num: int) = var it = n if num > 0: it.add nnkInfix.newTree(ident"*") it[1].addInfix(x, num - 1) while it.len < 3: it.add x result.addInfix(x, num - 2) let K = 4 * π * N_A * r_e^2 * m_e * c^2 # usually in: [MeV mol⁻¹ cm²] defUnit(cm³•g⁻¹) defUnit(J•m⁻¹) defUnit(cm⁻³) defUnit(g•mol⁻¹) defUnit(MeV•g⁻¹•cm²) defUnit(mol⁻¹) defUnit(keV•cm⁻¹) proc electronDensity(ρ: g•cm⁻³, Z, A: UnitLess): cm⁻³ = result = N_A * Z * ρ / (A * M_u.to(g•mol⁻¹)) proc I[T](z: float): T = ## approximation #result = 188.0.eV.to(T) # 188.0 eV from NIST table #10 * z * 1e-6 result = (10.eV * z).to(T) proc betheBloch(ρ: g•cm⁻³, z, Z, A, β: UnitLess): J•m⁻¹ = ## result in J / m let ec = e^2 / (4 * π * ε_0) var res1 = 4 * π / (m_e * c^2) * electronDensity(ρ, Z, A) * z^2 / (β^2) let lnArg = 2 * m_e * c^2 * β^2 / (I[Joule](Z) * (1 - β^2)) var res2 = ec^2 * ( ln(lnArg) - β^2 ) result = (res1 * res2).to(J•m⁻¹) proc calcβ(γ: UnitLess): UnitLess = result = sqrt(1.0 - 1.0 / (γ^2)) proc betheBlochPDG(z, Z: UnitLess, A: g•mol⁻¹, γ: UnitLess, M: kg): MeV•g⁻¹•cm² = ## result in MeV cm² g⁻¹ (normalized by density) ## z: charge of particle ## Z: charge of particles making up medium ## A: atomic mass of particles making up medium ## γ: Lorentz factor of particle ## M: mass of particle in MeV (or same mass as `m_e` defined as) let β = calcβ(γ) let W_max = 2 * m_e * c^2 * β^2 * γ^2 / (1 + 2 * γ * m_e / M + (m_e / M)^2) let lnArg = 2 * m_e * c^2 * β^2 * γ^2 * W_max / (I[Joule](Z)^2) result = (K * z^2 * Z / A * 1.0 / (β^2) * ( 0.5 * ln(lnArg) - β^2 )).to(MeV•g⁻¹•cm²) proc density(p: mbar, M: g•mol⁻¹, temp: Kelvin): g•cm⁻³ = ## returns the density of the gas for the given pressure. ## The pressure is assumed in `mbar` and the temperature (in `K`). ## The default temperature corresponds to BabyIAXO aim. ## Returns the density in `g / cm^3` let gasConstant = 8.314.J•K⁻¹•mol⁻¹ # joule K^-1 mol^-1 let pressure = p.to(Pa) # pressure in Pa # factor 1000 for conversion of M in g / mol to kg / mol result = (pressure * M / (gasConstant * temp)).to(g•cm⁻³) proc E_to_γ(E: GeV): UnitLess = result = E.to(Joule) / (m_μ * c^2) + 1 proc γ_to_E(γ: UnitLess): GeV = result = ((γ - 1) * m_μ * c^2).to(GeV) let muE = 1.0.GeV let muγ = E_to_γ(muE) echo muE #echo m_μ_eV * c^2 echo muγ let muβ = calcβ(muγ) type Element = object name: string Z: UnitLess M: g•mol⁻¹ A: UnitLess # numerically same as `M` ρ: g•cm⁻³ proc initElement(name: string, Z: UnitLess, M: g•mol⁻¹, ρ: g•cm⁻³): Element = Element(name: name, Z: Z, M: M, A: M.UnitLess, ρ: ρ) let M_Ar = 39.95.g•mol⁻¹ # molar mass. Numerically same as relative atomic mass let M_Xe = 131.293.g•mol⁻¹ # molar mass. Numerically same as relative atomic weight let ρAr = density(1050.mbar, M_Ar, temp = 293.15.K) let ρXe = density(1050.mbar, M_Xe, temp = 293.15.K) let ρPb = 11.34.g•cm⁻³ let ρFe = 7.874.g•cm⁻³ echo "Density ", ρAr let Argon = initElement("ar", 18.0.UnitLess, 39.95.g•mol⁻¹, ρAr) let Xenon = initElement("xe", 54.0.UnitLess, 131.293.g•mol⁻¹, ρXe) let Lead = initElement("pb", 82.0.UnitLess, 207.2.g•mol⁻¹, ρPb) let Iron = initElement("fe", 26.0.UnitLess, 55.845.g•mol⁻¹, ρFe) let Silicon = initElement("si", 14.0.UnitLess, 28.0855.g•mol⁻¹, 2.329.g•cm⁻³) let r = betheBloch(Argon.ρ, -1, Argon.Z, Argon.A, muβ) echo "-----------" echo r.to(keV•cm⁻¹), " at CAST setup" echo "-----------" let r2 = betheBlochPDG(-1, Argon.Z, Argon.M, muγ, m_μ) echo (r2 * Argon.ρ).to(keV•cm⁻¹), " at CAST setup" import seqmath, ggplotnim, sequtils, strformat proc computeBethe(e: Element): DataFrame = ## plots a bunch of different gammas for one set of gas let γs = linspace(1.2, 40.0, 1000) let βs = γs.mapIt(it.calcβ()) # convert both ⇒ keV/cm let ⟨dE_dx⟩_PDG = γs.mapIt((betheBlochPDG(-1, e.Z, e.M, it, m_μ) * e.ρ).to(keV•cm⁻¹).float) let ⟨dE_dx⟩_W = βs.mapIt((betheBloch(e.ρ, -1, e.Z, e.A, it).to(keV•cm⁻¹)).float) result = toDf(γs, ⟨dE_dx⟩_PDG, ⟨dE_dx⟩_W) .gather(["⟨dE_dx⟩_PDG", "⟨dE_dx⟩_W"], key = "Type", value = "⟨dE_dx⟩") result["Z"] = constantColumn(e.Z, result.len) var dfGas = computeBethe(Argon) dfGas.add computeBethe(Lead) proc plotGammas(df: DataFrame) = ggplot(df, aes("γs", "⟨dE_dx⟩", color = "Type", shape = "Z")) + geom_line() + ylab("⟨dE/dx⟩ [keV/cm]") + xlab("γ (Lorentz factor)") + scale_y_log10() + margin(top = 2) + # does not work currently #scale_x_continuous(secAxis = sec_axis(transFn = (proc(γ: float): float = (sqrt(1.0 - 1.0 / (γ^2)))), # invTransFn = (proc(x: float): float = sqrt(1.0 - 1.0 / (x^2))), # name = "β")) + ggtitle(&"Mean ionization energy of muons with γ in Ar at {Argon.ρ.float:.2e} g/cm³ and in Pb") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/bethe_bloch_gammas.pdf") plotGammas(dfGas) proc plotE_vs_γ() = let EsFloat = linspace(0.1, 100.0, 1000) let Es = EsFloat.mapIt(it.GeV) #γs.mapIt(it.γ_to_E().float) let γs = Es.mapIt((E_to_γ(it.to(GeV))).float) let df = toDf(EsFloat, γs) echo df ggplot(df, aes("EsFloat", "γs")) + geom_line() + xlab("E [GeV]") + ylab("γ") + scale_x_log10() + scale_y_log10() + ggtitle("Dependence of γ on energy in GeV") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/energy_vs_gamma.pdf") plotE_vs_γ() proc intBethe(e: Element, d_total: cm, E0: eV, dx = 1.μm): eV = ## integrated energy loss of bethe formula after `d` cm of matter ## and returns the energy remaining var γ: UnitLess = E_to_γ(E0.to(GeV)) var d: cm result = E0 var totalLoss = 0.eV while d < d_total and result > 0.eV: let E_loss: MeV = betheBlochPDG(-1, e.Z, e.M, γ, m_μ) * e.ρ * dx result = result - E_loss.to(eV) γ = E_to_γ(result.to(GeV)) #.to(GeV)) d = d + dx.to(cm) totalLoss = totalLoss + E_loss.to(eV) echo "Resulting d is ", d echo "And result ", result echo "Total loss in: ", totalLoss.to(keV) echo "----------\n" result = max(0.float, result.float).eV echo "Integrated bethe of Lead: ", intBethe(Lead, 20.cm, 10.GeV.to(eV)), " for 10 GeV μ" echo "Integrated bethe of Lead: ", intBethe(Lead, 20.cm, 1.GeV.to(eV)), " for 1 GeV μ" echo "------------------------\n" echo "Integrated bethe of Iron: ", intBethe(Iron, 20.cm, 10.GeV.to(eV)), " for 10 GeV μ" echo "Integrated bethe of Iron: ", intBethe(Iron, 20.cm, 1.GeV.to(eV)), " for 1 GeV μ" echo "Integrated bethe of Argon: ", intBethe(Argon, 3.cm, 1.GeV.to(eV)), " for 1 GeV μ" echo "Integrated bethe of Argon: ", intBethe(Argon, 3.cm, 5.GeV.to(eV)), " for 5 GeV μ" echo "Integrated bethe of Argon: ", intBethe(Argon, 3.cm, 10.GeV.to(eV)), " for 10 GeV μ" echo "Integrated bethe of Xenon: ", intBethe(Xenon, 3.cm, 10.GeV.to(eV)), " for 10 GeV μ" echo "Integrated bethe of Argon: ", intBethe(Argon, 3.cm, 100.GeV.to(eV)), " for 100 GeV μ" echo "Integrated bethe of Si 450μm: ", intBethe(Argon, 450.μm.to(cm), 100.GeV.to(eV)), " for 100 GeV μ" if true: quit() import strutils proc plotDetectorAbsorption(element: Element) = let E_float = logspace(-2, 2, 1000) let energies = E_float.mapIt(it.GeV) let E_loss = energies.mapIt((it.to(eV) - intBethe(element, 3.cm, it.to(eV))).to(keV).float) let df = toDf(E_float, E_loss) ggplot(df, aes("E_float", "E_loss")) + geom_line() + xlab("μ Energy [GeV]") + ylab("ΔE [keV]") + scale_x_log10() + scale_y_log10() + ggtitle(&"Energy loss of Muons in 3 cm {element.name.capitalizeAscii} at CAST conditions") + ggsave(&"/home/basti/org/Figs/statusAndProgress/muonStudies/{element.name}_energy_loss_cast.pdf") plotDetectorAbsorption(Argon) plotDetectorAbsorption(Xenon) echo "-----" echo E_to_γ(10.GeV) echo "Argon ", betheBlochPDG(-1, Argon.Z, Argon.M, E_to_γ(1.GeV), m_μ) * Argon.ρ echo "Xenon ", betheBlochPDG(-1, Xenon.Z, Xenon.M, E_to_γ(1.GeV), m_μ) * Argon.ρ if true: quit() #echo "An energy of 1 GeV is γ = ", E_to_γ(1.GeV) #echo "An energy of 10 GeV is γ = ", E_to_γ(10.GeV) #echo "An energy of 100 GeV is γ = ", E_to_γ(100.GeV)
- from: one long lead block
- back: one short lead block
Lead block dimensions
- (20, 10, 5) cm
Means: Assuming hit in center (4.5mm consider as single point), largest angle computed from 20cm of pipe to exit, another 5 cm to detector window and 3cm of gas. 28 cm from center chip to exit. Pipe ~8 cm diameter.
Compute max angle that can be passed w/o any lead.
At larger angles, we traverse more and more lead. Can discretize but very few angles probably between 0 and 20cm of lead (the max).
Given a distribution of muons and angles. Can compute resulting distribution after passing lead.
import math, unchained let α = arctan(4.cm / 28.cm).radToDeg echo α
Finally compute the effect on eccentricity for muons. Need to know the average width (can take from data; use X-ray in high range) vs 3 cm. Allows to estimate ecc. by comparing width to projected length if entering under angle. Something like tan(α) * 3 cm or so.
First compute the mean eccentricity given an incidence angle.
According to
the mean width of events in the 8-10 keV hump is 5 mm. So we use that
and model a track as a 5 mm wide cylinder.
Eccentricity computed from projection of tilted cylinder. Lowest point
to highest point along projection is "length".
import unchained, ggplotnim, strformat, sequtils let w = 5.mm # mean width of a track in 8-10keV hump let h = 3.cm # detector height proc computeLength(α: UnitLess): mm = ## todo: add degrees? ## α: float # Incidence angle var w_prime = w / cos(α) # projected width taking incidence angle into account # let L = h / cos(α)# track length given incidence angle let L_prime = tan(α) * h # projected `'length'` of track from center to center let L_full = L_prime + w_prime # full `'length'` is bottom to top, thus + w_prime result = L_full.to(mm) proc computeEccentricity(L_full, w: mm, α: UnitLess): UnitLess = let w_prime = w / cos(α) result = L_full / w_prime let αs = linspace(0.0, degToRad(25.0), 1000) let εs = αs.mapIt(it.computeLength.computeEccentricity(w, it).float) let αsDeg = αs.mapIt(it.radToDeg) let df = toDf(αsDeg, εs) # maximum eccentricity for text annotation let max_εs = max(εs) let max_αs = max(αsDeg) # compute the maximum angle under which `no` lead is seen let d_open = 28.cm # assume 28 cm from readout to end of lead shielding let h_open = 5.cm # assume open height is 10 cm, so 5 cm from center let α_limit = arctan(h_open / d_open).radToDeg # data for the limit of 8-10 keV eccentricity let ε_max_hump = 1.3 # 1.2 is more reasonable, but 1.3 is the absolute upper limit ggplot(df, aes("αsDeg", "εs")) + geom_line() + geom_linerange(data = df.head(1), aes = aes(x = α_limit, yMin = 1.0, yMax = max_εs), color = some(color(1.0, 0.0, 1.0))) + geom_linerange(data = df.head(1), aes = aes(y = ε_max_hump, xMin = 0, xMax = max_αs), color = some(color(0.0, 1.0, 1.0))) + geom_text(data = df.head(1), aes = aes(x = α_limit, y = max_εs + 0.1, text = "Maximum angle no lead traversed")) + geom_text(data = df.head(1), aes = aes(x = 17.5, y = ε_max_hump + 0.1, text = "Largest ε in 8-10 keV hump")) + xlab("α: Incidence angle [°]") + ylab("ε: Eccentricity") + ylim(1.0, 4.0) + ggtitle(&"Expected eccentricity for tracks of mean width {w}") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/exp_eccentricity_given_incidence_angle.pdf")
import math, unchained let R_Earth = 6371.km let R_over_d = 174.UnitLess let I₀ = 90.0.m⁻²•s⁻¹•sr⁻¹ let n = 3.0 let E₀ = 25.0.GeV let E_c = 1.GeV let ε = 2000.GeV proc distanceAtmosphere(ϑ: Radian, d: KiloMeter = 36.6149.km): UnitLess = ## NOTE: The default value for `d` is not to be understood as a proper height. It.s an ## approximation based on a fit to get `R_Earth / d = 174`! result = sqrt((R_Earth / d * cos(ϑ))^2 + 2 * R_Earth / d + 1) - R_Earth / d * cos(ϑ) defUnit(m⁻²•s⁻¹•sr⁻¹) proc muonFlux(E: GeV, ϑ: Radian, E₀, E_c: GeV, I₀: m⁻²•s⁻¹•sr⁻¹, ε: GeV): m⁻²•s⁻¹•sr⁻¹ = let N = (n - 1) * pow((E₀ + E_c).float, n - 1) result = I₀ * N * pow((E₀ + E).float, -n) * pow((1 + E / ε).float, -1) * pow(distanceAtmosphere(ϑ), -(n - 1)) echo muonFlux(1.5.GeV, 0.Radian, E₀, E_c, I₀, ε) import ggplotnim, sequtils proc plotE_vs_flux(ϑ: Radian, E₀, E_c: GeV, I₀: m⁻²•s⁻¹•sr⁻¹, ε: GeV) = let energies = linspace(0.5, 100.0, 1000) let E = energies.mapIt(it.GeV) let flux = E.mapIt(muonFlux(it, 0.Radian, E₀, E_c, I₀, ε).float) let df = toDf(energies, flux) ggplot(df, aes("energies", "flux")) + geom_line() + xlab("Energy [GeV]") + ylab("Flux [m⁻²•s⁻¹•sr⁻¹]") + scale_x_log10() + scale_y_log10() + ggtitle("Flux dependency on the energy of muons at ϑ = 0°") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/energy_vs_flux_cosmic_muons.pdf") plotE_vs_flux(0.Radian, 4.29.GeV, 0.5.GeV, 70.7.m⁻²•s⁻¹•sr⁻¹, 854.GeV) proc plotFlux_vs_ϑ() = let thetas = linspace(0.0, π/2.0, 1000) let ϑs = thetas.mapIt(it.Radian) let flux = ϑs.mapIt(muonFlux(5.GeV, it, E₀, E_c, I₀, ε).float) let df = toDf(thetas, flux) ggplot(df, aes("thetas", "flux")) + geom_line() + xlab("Zenith angle ϑ [Rad]") + ylab("Flux [m⁻²•s⁻¹•sr⁻¹]") + scale_y_log10() + ggtitle("Flux dependency on the zenith angle ϑ at 5 GeV") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/flux_vs_zenith_angle_cosmic_muons.pdf") plotFlux_vs_ϑ() proc plotFlux_at_CAST() = let energies = linspace(0.5, 100.0, 1000) let E = energies.mapIt(it.GeV) let flux = E.mapIt(muonFlux(it, 88.0.degToRad.Radian, E₀, E_c, I₀, ε).float) let df = toDf(energies, flux) ggplot(df, aes("energies", "flux")) + geom_line() + xlab("Energy [GeV]") + ylab("Flux [m⁻²•s⁻¹•sr⁻¹]") + scale_x_log10() + scale_y_log10() + ggtitle("Flux dependency on the energy at ϑ = 88° at CAST altitude") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/flux_at_cast_88_deg.pdf") plotFlux_at_CAST() proc plotFluxAdjEnergyLoss() = let energies = linspace(0.5, 100.0, 50000) let E = energies.mapIt(it.GeV) let flux = E.mapIt(muonFlux(it, 88.0.degToRad.Radian, E₀, E_c, I₀, ε).float) let E_loss = E.mapIt((it.to(eV) - intBethe(Argon, 3.cm, it.to(eV))).to(keV).float) let fluxSum = flux.sum let EWidth = energies[1] - energies[0] let df = toDf(energies, E_loss, flux) .mutate(f{"flux" ~ `flux` / fluxSum}, f{"AdjFlux" ~ `E_loss` * `flux`}) echo df["AdjFlux", float].sum ggplot(df, aes("flux", "E_loss")) + geom_line() + xlab("Flux [m⁻²•s⁻¹•sr⁻¹]") + ylab("⟨ΔE⟩ in 3 cm Ar [keV]") + margin(top = 2) + ggtitle("Energy loss in Ar at CAST conditions flux adjusted for muons at ϑ = 88°") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/flux_adjusted_energy_loss_cast.pdf") ggplot(df, aes("AdjFlux")) + geom_histogram() + #xlab("Flux [m⁻²•s⁻¹•sr⁻¹]") + ylab("⟨ΔE⟩ in 3 cm Ar [keV]") + margin(top = 2) + #ggtitle("Energy loss in Ar at CAST conditions flux adjusted for muons at ϑ = 88°") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/flux_adj_histogram.pdf") ggplot(df, aes("flux", "E_loss")) + geom_line() + #xlab("Flux [m⁻²•s⁻¹•sr⁻¹]") + ylab("⟨ΔE⟩ in 3 cm Ar [keV]") + margin(top = 2) + #ggtitle("Energy loss in Ar at CAST conditions flux adjusted for muons at ϑ = 88°") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/flux_adj_vs_loss.pdf") ## TODO: compute the loss for each possible energy ## then compute histogram of ~100_000 elements using `flux` as weight! let dHisto = histogram(E_loss, weights = flux, bins = 100) let df2 = toDf({"Energy" : dHisto[1], "Flux" : dHisto[0]}) echo df2 ggplot(df2, aes("Energy", "Flux")) + geom_histogram(stat = "identity") + xlim(5.0, 15.0) + ggtitle("Energy loss of muons at CAST") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/energy_loss_at_cast_spectrum.pdf") plotFluxAdjEnergyLoss()
23.1. Full computation of expected muon energy loss
This was written as a combined study of the muon loss and is essentially a cleaned up version of the above section and is found in ./../Mails/KlausUpdates/klaus_update_23_02_21.html.
23.1.1. What kind of energy deposition can we expect from muons?
For reference the current background rate in fig. 344. There is a hump between 8-10 keV with a peak at ~9.3-9.4 keV.
Let's try to compute what kind of energy deposition we can actually expect from muons at CAST.
The energy deposition of muons can be computed using the Bethe formula. However, this only gives us a mean energy loss for muons of a specific γ. That means we need to understand the muon flux at different energies / γs, which depends on the zenith angle ϑ and the altitude of the location.
Using https://arxiv.org/pdf/1606.06907.pdf we can get some analytical approximation for the flux at different ϑ and altitudes.
On the other hand we need to know the actual path length a muon sees through our detector to compute the real path length (based on a 3 cm long / high gas volume). We also need to have an understanding of what the flux looks like that actually enters our detector, due to the lead shielding around the detector. For that we need an estimate on the maximum allowed angle that can be seen through opening towards the telescope without passing through any lead as well as correlating that with the eccentricities we actually see in our background data in the 8-10 keV hump (anything that has a larger eccentricity does not have to worry us).
- Energy loss of muons in Ar at CAST conditions
First, let's compute the mean energy loss muons see traversing Argon at the conditions under which we used the detector and plot the energy loss over a distance of 3 cm for different incoming muon energies.
import math, macros, unchained import seqmath, ggplotnim, sequtils, strformat let K = 4 * π * N_A * r_e^2 * m_e * c^2 # usually in: [MeV mol⁻¹ cm²] defUnit(cm³•g⁻¹) defUnit(J•m⁻¹) defUnit(cm⁻³) defUnit(g•mol⁻¹) defUnit(MeV•g⁻¹•cm²) defUnit(mol⁻¹) defUnit(keV•cm⁻¹) defUnit(g•cm⁻³) proc electronDensity(ρ: g•cm⁻³, Z, A: UnitLess): cm⁻³ = result = N_A * Z * ρ / (A * M_u.to(g•mol⁻¹)) proc I[T](z: float): T = ## approximation #result = 188.0.eV.to(T) # 188.0 eV from NIST table #10 * z * 1e-6 result = (10.eV * z).to(T) proc betheBloch(ρ: g•cm⁻³, z, Z, A, β: UnitLess): J•m⁻¹ = ## result in J / m let ec = e^2 / (4 * π * ε_0) var res1 = 4 * π / (m_e * c^2) * electronDensity(ρ, Z, A) * z^2 / (β^2) let lnArg = 2 * m_e * c^2 * β^2 / (I[Joule](Z) * (1 - β^2)) var res2 = ec^2 * ( ln(lnArg) - β^2 ) result = res1 * res2 proc calcβ(γ: UnitLess): UnitLess = result = sqrt(1.0 - 1.0 / (γ^2)) proc betheBlochPDG(z, Z: UnitLess, A: g•mol⁻¹, γ: UnitLess, M: kg): MeV•g⁻¹•cm² = ## result in MeV cm² g⁻¹ (normalized by density) ## z: charge of particle ## Z: charge of particles making up medium ## A: atomic mass of particles making up medium ## γ: Lorentz factor of particle ## M: mass of particle in MeV (or same mass as `m_e` defined as) let β = calcβ(γ) let W_max = 2 * m_e * c^2 * β^2 * γ^2 / (1 + 2 * γ * m_e / M + (m_e / M)^2) let lnArg = 2 * m_e * c^2 * β^2 * γ^2 * W_max / (I[Joule](Z)^2) result = (K * z^2 * Z / A * 1.0 / (β^2) * ( 0.5 * ln(lnArg) - β^2 )).to(MeV•g⁻¹•cm²) proc density(p: mbar, M: g•mol⁻¹, temp: Kelvin): g•cm⁻³ = ## returns the density of the gas for the given pressure. ## The pressure is assumed in `mbar` and the temperature (in `K`). ## The default temperature corresponds to BabyIAXO aim. ## Returns the density in `g / cm^3` let gasConstant = 8.314.J•K⁻¹•mol⁻¹ # joule K^-1 mol^-1 let pressure = p.to(Pa) # pressure in Pa # factor 1000 for conversion of M in g / mol to kg / mol result = (pressure * M / (gasConstant * temp)).to(g•cm⁻³) proc E_to_γ(E: GeV): UnitLess = result = E.to(Joule) / (m_μ * c^2) + 1 proc γ_to_E(γ: UnitLess): GeV = result = ((γ - 1) * m_μ * c^2).to(GeV) type Element = object Z: UnitLess M: g•mol⁻¹ A: UnitLess # numerically same as `M` ρ: g•cm⁻³ proc initElement(Z: UnitLess, M: g•mol⁻¹, ρ: g•cm⁻³): Element = Element(Z: Z, M: M, A: M.UnitLess, ρ: ρ) # molar mass. Numerically same as relative atomic mass let M_Ar = 39.95.g•mol⁻¹ let ρAr = density(1050.mbar, M_Ar, temp = 293.15.K) let Argon = initElement(18.0.UnitLess, 39.95.g•mol⁻¹, ρAr) proc intBethe(e: Element, d_total: cm, E0: eV, dx = 1.μm): eV = ## integrated energy loss of bethe formula after `d` cm of matter ## and returns the energy remaining var γ: UnitLess = E_to_γ(E0.to(GeV)) var d: cm result = E0 var totalLoss = 0.eV while d < d_total and result > 0.eV: let E_loss: MeV = betheBlochPDG(-1, e.Z, e.M, γ, m_μ) * e.ρ * dx result = result - E_loss.to(eV) γ = E_to_γ(result.to(GeV)) d = d + dx.to(cm) totalLoss = totalLoss + E_loss.to(eV) result = max(0.float, result.float).eV proc plotDetectorAbsorption() = let E_float = logspace(-2, 2, 1000) let energies = E_float.mapIt(it.GeV) let E_loss = energies.mapIt( (it.to(eV) - intBethe(Argon, 3.cm, it.to(eV))).to(keV).float ) let df = toDf(E_float, E_loss) ggplot(df, aes("E_float", "E_loss")) + geom_line() + xlab("μ Energy [GeV]") + ylab("ΔE [keV]") + scale_x_log10() + scale_y_log10() + ggtitle("Energy loss of Muons in 3 cm Ar at CAST conditions") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/ar_energy_loss_cast.pdf") plotDetectorAbsorption()
This results in fig. 345 for the energy loss of muons in our detector along 3 cm of gas depending on the energy of the incoming muon. The energy loss is computed by numerically integrating the Bethe formula with a step size of 1 μm.
Figure 345: Energy loss in Ar at 1050 mbar along 3 cm for muons of different energies. - Model muon flux
Second, let's look at the muon flux for a set of parameters shown in the paper to see if we copied the equation correctly.
import math, unchained, ggplotnim, sequtils let R_Earth = 6371.km let R_over_d = 174.UnitLess let n = 3.0 let E₀ = 25.0.GeV let I₀ = 90.0.m⁻²•s⁻¹•sr⁻¹ let E_c = 1.GeV let ε = 2000.GeV func distanceAtmosphere(ϑ: Radian, d: KiloMeter = 36.6149.km): UnitLess = ## NOTE: The default value for `d` is not to be understood as a proper height. It.s an ## approximation based on a fit to get `R_Earth / d = 174`! let R_Earth = 6371.km result = sqrt((R_Earth / d * cos(ϑ))^2 + 2 * R_Earth / d + 1) - R_Earth / d * cos(ϑ) #debugecho "ϑ = ", ϑ, " d = ", d, " result = ", result defUnit(m⁻²•s⁻¹•sr⁻¹) proc muonFlux(E: GeV, ϑ: Radian, E₀, E_c: GeV, I₀: m⁻²•s⁻¹•sr⁻¹, ε: GeV): m⁻²•s⁻¹•sr⁻¹ = let N = (n - 1) * pow((E₀ + E_c).float, n - 1) result = I₀ * N * pow((E₀ + E).float, -n) * pow((1 + E / ε).float, -1) * pow(distanceAtmosphere(ϑ), -(n - 1)) proc plotE_vs_flux(ϑ: Radian, E₀, E_c: GeV, I₀: m⁻²•s⁻¹•sr⁻¹, ε: GeV) = let energies = linspace(0.5, 100.0, 1000) let E = energies.mapIt(it.GeV) let flux = E.mapIt(muonFlux(it, 0.Radian, E₀, E_c, I₀, ε).float) let df = toDf(energies, flux) ggplot(df, aes("energies", "flux")) + geom_line() + xlab("Energy [GeV]") + ylab("Flux [m⁻²•s⁻¹•sr⁻¹]") + scale_x_log10() + scale_y_log10() + ggtitle("Flux dependency on the energy of muons at ϑ = 0°") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/energy_vs_flux_cosmic_muons.pdf") plotE_vs_flux(0.Radian, 4.29.GeV, 0.5.GeV, 70.7.m⁻²•s⁻¹•sr⁻¹, 854.GeV)
yields fig. 346, which matches nicely figure 3 from the linked paper.
Figure 346: Flux of muons at sea level and ϑ = 0°, matching fig. 3 from the paper. From here we can adjust the zenith angle ϑ and the altitude (by guessing some parameters interpolating between the values from tab. 1 in the paper) to compute reasonable values for CAST:
proc plotFlux_at_CAST() = let energies = linspace(0.5, 100.0, 1000) let E = energies.mapIt(it.GeV) let flux = E.mapIt(muonFlux(it, 88.0.degToRad.Radian, E₀, E_c, I₀, ε).float) let df = toDf(energies, flux) ggplot(df, aes("energies", "flux")) + geom_line() + xlab("Energy [GeV]") + ylab("Flux [m⁻²•s⁻¹•sr⁻¹]") + scale_x_log10() + scale_y_log10() + ggtitle("Flux dependency on the energy at ϑ = 88° at CAST altitude") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/flux_at_cast_88_deg.pdf") plotFlux_at_CAST()
which gives us fig. 347 as a reasonable approximation for the expected muon flux at CAST.
Figure 347: Expected muon flux at CAST for ϑ = 88°. Why was ϑ = 88° chosen?
- Model maximum allowed angle
The reason ϑ = 88° was chosen is due to the restriction on the maximum allowed eccentricity for a cluster to still end up as a possible cluster in our 8-10 keV hump. See the
eccentricitysubplot in fig. 348.Figure 348: See the eccentricity subplot for an upper limit on the allowed eccentricity for events in the 8-10 keV hump. Values should not be above ε = 1.3. From this we can deduce the eccentricity should be smaller than ε = 1.3. What does this imply for the largest possible angles allowed in our detector? And how does the opening of the "lead pipe window" correspond to this?
Let's compute by modeling a muon track as a cylinder. Reading off the mean
widthfrom the above fig. to w = 5 mm and taking into account the detector height of 3 cm we can compute the relation between different angles and corresponding eccentricities.In addition we will compute the largest possible angle a muon (from the front of the detector of course) can enter, under which it does not see the lead shielding.
import unchained, ggplotnim, strformat, sequtils let w = 5.mm # mean width of a track in 8-10keV hump let h = 3.cm # detector height proc computeLength(α: UnitLess): mm = ## todo: add degrees? ## α: float # Incidence angle var w_prime = w / cos(α) # projected width taking incidence # angle into account let L_prime = tan(α) * h # projected `'length'` of track # from center to center let L_full = L_prime + w_prime # full `'length'` is bottom to top, thus # + w_prime result = L_full.to(mm) proc computeEccentricity(L_full, w: mm, α: UnitLess): UnitLess = let w_prime = w / cos(α) result = L_full / w_prime let αs = linspace(0.0, degToRad(25.0), 1000) let εs = αs.mapIt(it.computeLength.computeEccentricity(w, it).float) let αsDeg = αs.mapIt(it.radToDeg) let df = toDf(αsDeg, εs) # maximum eccentricity for text annotation let max_εs = max(εs) let max_αs = max(αsDeg) # compute the maximum angle under which `no` lead is seen let d_open = 28.cm # assume 28 cm from readout to end of lead shielding let h_open = 5.cm # assume open height is 10 cm, so 5 cm from center let α_limit = arctan(h_open / d_open).radToDeg # data for the limit of 8-10 keV eccentricity let ε_max_hump = 1.3 # 1.2 is more reasonable, but 1.3 is the # absolute upper limit ggplot(df, aes("αsDeg", "εs")) + geom_line() + geom_linerange(data = df.head(1), aes = aes(x = α_limit, yMin = 1.0, yMax = max_εs), color = some(color(1.0, 0.0, 1.0))) + geom_linerange(data = df.head(1), aes = aes(y = ε_max_hump, xMin = 0, xMax = max_αs), color = some(color(0.0, 1.0, 1.0))) + geom_text(data = df.head(1), aes = aes(x = α_limit, y = max_εs + 0.1, text = "Maximum angle no lead traversed")) + geom_text(data = df.head(1), aes = aes(x = 17.5, y = ε_max_hump + 0.1, text = "Largest ε in 8-10 keV hump")) + xlab("α: Incidence angle [°]") + ylab("ε: Eccentricity") + ylim(1.0, 4.0) + ggtitle(&"Expected eccentricity for tracks of mean width {w}") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/exp_eccentricity_given_incidence_angle.pdf")
Resulting in fig. 349.
Figure 349: Relationship between incidence angle of muons of a width of 5 mm and their expected mean eccentricity. Drawn as well are the maximum angle under which no lead is seen (from the front) as well as the larges ε seen in the data. This leads to an upper bound of ~3° from the horizontal. Hence the (somewhat arbitrary choice) of 88° for the ϑ angle above.
- Putting it all together
Using the knowledge of the maximum angle of muons entering our detector, the muon flux at ϑ = 88° at CAST and the energy loss of different muons, we can compute an expected mean value for the energy deposition.
We can do this by computing the relative flux of muons we expect, scaling the energy loss of the muons by their flux and summing those values.
proc computeMeanEnergyLoss() = let energies = linspace(0.5, 100.0, 1000) let E = energies.mapIt(it.GeV) let flux = E.mapIt(muonFlux( it, 88.0.degToRad.Radian, E₀, E_c, I₀, ε).float ) let E_loss = E.mapIt( (it.to(eV) - intBethe(Argon, 3.cm, it.to(eV))).to(keV).float ) let fluxSum = flux.sum let df = toDf(energies, E_loss, flux) .mutate(f{"flux" ~ `flux` / fluxSum}, f{"AdjFlux" ~ `E_loss` * `flux`}) echo "Mean energy loss: ", df["AdjFlux", float].sum computeMeanEnergyLoss()
which results in a value (given these assumptions here) of: ⟨ΔE⟩ = 11.63 keV
old value based on muon mass 10 times heavier…:
which results in a value (given these assumptions here) of:⟨ΔE⟩ = 9.221 keV - Compute the expected spectrum
We can compute the expected spectrum at CAST by calculating the expected flux as well as the expected energy loss for many different muon energies. Then we compute the histogram of all energy losses and use the fluxes for each muon as its weight. This should (assuming linearly spaced energies) yield the expected histogram for the spectrum.
proc computeCASTspectrum() = ## TODO: compute the loss for each possible energy ## then compute histogram of ~100_000 elements using `flux` as weight! let energies = linspace(0.5, 100.0, 50000) let E = energies.mapIt(it.GeV) let flux = E.mapIt(muonFlux(it, 88.0.degToRad.Radian, E₀, E_c, I₀, ε).float) let E_loss = E.mapIt((it.to(eV) - intBethe(Argon, 3.cm, it.to(eV))).to(keV).float) let dHisto = histogram(E_loss, weights = flux, bins = 100) let df = toDf({"Energy" : dHisto[1], "Flux" : dHisto[0]}) echo df ggplot(df, aes("Energy", "Flux")) + geom_histogram(stat = "identity", hdKind = hdOutline) + xlim(5.0, 15.0) + ggtitle("Energy loss of muons at CAST") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/energy_loss_at_cast_spectrum_light_muon.pdf") #computeCASTspectrum()
Figure 350: The expected muon spectrum (expected losses of all muons) at CAST under an angle of ϑ = 88°. Figure 351: The same plot as the above but for a muon that is 10 times heavier than the real muon (~1.8e-27 kg instead of ~1.8e-28 kg) - Computing the expected spectrum
Let's sample from the flux and put compute the expected mean energy loss for each element.
Before we can do that, we need to compute the lower limit of the cutoff energy given muons under the angle we look at. Under ϑ = 88° the path through atmosphere is already pretty long, which means the energy of muons is already quite high.
proc computeHeight(S: Meter, ϑ: Radian): KiloMeter = ## For given remaining distance distance along the path of a muon ## `S` (see fig. 1 in 1606.06907) computes the remaining height above ## ground. Formula is the result of inverting eq. 7 to `d` using quadratic ## formula. Positive result, because negative is negative. result = (-1.0 * R_Earth + sqrt(R_Earth^2 + S^2 + 2 * S * R_Earth * cos(ϑ)).m).to(km) import algorithm defUnit(K•m⁻¹) proc barometricFormula(h: KiloMeter): g•cm⁻³ = let hs = @[0.0.km, 11.0.km] let ρs = @[1.225.kg•m⁻³, 0.36391.kg•m⁻³] let Ts = @[288.15.K, 216.65.K] let Ls = @[-1.0 * 0.0065.K•m⁻¹, 0.0.K•m⁻¹] let M_air = 0.0289644.kg•mol⁻¹ let R = 8.3144598.N•m•mol⁻¹•K⁻¹ let g_0 = 9.80665.m•s⁻² let idx = hs.mapIt(it.float).lowerBound(h.float) - 1 case idx of 0: # in Troposphere, using regular barometric formula for denities let expArg = g_0 * M_air / (R * Ls[idx]) result = (ρs[idx] * pow(Ts[idx] / (Ts[idx] + Ls[idx] * (h - hs[idx])), expArg)).to(g•cm⁻³) of 1: # in Tropopause, use equation valid for L_b = 0 result = (ρs[idx] * exp(-1.0 * g_0 * M_air * (h - hs[idx]) / (R * Ts[idx]))).to(g•cm⁻³) else: doAssert false, "Invalid height! Outside of range!" proc intBetheAtmosphere(E: GeV, ϑ: Radian, dx = 1.m): eV = ## integrated energy loss using Bethe formula for muons generated at ## `15.km` under an angle of `ϑ` to the observer for a muon of energy ## `E`. # Nitrogen. Placeholder for full atomsphere let e = initElement(7.0.UnitLess, 14.006.g•mol⁻¹, 1.2506.g•dm⁻³.to(g•cm⁻³)) var γ: UnitLess = E_to_γ(E.to(GeV)) result = E.to(eV) var totalLoss = 0.eV let h_muon = 15.km # assume creation happens in `15.km` let S = h_muon.to(m) * distanceAtmosphere(ϑ.rad, d = h_muon) #echo "THTA ϑ = ", ϑ #echo distanceAtmosphere(ϑ.rad, d = h_muon) #echo distanceAtmosphere(ϑ.rad) #if true: quit() echo "S to pass through ", S.to(km) var S_prime = S while S_prime > 0.m and result > 0.eV: #echo "S prime ", S_prime #echo "HEIGHT ", computeHeight(15_000.0.m, 0.Radian) let h = computeHeight(S_prime, ϑ) let ρ_at_h = barometricFormula(h) #echo "h ", h, " ρ ", ρ_at_h let E_loss: MeV = betheBlochPDG(-1, e.Z, e.M, γ, m_μ) * ρ_at_h * dx result = result - E_loss.to(eV) S_prime = S_prime - dx γ = E_to_γ(result.to(GeV)) totalLoss = totalLoss + E_loss.to(eV) echo "total Loss ", totalLoss.to(GeV) result = max(0.float, result.float).eV block MuonLimits: let τ_μ = 2.1969811.μs # naively this means given some distance `s` the muon can # traverse `s = c • τ_μ` (approximating its speed by `c`) before # it has decayed with a 1/e chance # due to special relativity this is extended by γ let s = c * τ_μ echo s # given production in 15 km, means let h = 15.km echo h / s # so a reduction of (1/e)^22. So 0. # now it's not 15 km but under an angle `ϑ = 88°`. let R_Earth = 6371.km let R_over_d = 174.UnitLess let n = 3.0 let E₀ = 25.0.GeV let I₀ = 90.0.m⁻²•s⁻¹•sr⁻¹ let E_c = 1.GeV let ε = 2000.GeV #proc distanceAtmosphere(ϑ: Radian): UnitLess = # result = sqrt((R_over_d * cos(ϑ))^2 + 2 * R_over_d + 1) - R_over_d * cos(ϑ) # distance atmospher gives S / d, where `d` corresponds to our `h` up there let S = h * distanceAtmosphere(88.0.degToRad.rad) echo "S = ", S echo "S 2 = ", h * distanceAtmosphere(88.0.degToRad.rad, d = 15.0.km) echo "DDDDDD ", distanceAtmosphere(88.0.degToRad.rad, d = 15.0.km) echo "ϑϑϑ ", 88.0.degToRad.rad echo "h ", h # so about 203 km # so let's say 5 * mean distance is ok, means we ned let S_max = S / 5.0 # so need a `γ` such that `s` is stretched to `S_max` let γ = S_max / s echo γ # ouch. Something has to be wrong. γ of 61? # corresponds to an energy loss of what? let Nitrogen = initElement(7.0.UnitLess, 14.006.g•mol⁻¹, 1.2506.g•dm⁻³.to(g•cm⁻³)) echo "Energy left: ", intBethe(Nitrogen, S.to(cm), 6.GeV.to(eV), dx = 1.m.to(μm)).to(GeV) echo intBetheAtmosphere(6.GeV, ϑ = 0.Radian).to(GeV) echo intBetheAtmosphere(200.GeV, ϑ = 88.0.degToRad.Radian).to(GeV) echo "S@75° = ", h * distanceAtmosphere(75.0.degToRad.rad, d = 15.0.km) echo intBetheAtmosphere(100.GeV, ϑ = 75.0.degToRad.Radian).to(GeV) echo E_to_γ(4.GeV) echo E_to_γ(0.GeV)
Compute the energy loss through atmosphere!
Then compute the histogram of each:
import random, algorithm proc sampleFlux(samples = 1_000_000): DataFrame = randomize(1337) let energies = linspace(0.5, 100.0, 10000) let E = energies.mapIt(it.GeV) let flux = E.mapIt(muonFlux(it, 88.0.degToRad.Radian, E₀, E_c, I₀, ε).float) # given flux compute CDF let fluxCS = flux.cumSum() let fluxCDF = fluxCS.mapIt(it / fluxCS[^1]) var losses = newSeq[float]() var energySamples = newSeq[float]() for i in 0 ..< samples: # given the fluxCDF sample different energies, which correspond to the # distribution expected at CAST let idx = fluxCdf.lowerBound(rand(1.0)) let E_element = E[idx] # given this energy `E` compute the loss let loss = (E_element.to(eV) - intBethe(Argon, 3.cm, E_element.to(eV), dx = 50.μm)).to(keV).float losses.add loss #echo "Index ", i, " yields energy ", E_element, " and loss ", loss energySamples.add E_element.float let df = toDf(energySamples, losses) ggplot(df, aes("losses")) + geom_histogram(bins = 300) + margin(top = 2) + xlim(5, 15) + ggtitle(&"Energy loss of muon flux at CAST based on MC sampling with {samples} samples") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/sampled_energy_loss.pdf") ggplot(df, aes("energySamples")) + geom_histogram(bins = 300) + margin(top = 2) + ggtitle(&"Sampled energies for energy loss of muon flux at CAST") + ggsave("/home/basti/org/Figs/statusAndProgress/muonStudies/sampled_energy_for_energy_loss.pdf") discard sampleFlux()
Figure 352: Histogram of the energy samples via sampling using inverse CDF method from the flux as computed in fig. 347. Plotting a log-log plot of this reproduces the flux. - Caveats
This of course does not take into account the following three things:
- muons have to traverse a bunch of concrete, steel, copper… to get to the detector
- muons from the back pass through ~20 cm of lead
- the flux is not exact, in particular the effect of lower energy muons than 500 MeV (and how many of these actually pass through the above two)
23.2. TODO Landau distribution
Can we compute the Landau distribution for our CAST detector from theory?
24. TODO Uncertainties (stat. / syst.) on limit calculation
NOTE: This is related to section 29.1!
In general: what is effect of the uncertainty on limit likelihood: on S and B?
All things that should result in a correlated / uncorrelated "number" that goes into S or B.
UPDATE: :
[ ]Think about whether we ideally really should have a systematic for the z position of the detector. I.e. varying it change the size of the axion image. This is in relation to finding out about the fact that the axion image is actually produced in the center of the chamber instead of the readout plane! Our lack of knowledge about this implies that we should try to account for it.
24.1. List of different uncertainties
Table of the overview.
| Uncertainty | signal or background? | rel. σ [%] | bias? | note | reference |
|---|---|---|---|---|---|
| Earth <-> Sun distance | signal | 0.7732 | Likely to larger values, due to data taking time | 24.1.4.1 | |
| Window thickness (± 10nm) | signal | 0.5807 | none | 24.1.4.2 | |
| Solar models | signal | < 1 | none | unclear from plot, need to look at code | 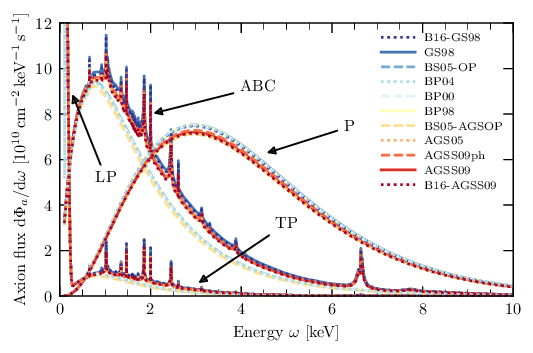 |
| Magnet length (- 1cm) | signal | 0.2159 | likely 9.26m | 24.1.4.3 | |
| Magnet bore diameter (± 0.5mm) | signal | 2.32558 | have measurements indicating 42.x - 43 | 24.1.4.3 | |
| Window rotation (30° ± 0.5°) | signal | 0.18521 | none | rotation seems to be same in both data takings | 24.1.4.4 |
| Nuisance parameter integration routine | For performance reasons less precise integrations. | ||||
| Software efficiency | signal | ~2 | none | Eff. εphoto < 2, but εescape > 3% (less reliable). Choose! | 24.1.7.4 |
| Gas gain time binning | background | 0.26918 | to 0 | Computed background clusters for different gas gain binnings | 24.1.7.1 |
| Reference dist interp (CDL morphing) | background | 0.0844 | none | 24.1.7.2 | |
| Gas gain variation | ? | Partially encoded / fixed w/ gas gain time binning. | |||
| Random coincidences in septem/line veto | 24.1.5.1 | ||||
| Background interpolation (params & shape) | background | ? | none | From error prop. But unclear interpretation. Statistical. | 24.1.6.1 |
| Energy calibration | 24.1.7.3 | ||||
| Alignment (signal, related mounting) | signal (position) | 0.5 mm | none | From X-ray finger & laser alignment | 24.1.4.4 |
| Detector mounting precision (±0.25mm) | signal (position) | 0.25 mm | M6 screws in 6.5mm holes. Results in misalignment, above. | ||
| Gas gain vs charge calib fit | ? | none |
24.1.1. Computing the combined uncertainties
import math let ss = [0.77315941, # based on real tracking dates # 3.3456, <- old number for Sun ⇔ Earth using min/max perihelion/aphelion 0.5807, 1.0, 0.2159, 2.32558, 0.18521] #1.727] # software efficiency of LnL method. Included in `mcmc_limit` directly! let bs = [0.26918, 0.0844] proc total(vals: openArray[float]): float = for x in vals: result += x * x result = sqrt(result) echo "Combined uncertainty signal: ", total(ss) / 100.0 echo "Combined uncertainty background: ", total(bs) / 100.0 echo "Position: ", sqrt(pow((0.5 / 7.0), 2) + pow((0.25 / 7.0), 2))
Compared to 4.582 % we're now down to 3.22%! (in each case including
already the software efficiency, which we don't actually include
anymore here, but in mcmc_limit).
Without the software efficiency we're down to 2.7%!
- Old results
These were the numbers that still used the Perihelion/Aphelion based distances for the systematic of Sun ⇔ Earth distance.
Combined uncertainty signal: 0.04582795952309026 Combined uncertainty background: 0.002821014576353691 Position: 0.07985957062499248 NOTE: The value used here is not the one that was used in most mcmc limit calculations. There we used:
σ_sig = 0.04692492913207222,
which comes out from assuming 2% uncertainty for the software efficiency instead of the
1.727that now show up in the code!
24.1.2. Computed expected limit with above parameters
(technically using position 0.05, because the numbers there are not 1σ, but maxima)
Expected limit: gae² = 5.845913928155455e-21 @gaγ = 1e-12
which implies:
gae gaγ = √gae² * gaγ = 7.645855e-11 * 1e-12 = 7.645855e-23
24.1.3. Signal [0/3]
[ ]signal position (i.e. the spot of the raytracing result)- to be implemented as a nuisance parameter (actually 2) in the limit calculation code.
[ ]pointing precision of the CAST magnet- check the reports of the CAST sun filming. That should give us a good number for the alignment accuracy
[ ]detector and telescope alignment- detector alignment goes straight into the signal position one. The
telescope alignment can be estimated maybe from the geometer
measurements. In any case that will also directly impact the
placement / shape of the axion image. So this should be
redundant. Still need to check the geometer measurements to get a
good idea here.
[X]compute center based on X-ray finger run[X]find image of laser alignment with plastic target[ ]find geometer measurements and see where they place us (good for relative from 2017/18 to end of 2018)
- detector alignment goes straight into the signal position one. The
telescope alignment can be estimated maybe from the geometer
measurements. In any case that will also directly impact the
placement / shape of the axion image. So this should be
redundant. Still need to check the geometer measurements to get a
good idea here.
24.1.4. Signal rate & efficiency [5/7]
[ ](solar model)[X]look into the work by Lennert & Sebastian. What does their study of different solar models imply for different fluxes?[ ]check absolute number for
[X]axion rate as a function of distance Earth ⇔ Sun (depends on time data was taken)[X]simple: compute different rate based on perihelion & aphelion. Difference is measure for > 1σ uncertainty on flux[ ]more complex: compute actual distance at roughly times when data taking took place. Compare those numbers with the AU distance used in the ray tracer & in axion flux (expRatein code).
[X]telescope and window efficiencies[X]window: especially uncertainty of window thickness: Yevgen measured thickness of 3 samples using ellipsometry and got values O(350 nm)! Norcada themselves say 300 ± 10 nm- compute different absorptions for the 300 ± 10 nm case (integrated over some energy range) and for the extrema (Yevgen). That should give us a number in flux one might lose / gain.
[X]window rotation (position of the strongbacks), different for two run periods & somewhat uncertain[X]measurement: look at occupancy of calibration runs. This should give us a well defined orientation for the strongback. From that we can adjust the raytracing. Ideally this does not count as a systematic as we can measure it (I think, but need to do!)[X]need to look at X-ray finger runs reconstructed & check occupancy to compare with occupancies of the calibration data[X]determine the actual loss based on the rotation uncertainty if plugged into raytracer & computed total signal?
[X]magnet length, diameter and field strength (9 T?)- magnet length sometimes reported as 9.25 m, other times as 9.26
[X]compute conversion probability for 9.26 ± 0.01 m. Result affects signal. Get number.
- diameter sometimes reported as 43 mm, sometimes 42.5 (iirc, look
up again!), but numbers given by Theodoros from a measurement for
CAPP indicated essentially 43 (with some measured uncertainty!)
[X]treated the same way as magnet length. Adjust area accordingly & get number for the possible range.
- magnet length sometimes reported as 9.25 m, other times as 9.26
[ ]Software signal efficiency due to linear logL interpolation, for classification signal / background[ ]what we already did: took two bins surrounding a center bin and interpolated the middle one. -> what is difference between interpolated and real? This is a measure for its uncertainty.
[X]detector mounting precision:[X]6 mounting holes, a M6. Hole size 6.5 mm. Thus, easily 0.25mm variation is possible (discussed with Tobi).[X]plug can be moved about ±0.43mm away from the center. On septemboard variance of plugs is ±0.61mm.
- Distance Earth ⇔ Sun
The distance between Earth and the Sun varies between:
Aphelion: 152100000 km Perihelion: 147095000 km Semi-major axis: 149598023 km
which first of all is a variation of a bit more than 3% or about ~1.5% from one AU. The naive interpretation of the effect on the signal variation would then be 1 / (1.015²) = ~0.971, a loss of about 3% for the increase from the semi-major axis to the aphelion (or the inverse for an increase to the aphelion).
In more explicit numbers:
import math proc flux(r: float): float = result = 1 / (r * r) let f_au = flux(149598023) let f_pe = flux(147095000) let f_ap = flux(152100000) echo "Flux at 1 AU: ", f_au echo "Flux at Perihelion: ", f_pe echo "Flux at Aphelion: ", f_ap echo "Flux decrease from 1 AU to Perihelion: ", f_au / f_pe echo "Flux increase from 1 AU to Aphelion: ", f_au / f_ap echo "Mean of increase & decrease: ", (abs(1.0 - f_au / f_pe) + abs(1.0 - f_au / f_ap)) / 2.0 echo "Total flux difference: ", f_pe / f_ap
- UPDATE:
In section [BROKEN LINK: sec:journal:01_07_23_sun_earth_dist] of the
journal.orgwe discuss the real distances during the CAST trackings. The numbers we actually need to care about are the following:Mean distance during trackings = 0.9891144450781392 Variance of distance during trackings = 1.399449924353128e-05 Std of distance during trackings = 0.003740922245052853
referring to the CSV file: ./../resources/sun_earth_distance_cast_solar_trackings.csv
where the numbers are in units of 1 AU.
So the absolute numbers come out to:
import unchained const mean = 0.9891144450781392 echo "Actual distance = ", mean.AU.to(km)
This means an improvement in flux, following the code snippet above:
import math, unchained, measuremancer proc flux[T](r: T): T = result = 1 / (r * r) let mean = 0.9891144450781392.AU.to(km).float ± 0.003740922245052853.AU.to(km).float echo "Flux increase from 1 AU to our actual mean: ", pretty(flux(mean) / flux(1.AU.to(km).float), precision = 8)
Which comes out to be an equivalent of 0.773% for the signal uncertainty now!
This is a really nice improvement from the 3.3% we had before! It should bring the signal uncertainty from ~4.5% down to close to 3% probably.
This number was reproduced using
readOpacityFileas well by (seejournal.orgon for more details):import ggplotnim let df1 = readCsv("~/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_1AU.csv") .filter(f{`type` == "Total flux"}) let df2 = readCsv("~/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv") .filter(f{`type` == "Total flux"}) let max1AU = df1["diffFlux", float].max let max0989AU = df2["diffFlux", float].max echo "Ratio of 1 AU to 0.989 AU = ", max0989AU / max1AU
Bang on!
- UPDATE:
- Variation of window thickness
The thickness of the SiN windows will vary somewhat. Norcada says they are within 10nm of 300nm thickness. Measurements done by Yevgen rather imply variations on the O(50 nm). Difficult to know which numbers to trust. The thickness goes into the transmission according to Beer-Lambert's law. Does this imply quadratically?
I'm a bit confused playing around with the Henke tool.
TODO: get a data file for 1 μm and for 2 μm and check what the difference is.
import ggplotnim let df1 = readCsv("/home/basti/org/resources/si_nitride_1_micron_5_to_10_kev.txt", sep = ' ') .mutate(f{"TSq" ~ `Transmission` * `Transmission`}) let df2 = readCsv("/home/basti/org/resources/si_nitride_2_micron_5_to_10_kev.txt", sep = ' ') let df = bind_rows(df1, df2, id = "id") ggplot(df, aes("Energy[eV]", "Transmission", color = "id")) + geom_line() + geom_line(data = df1, aes = aes(y = "TSq"), color = "purple", lineType = ltDashed) + ggsave("/tmp/transmissions.pdf") # compute the ratio let dfI = inner_join(df1.rename(f{"T1" <- "Transmission"}), df2.rename(f{"T2" <- "Transmission"}), by = "Energy[eV]") .mutate(f{"Ratio" ~ `T1` / `T2`}) echo dfI ggplot(dfI, aes("Energy[eV]", "Ratio")) + geom_line() + ggsave("/tmp/ratio_transmissions_1_to_2_micron.pdf")
The resulting
Ratiohere kind of implies that we're missing something…. Ah, no. TheRatiothing was a brain fart. Just squaring the 1μm thing does indeed reproduce the 2μm case! All good here.So how do we get the correct value then for e.g. 310nm when having 300nm?
If my intuition is correct (we'll check with a few other numbers in a minute) then essentially the following holds:
\[ T_{xd} = (T_d)^x \]
where
T_dis the transmission of the material at thicknessdand we get the correct transmission for a different thickness that is a multiplexofdby the given power-law relation.Let's apply this to the files we have for the 300nm window and see what we get if we also add 290 and 300 nm.
import ggplotnim, strformat, math proc readFile(fname: string): DataFrame = result = readCsv(fname, sep = ' ') .rename(f{"Energy / eV" <- "PhotonEnergy(eV)"}) .mutate(f{"E / keV" ~ c"Energy / eV" / 1000.0}) let sinDf = readFile("../resources/Si3N4_density_3.44_thickness_0.3microns.txt") .mutate(f{float: "T310" ~ pow(`Transmission`, 310.0 / 300.0)}) .mutate(f{float: "T290" ~ pow(`Transmission`, 290.0 / 300.0)}) var sin1Mu = readFile("../resources/Si3N4_density_3.44_thickness_1microns.txt") .mutate(f{float: "Transmission" ~ pow(`Transmission`, 0.3 / 1.0)}) sin1Mu["Setup"] = "T300_from1μm" var winDf = sinDf.gather(["Transmission", "T310", "T290"], key = "Setup", value = "Transmission") ggplot(winDf, aes("E / keV", "Transmission", color = "Setup")) + geom_line() + geom_line(data = sin1Mu, lineType = ltDashed, color = "purple") + xlim(0.0, 3.0, outsideRange = "drop") + xMargin(0.02) + yMargin(0.02) + margin(top = 1.5) + ggtitle("Impact of 10nm uncertainty on window thickness. Dashed line: 300nm transmission computed " & "from 1μm via power law T₃₀₀ = T₁₀₀₀^{0.3/1}") + ggsave("/home/basti/org/Figs/statusAndProgress/window_uncertainty_transmission.pdf", width = 853, height = 480)
Plot
shows us the impact on the transmission of the uncertainty on the window thickness. In terms of such transmission the impact seems almost negligible as long as it's small. However, to get an accurate number, we should check the integrated effect on the axion flux after conversion & going through the window. That then takes into account the energy dependence and thus gives us a proper number of the impact on the signal.
import sequtils, math, unchained, datamancer import numericalnim except linspace, cumSum # import ./background_interpolation defUnit(keV⁻¹•cm⁻²) type Context = object integralBase: float efficiencySpl: InterpolatorType[float] defUnit(keV⁻¹•cm⁻²•s⁻¹) defUnit(keV⁻¹•m⁻²•yr⁻¹) defUnit(cm⁻²) defUnit(keV⁻¹•cm⁻²) proc readAxModel(): DataFrame = let upperBin = 10.0 proc convert(x: float): float = result = x.keV⁻¹•m⁻²•yr⁻¹.to(keV⁻¹•cm⁻²•s⁻¹).float result = readCsv("/home/basti/CastData/ExternCode/AxionElectronLimit/axion_diff_flux_gae_1e-13_gagamma_1e-12.csv") .mutate(f{"Energy / keV" ~ c"Energy / eV" / 1000.0}, f{float: "Flux / keV⁻¹•cm⁻²•s⁻¹" ~ convert(idx("Flux / keV⁻¹ m⁻² yr⁻¹"))}) .filter(f{float: c"Energy / keV" <= upperBin}) proc detectionEff(spl: InterpolatorType[float], energy: keV): UnitLess = # window + gas if energy < 0.001.keV or energy > 10.0.keV: return 0.0 result = spl.eval(energy.float) proc initContext(thickness: NanoMeter): Context = let combEffDf = readCsv("/home/basti/org/resources/combined_detector_efficiencies.csv") .mutate(f{float: "Efficiency" ~ pow(idx("300nm SiN"), thickness / 300.nm)}) ## no-op if input is also 300nm let effSpl = newCubicSpline(combEffDf["Energy [keV]", float].toRawSeq, combEffDf["Efficiency", float].toRawSeq) # effective area included in raytracer let axData = readAxModel() let axModel = axData .mutate(f{"Flux" ~ idx("Flux / keV⁻¹•cm⁻²•s⁻¹") * detectionEff(effSpl, idx("Energy / keV").keV) }) let integralBase = simpson(axModel["Flux", float].toRawSeq, axModel["Energy / keV", float].toRawSeq) result = Context(integralBase: integralBase, efficiencySpl: effSpl) defUnit(cm²) defUnit(keV⁻¹) func conversionProbability(): UnitLess = ## the conversion probability in the CAST magnet (depends on g_aγ) ## simplified vacuum conversion prob. for small masses let B = 9.0.T let L = 9.26.m let g_aγ = 1e-12.GeV⁻¹ # ``must`` be same as reference in Context result = pow( (g_aγ * B.toNaturalUnit * L.toNaturalUnit / 2.0), 2.0 ) defUnit(cm⁻²•s⁻¹) defUnit(m⁻²•yr⁻¹) proc expRate(integralBase: float): UnitLess = let trackingTime = 190.h let areaBore = π * (2.15 * 2.15).cm² result = integralBase.cm⁻²•s⁻¹ * areaBore * trackingTime.to(s) * conversionProbability() let ctx300 = initContext(300.nm) let rate300 = expRate(ctx300.integralBase) let ctx310 = initContext(310.nm) let rate310 = expRate(ctx310.integralBase) let ctx290 = initContext(290.nm) let rate290 = expRate(ctx290.integralBase) echo "Decrease: 300 ↦ 310 nm: ", rate310 / rate300 echo "Increase: 300 ↦ 290 nm: ", rate290 / rate300 echo "Total change: ", rate290 / rate310 echo "Averaged difference: ", (abs(1.0 - rate310 / rate300) + abs(1.0 - rate290 / rate300)) / 2.0
- Magnet length & bore diameter
Length was reported to be 9.25m in the original CAST proposal, compared to the since then reported 9.26m.
Conversion probability scales by length quadratically, so the change in flux should thus also just be quadratic.
The bore diameter was also given as 42.5mm (iirc) initially, but later as 43mm. The amount of flux scales by the area.
import math echo 9.25 / 9.26 # Order 0.1% echo pow(42.5 / 2.0, 2.0) / pow(43 / 2.0, 2.0) # Order 2.3%
With the conversion probability:
\[ P_{a↦γ, \text{vacuum}} = \left(\frac{g_{aγ} B L}{2} \right)^2 \left(\frac{\sin\left(\delta\right)}{\delta}\right)^2 \]
The change in conversion probability from a variation in magnet length is thus (using the simplified form if δ is small:
import unchained, math func conversionProbability(L: Meter): UnitLess = ## the conversion probability in the CAST magnet (depends on g_aγ) ## simplified vacuum conversion prob. for small masses let B = 9.0.T let g_aγ = 1e-12.GeV⁻¹ # ``must`` be same as reference in Context result = pow( (g_aγ * B.toNaturalUnit * L.toNaturalUnit / 2.0), 2.0 ) let P26 = conversionProbability(9.26.m) let P25 = conversionProbability(9.25.m) let P27 = conversionProbability(9.25.m) echo "Change from 9.26 ↦ 9.25 m = ", P26 / P25 echo "Change from 9.25 ↦ 9.27 m = ", P27 / P25 echo "Relative change = ", (abs(1.0 - P27 / P26) + abs(1.0 - P25 / P26)) / 2.0
And now for the area:
As it only goes into the expected rate by virtue of, well, being the area we integrate over, we simply need to look at the change in area from a change in bore radius.
proc expRate(integralBase: float): UnitLess = let trackingTime = 190.h let areaBore = π * (2.15 * 2.15).cm² result = integralBase.cm⁻²•s⁻¹ * areaBore * trackingTime.to(s) * conversionProbability()
import unchained, math defUnit(MilliMeter²) proc boreArea(diameter: MilliMeter): MilliMeter² = result = π * (diameter / 2.0)^2 let areaD = boreArea(43.mm) let areaS = boreArea(42.5.mm) let areaL = boreArea(43.5.mm) echo "Change from 43 ↦ 42.5 mm = ", areaS / areaD echo "Change from 43 ↦ 43.5 mm = ", areaL / areaD echo "Relative change = ", (abs(1.0 - areaL / areaD) + abs(1.0 - areaS / areaD)) / 2.0
- Window rotation & alignment precision
[2/2]
Rotation of the window. Initially we assumed that the rotation was different in the two different data taking periods.
We can check the rotation by looking at the occupancy runs taken in the 2017 dataset and in the 2018 dataset.
The 2017 occupancy (filtered to only use events in eccentricity 1 - 1.4) is
and for 2018:
They imply that the angle was indeed the same (compare with the sketch of our windows in fig. 2). However, there seems to be a small shift in y between the two, which seems hard to explain. Such a shift only makes sense (unless I'm missing something!) if there is a shift between the chip and the window, but not for any kind of installation shift or shift in the position of the 55Fe source. I suppose a slight change in how the window is mounted on the detector can already explain it? This is < 1mm after all.
In terms of the rotation angle, we'll just read it of using Inkscape.
It comes out to pretty much exactly 30°, see fig. 354. I suppose this makes sense given the number of screws (6?). Still, this implies that the window was mounted perfectly aligned with some line relative to 2 screws. Not that it matters.
Figure 354: Measurement of the rotation angle of the window in 2018 data taking (2017 is the same) using Inkscape. Comes out to ~30° (with maybe 0.5° margin for error, aligned for exactly 30° for the picture, but some variation around that all looks fine). Need to check the number used in the raytracing code. There we have (also see discussion with Johanna in Discord):
case wyKind of wy2017: result = degToRad(10.8) of wy2018: result = degToRad(71.5) of wyIAXO: result = degToRad(20.0) # who knows
so an angle of 71.5 (2018) and 10.8 (2017). Very different from the number we get in Inkscape based on the calibration runs.
She used the following plot:
to extract the angles.
The impact of this on the signal only depends on where the strongbacks are compared to the axion image.
Fig. 355 shows the axion image for the rotation of 71.5° (Johanna from X-ray finger) and fig. 356 shows the same for a rotation of 30° (our measurement). The 30° case matches nicely with the extraction of fig. 354.

Figure 355: Axion image for a window setup rotated to 71.5° (the number Johanna read off from the X-ray finger run). 
Figure 356: Axion image for a window setup rotated to 30° (the number we read off from from the calibration runs). From here there are 2 things to do:
[X]reconstruct the X-ray finger runs & check the rotation of those again using the same occupancy plots as for the calibration runs.[X]compute the integrated signal for the 71.5°, 30° and 30°±0.5° cases and see how the signal differs. The latter will be the number for the systematic we'll use. We do that by just summing the raytracing output.
To do the latter, we need to add an option to write the CSV files in the raytracer first.
import datamancer proc print(fname: string): float = let hmap = readCsv(fname) result = hmap["photon flux", float].sum let f71 = print("/home/basti/org/resources/axion_images_systematics/axion_image_2018_71_5deg.csv") let f30 = print("/home/basti/org/resources/axion_images_systematics/axion_image_2018_30deg.csv") let f29 = print("/home/basti/org/resources/axion_images_systematics/axion_image_2018_29_5deg.csv") let f31 = print("/home/basti/org/resources/axion_images_systematics/axion_image_2018_30_5deg.csv") echo f71 echo f30 echo "Ratio : ", f30 / f71 echo "Ratio f29 / f31 ", f29 / f31 echo "Difference ", (abs(1.0 - (f29/f30)) + abs(1.0 - (f31/f30))) / 2.0
Now on to the reconstruction of the X-ray finger run.
I copied the X-ray finger runs from tpc19 over to ./../../CastData/data/XrayFingerRuns/. The run of interest is mainly the run 189, as it's the run done with the detector installed as in 2017/18 data taking.
cd /dev/shm # store here for fast access & temporary cp ~/CastData/data/XrayFingerRuns/XrayFingerRun2018.tar.gz . tar xzf XrayFingerRun2018.tar.gz raw_data_manipulation -p Run_189_180420-09-53 --runType xray --out xray_raw_run189.h5 reconstruction -i xray_raw_run189.h5 --out xray_reco_run189.h5 # make sure `config.toml` for reconstruction uses `default` clustering! plotData \ --h5file xray_reco_run189.h5 \ --runType=rtBackground \ -b bGgPlot \ --h5Compare ~/CastData/data/2018/reco_186.h5 \ --ingrid \ --occupancy \ --config /home/basti/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml
which gives us the following plot:
Figure 357: Occupancies of cluster centers of the X-ray finger run (189) in 2018. Shows the same rotation as the calibration runs here. Using
TimepixAnalysis/Tools/printXyDatasetwe can now compute the center of the X-ray finger run.cd ~/CastData/ExternCode/TimepixAnalysis/Tools/ ./printXyDataset -f /dev/shm/xray_reco_run189.h5 -c 3 -r 189 \ --dset centerX --reco \ --cuts '("eccentricity", 0.9, 1.4)' \ --cuts '("centerX", 3.0, 11.0)' \ --cuts '("centerY", 3.0, 11.0)' ./printXyDataset -f /dev/shm/xray_reco_run189.h5 -c 3 -r 189 \ --dset centerY --reco \ --cuts '("eccentricity", 0.9, 1.4)' \ --cuts '("centerX", 3.0, 11.0)' \ --cuts '("centerY", 3.0, 11.0)'
So we get a mean of:
- centerX: 7.658
- centerY: 6.449
meaning we are ~0.5 mm away from the center in either direction. Given that there is distortion due to the magnet optic, uncertainty about the location of X-ray finger & emission characteristic, using a variation of 0.5mm seems reasonable.
This also matches more or less the laser alignment we did initially, see fig. 358.

Figure 358: Laser alignment using target on flange at CAST. Visible deviation is ~0.5mm more or less. - TODO Question about signal & window
One thing we currently do not take into account is that when varying the signal position using the nuisance parameters, we move the window strongback with the position…
In principle we're not allowed to do that. The strongbacks are part of the detector & not the signal (but are currently convolved into the image).
The strongback position depends on the detector mounting precision only.
So if the main peak was exactly on the strongback, we'd barely see anything!
- TODO Graphite spacer rotation (telescope rotation) via X-ray finger run
[X]Determine the rotation angle of the graphite spacer from the X-ray finger data -> do now. X-ray finger run:->
 -> It comes out to 14.17°!
But for run 21 (between which detector was dismounted of course):
-> It comes out to 14.17°!
But for run 21 (between which detector was dismounted of course):
 -> Only 11.36°!
That's a huge uncertainty given the detector was only dismounted!
3°.
-> Only 11.36°!
That's a huge uncertainty given the detector was only dismounted!
3°.[ ]This variance has a big impact on the systematic uncertainty here!
- Integration routines for nuisance parameters
For performance reasons we cannot integrate out the nuisance parameters using the most sophisticated algorithms. Maybe in the end we could assign a systematic by computing a few "accurate" integrations (e.g. integrating out \(θ_x\) and \(θ_y\)) with adaptive gauss and then with our chosen method and compare the result on the limit? Could just be a "total" uncertainty on the limit w/o changing any parameters.
24.1.5. Detector behavior [0/1]
[ ]drift in # hits in ⁵⁵Fe. "Adaptive gas gain" tries to minimize this, maybe variation of mean energy over time after application a measure for uncertainty? -> should mainly have an effect on software signal efficiency.- goes into S of limit likelihood (ε), which is currently assumed a constant number
[ ]veto random coincidences
24.1.6. Background [0/2]
[ ]background interpolation- we already did: study of statistical uncertainty (both MC as well
as via error propagation)
[X]extract from error propagation code unclear what to do with these numbers!
- we already did: study of statistical uncertainty (both MC as well
as via error propagation)
[ ]septem veto can suffer from uncertainties due to possible random coincidences of events on outer chip that veto a center event, which are not actually correlated. In our current application of it, this implies a) a lower background rate, but b) a lower software signal efficiency as we might also remove real photons. So its effect is on ε as ell.[ ]think about random coincidences, derive some formula similar to lab course to compute chance
- Background interpolation
[0/1]
Ref: 29.1.3.5 and
ntanglethis file and run/tmp/background_interpolation_error_propagation.nimfor the background interpolation withMeasuremancererror propagation.For an input of 8 clusters in a search radius around a point we get numbers such as:
Normalized value (gauss) : 6.08e-06 ± 3.20e-06 CentiMeter⁻²•Second⁻¹so an error that is almost 50% of the input.However, keep in mind that this is for a small area around the specific point. Just purely from Poisson statistics we expect an uncertainty of 2.82 for 8 events \[ ΔN = √8 = 2.82 \]
As such this makes sense (the number is larger due to the gaussian nature of the distance calculation etc.) and just being a weighted sum of
1 ± 1terms error propagated.If we compute the same for a larger number of points, the error should go down, which can be seen comparing fig. 431 with fig. 432 (where the latter has artificially increased statistics).
As this is purely a statistical effect, I'm not sure how to quantify any kind of systematic errors.
The systematics come into play, due to the:
- choice of radius & sigma
- choice of gaussian weighting
- choice of "energy radius"
[ ]look at background interpolation uncertainty section linked above. Modify to also include a section about a flat model that varies the different parameters going into the interpolation.[ ]use existing code to compute a systematic based on the kind of background model. Impact of background hypothesis?
24.1.7. Energy calibration, likelihood method [0/1]
[ ]the energy calibration as a whole has many uncertainties (due to detector variation, etc.)- gas gain time binning:
[ ]compute everything up to background rate for no time binning, 90 min and maybe 1 or 2 other values. Influence on σb is the change in background that we see from this (will be a lot of work, but useful to make things more reproducible).
[ ]compute energy of 55Fe peaks after energy calibration. Variation gives indication for systematic influence.
- gas gain time binning:
- Gas gain time binning
We need to investigate the impact of the gas gain binning on the background rate. How do we achieve that?
Simplest approach:
- Compute gas gain slices for different cases (no binning, 30 min binning, 90 min binning, 240 min binning ?)
- calculate energy based on the used gas gain binning
- compute the background rate for each case
- compare amount of background after that.
Question: Do we need to recompute the gas gain for the calibration data as well? Yes, as the gas gain slices directly go into the 'gain fit' that needs to be done in order to compute the energy for any cluster.
So, the whole process is only made complicated by the fact that we need to change the
config.tomlfile in between runs. In the future this should be a CL argument. For the time being, we can use the same approach as in/home/basti/CastData/ExternCode/TimepixAnalysis/Tools/backgroundRateDifferentEffs/backgroundRateDifferentEfficiencies.nimwhere we simply read the toml file, rewrite the single line and write it back.Let's write a script that does mainly steps 1 to 3 for us.
import shell, strformat, strutils, sequtils, os # an interval of 0 implies _no_ gas gain interval, i.e. full run const intervals = [0, 30, 90, 240] const Tmpl = "$#Runs$#_Reco.h5" const Path = "/home/basti/CastData/data/systematics/" const TomlFile = "/home/basti/CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/config.toml" proc rewriteToml(path: string, interval: int) = ## rewrites the given TOML file in the `path` to use the `interval` ## instead of the existing value var data = readFile(path).splitLines for l in mitems(data): if interval == 0 and l.startsWith("fullRunGasGain"): l = "fullRunGasGain = true" elif interval != 0 and l.startsWith("fullRunGasGain"): l = "fullRunGasGain = false" elif interval != 0 and l.startsWith("gasGainInterval"): l = "gasGainInterval = " & $interval writeFile(path, data.join("\n")) proc computeGasGainSlices(fname: string, interval: int) = let (res, err, code) = shellVerboseErr: one: cd ~/CastData/data/systematics reconstruction ($fname) "--only_gas_gain" if code != 0: raise newException(Exception, "Error calculating gas gain for interval " & $interval) proc computeGasGainFit(fname: string, interval: int) = let (res, err, code) = shellVerboseErr: one: cd ~/CastData/data/systematics reconstruction ($fname) "--only_gain_fit" if code != 0: raise newException(Exception, "Error calculating gas gain fit for interval " & $interval) proc computeEnergy(fname: string, interval: int) = let (res, err, code) = shellVerboseErr: one: cd ~/CastData/data/systematics reconstruction ($fname) "--only_energy_from_e" if code != 0: raise newException(Exception, "Error calculating energy for interval " & $interval) proc computeLikelihood(f, outName: string, interval: int) = let args = { "--altCdlFile" : "~/CastData/data/CDL_2019/calibration-cdl-2018.h5", "--altRefFile" : "~/CastData/data/CDL_2019/XrayReferenceFile2018.h5", "--cdlYear" : "2018", "--region" : "crGold"} let argStr = args.mapIt(it[0] & " " & it[1]).join(" ") let (res, err, code) = shellVerboseErr: one: cd ~/CastData/data/systematics likelihood ($f) "--h5out" ($outName) ($argStr) if code != 0: raise newException(Exception, "Error computing likelihood cuts for interval " & $interval) #proc plotBackgroundRate(f1, f2: string, eff: float) = # let suffix = &"_eff_{eff}" # let (res, err, code) = shellVerboseErr: # one: # cd ~/CastData/ExternCode/TimepixAnalysis/Plotting/plotBackgroundRate # ./plotBackgroundRate ($f1) ($f2) "--suffix" ($suffix) # ./plotBackgroundRate ($f1) ($f2) "--separateFiles --suffix" ($suffix) # if code != 0: # raise newException(Exception, "Error plotting background rate for eff " & $eff) let years = [2017, 2018] let calibs = years.mapIt(Tmpl % ["Calibration", $it]) let backs = years.mapIt(Tmpl % ["Data", $it]) copyFile(TomlFile, "/tmp/toml_file.backup") for interval in intervals: ## rewrite toml file rewriteToml(TomlFile, interval) ## compute new gas gain for new interval for all files for f in concat(calibs, backs): computeGasGainSlices(f, interval) ## use gas gain slices to compute gas gain fit for f in calibs: computeGasGainFit(f, interval) ## compute energy based on new gain fit for f in concat(calibs, backs): computeEnergy(f, interval) ## compute likelihood based on new energies var logFs = newSeq[string]() for b in backs: let yr = if "2017" in b: "2017" else: "2018" let fname = &"out/lhood_{yr}_interval_{interval}.h5" logFs.add fname ## XXX: need to redo likelihood computation!! computeLikelihood(b, fname, interval) ## plot background rate for all combined? or just plot cluster centers? can all be done later... #plotBackgroundRate(log, eff)
import shell, strformat, strutils, sequtils, os # an interval of 0 implies _no_ gas gain interval, i.e. full run const intervals = [0, 30, 90, 240] const Tmpl = "$#Runs$#_Reco.h5" echo (Tmpl % ["Data", "2017"]).extractFilename
The resulting files are found in ./../../CastData/data/systematics/out/ or ./../../CastData/data/systematics/ on my laptop.
Let's extract the number of clusters found on the center chip (gold region) for each of the intervals:
cd ~/CastData/data/systematics for i in 0 30 90 240 do echo Inteval: $i extractClusterInfo -f lhood_2017_interval_$i.h5 --short --region crGold extractClusterInfo -f lhood_2018_interval_$i.h5 --short --region crGold done
The numbers pretty much speak for themselves.
let nums = { 0 : 497 + 244, 30 : 499 + 244, 90 : 500 + 243, 240 : 497 + 244 } # reference is 90 let num90 = nums[2][1] var minVal = Inf var maxVal = 0.0 for num in nums: let rat = num[1] / num90 echo "Ratio of ", num, " = ", rat minVal = min(minVal, rat) maxVal = max(maxVal, rat) echo "Deviation: ", maxVal - minVal
NOTE: The one 'drawback' of this approach taken here is the following: the CDL data was not reconstructed using the changed gas gain data. However that is much less important, as we assume constant gain over the CDL runs anyway more or less / want to pick the most precise description of our data!
- Interpolation of reference distributions (CDL morphing)
[/]
We already did the study of the variation in the interpolation for the reference distributions. To estimate the systematic uncertainty related to that, we should simply look at the computation of the "intermediate" distributions again and compare the real numbers to the interpolated ones. The deviation can be done per bin. The average & some quantiles should be a good number to refer to as a systematic.
The
cdlMorphingtool ./../../CastData/ExternCode/TimepixAnalysis/Tools/cdlMorphing/cdlMorphing.nim is well suited to this. We will compute the difference between the morphed and real data for each bin & sum the squares for each target/filter (those that are morphed, so not the outer two of course).Running the tool now yields the following output:
Target/Filter: Cu-EPIC-0.9kV = 0.0006215219861090395 Target/Filter: Cu-EPIC-2kV = 0.0007052150065674744 Target/Filter: Al-Al-4kV = 0.001483398679126846 Target/Filter: Ag-Ag-6kV = 0.001126063558474516 Target/Filter: Ti-Ti-9kV = 0.0006524420692883554 Target/Filter: Mn-Cr-12kV = 0.0004757207676502019 Mean difference 0.0008440603445360723
So we really have a miniscule difference there.
[ ]also compute the background rate achieved using no CDL morphing vs using it.
- Energy calibration
[/]
[ ]compute peaks of 55Fe energy. What is variation?
- Software efficiency systematic
[/]
In order to guess at the systematic uncertainty of the software efficiency, we can push all calibration data through the likelihood cuts and evaluate the real efficiency that way.
This means the following:
- compute likelihood values for all calibration runs
- for each run, remove extreme outliers using rough RMS transverse & eccentricity cuts
- filter to 2 energies (essentially a secondary cut), the photopeak and escape peak
- for each peak, push through likelihood cut. # after / # before is software efficiency at that energy
The variation we'll see over all runs tells us something about the systematic uncertainty & potential bias.
UPDATE: The results presented below the code were computed with the code snippet here as is (and multiple arguments of course, check
zsh_historyat home for details). A modified version now also lives at ./../../CastData/ExternCode/TimepixAnalysis/Tools/determineEffectiveEfficiency.nimUPDATE2 : While working on the below code for the script mentioned in the first update, I noticed a bug in the
filterEventsfunction:of "Escapepeak": let dset = 5.9.toRefDset() let xrayCuts = xrayCutsTab[dset] result.add applyFilters(df) of "Photopeak": let dset = 2.9.toRefDset() let xrayCuts = xrayCutsTab[dset] result.add applyFilters(df)
the energies are exchanged and
applyFiltersis applied todfand notsubDfas it should here![ ]Investigate the effect for the systematics of CAST! -> : I just had a short look at this. It seems like this is the correct output:
DataFrame with 3 columns and 67 rows: Idx Escapepeak Photopeak RunNumber dtype: float float int 0 0.6579 0.7542 83 1 0.6452 0.787 88 2 0.6771 0.7667 93 3 0.7975 0.7599 96 4 0.799 0.7605 102 5 0.8155 0.7679 108 6 0.7512 0.7588 110 7 0.8253 0.7769 116 8 0.7766 0.7642 118 9 0.7752 0.7765 120 10 0.7556 0.7678 122 11 0.7788 0.7711 126 12 0.7749 0.7649 128 13 0.8162 0.7807 145 14 0.8393 0.7804 147 15 0.7778 0.78 149 16 0.8153 0.778 151 17 0.7591 0.7873 153 18 0.8229 0.7819 155 19 0.8341 0.7661 157 20 0.7788 0.7666 159 21 0.7912 0.7639 161 22 0.8041 0.7675 163 23 0.7884 0.777 165 24 0.8213 0.7791 167 25 0.7994 0.7833 169 26 0.8319 0.7891 171 27 0.8483 0.7729 173 28 0.7973 0.7733 175 29 0.834 0.7771 177 30 0.802 0.773 179 31 0.7763 0.7687 181 32 0.8061 0.766 183 33 0.7916 0.7799 185 34 0.8131 0.7745 187 35 0.8366 0.8256 239 36 0.8282 0.8035 241 37 0.8072 0.8045 243 38 0.851 0.8155 245 39 0.7637 0.8086 247 40 0.8439 0.8135 249 41 0.8571 0.8022 251 42 0.7854 0.7851 253 43 0.8159 0.7843 255 44 0.815 0.7827 257 45 0.8783 0.8123 259 46 0.8354 0.8094 260 47 0.8 0.789 262 48 0.8038 0.8097 264 49 0.7926 0.7937 266 50 0.8275 0.7961 269 51 0.8514 0.8039 271 52 0.8089 0.7835 273 53 0.8134 0.7789 275 54 0.8168 0.7873 277 55 0.8198 0.7886 280 56 0.8447 0.7833 282 57 0.7876 0.7916 284 58 0.8093 0.8032 286 59 0.7945 0.8059 288 60 0.8407 0.7981 290 61 0.7824 0.78 292 62 0.7885 0.7869 294 63 0.7933 0.7823 296 64 0.837 0.7834 300 65 0.7594 0.7826 302 66 0.8333 0.7949 304
Std Escape = 0.04106537728575545 Std Photo = 0.01581231947284212 Mean Escape = 0.8015071105396809 Mean Photo = 0.7837728948033928
So a bit worse than initially thought…
import std / [os, strutils, random, sequtils, stats, strformat] import nimhdf5, cligen import numericalnim except linspace import ingrid / private / [likelihood_utils, hdf5_utils, ggplot_utils, geometry, cdl_cuts] import ingrid / calibration import ingrid / calibration / [fit_functions] import ingrid / ingrid_types import ingridDatabase / [databaseRead, databaseDefinitions, databaseUtils] # cut performed regardless of logL value on the data, since transverse # rms > 1.5 cannot be a physical photon, due to diffusion in 3cm drift # distance const RmsCleaningCut = 1.5 let CdlFile = "/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5" let RefFile = "/home/basti/CastData/data/CDL_2019/XrayReferenceFile2018.h5" proc drawNewEvent(rms, energy: seq[float]): int = let num = rms.len - 1 var idx = rand(num) while rms[idx] >= RmsCleaningCut or (energy[idx] <= 4.5 or energy[idx] >= 7.5): idx = rand(num) result = idx proc computeEnergy(h5f: H5File, pix: seq[Pix], group: string, a, b, c, t, bL, mL: float): float = let totalCharge = pix.mapIt(calibrateCharge(it.ch.float, a, b, c, t)).sum # compute mean of all gas gain slices in this run (most sensible) let gain = h5f[group / "chip_3/gasGainSlices", GasGainIntervalResult].mapIt(it.G).mean let calibFactor = linearFunc(@[bL, mL], gain) * 1e-6 # now calculate energy for all hits result = totalCharge * calibFactor proc generateFakeData(h5f: H5File, nFake: int, energy = 3.0): DataFrame = ## For each run generate `nFake` fake events let refSetTuple = readRefDsets(RefFile, yr2018) result = newDataFrame() for (num, group) in runs(h5f): # first read all x / y / tot data echo "Run number: ", num let xs = h5f[group / "chip_3/x", special_type(uint8), uint8] let ys = h5f[group / "chip_3/y", special_type(uint8), uint8] let ts = h5f[group / "chip_3/ToT", special_type(uint16), uint16] let rms = h5f[group / "chip_3/rmsTransverse", float] let cX = h5f[group / "chip_3/centerX", float] let cY = h5f[group / "chip_3/centerY", float] let energyInput = h5f[group / "chip_3/energyFromCharge", float] let chipGrp = h5f[(group / "chip_3").grp_str] let chipName = chipGrp.attrs["chipName", string] # get factors for charge calibration let (a, b, c, t) = getTotCalibParameters(chipName, num) # get factors for charge / gas gain fit let (bL, mL) = getCalibVsGasGainFactors(chipName, num, suffix = $gcIndividualFits) var count = 0 var evIdx = 0 when false: for i in 0 ..< xs.len: if xs[i].len < 150 and energyInput[i] > 5.5: # recompute from data let pp = toSeq(0 ..< xs[i].len).mapIt((x: xs[i][it], y: ys[i][it], ch: ts[i][it])) let newEnergy = h5f.computeEnergy(pp, group, a, b, c, t, bL, mL) echo "Length ", xs[i].len , " w/ energy ", energyInput[i], " recomp ", newEnergy let df = toDf({"x" : pp.mapIt(it.x.int), "y" : pp.mapIt(it.y.int), "ch" : pp.mapIt(it.ch.int)}) ggplot(df, aes("x", "y", color = "ch")) + geom_point() + ggtitle("funny its real") + ggsave("/tmp/fake_event_" & $i & ".pdf") sleep(200) if true: quit() # to store fake data var energies = newSeqOfCap[float](nFake) var logLs = newSeqOfCap[float](nFake) var rmss = newSeqOfCap[float](nFake) var eccs = newSeqOfCap[float](nFake) var ldivs = newSeqOfCap[float](nFake) var frins = newSeqOfCap[float](nFake) var cxxs = newSeqOfCap[float](nFake) var cyys = newSeqOfCap[float](nFake) var lengths = newSeqOfCap[float](nFake) while count < nFake: # draw index from to generate a fake event evIdx = drawNewEvent(rms, energyInput) # draw number of fake pixels # compute ref # pixels for this event taking into account possible double counting etc. let basePixels = (energy / energyInput[evIdx] * xs[evIdx].len.float) let nPix = round(basePixels + gauss(sigma = 10.0)).int # ~115 pix as reference in 3 keV (26 eV), draw normal w/10 around if nPix < 4: echo "Less than 4 pixels: ", nPix, " skipping" continue var pix = newSeq[Pix](nPix) var seenPix: set[uint16] = {} let evNumPix = xs[evIdx].len if nPix >= evNumPix: echo "More pixels to draw than available! ", nPix, " vs ", evNumPix, ", skipping!" continue if not inRegion(cX[evIdx], cY[evIdx], crSilver): echo "Not in silver region. Not a good basis" continue var pIdx = rand(evNumPix - 1) for j in 0 ..< nPix: # draw pix index while pIdx.uint16 in seenPix: pIdx = rand(evNumPix - 1) seenPix.incl pIdx.uint16 pix[j] = (x: xs[evIdx][pIdx], y: ys[evIdx][pIdx], ch: ts[evIdx][pIdx]) # now draw when false: let df = toDf({"x" : pix.mapIt(it.x.int), "y" : pix.mapIt(it.y.int), "ch" : pix.mapIt(it.ch.int)}) ggplot(df, aes("x", "y", color = "ch")) + geom_point() + ggsave("/tmp/fake_event.pdf") sleep(200) # reconstruct event let inp = (pixels: pix, eventNumber: 0, toa: newSeq[uint16](), toaCombined: newSeq[uint64]()) let recoEv = recoEvent(inp, -1, num, searchRadius = 50, dbscanEpsilon = 65, clusterAlgo = caDefault) if recoEv.cluster.len > 1 or recoEv.cluster.len == 0: echo "Found more than 1 or 0 cluster! Skipping" continue # compute charge let energy = h5f.computeEnergy(pix, group, a, b, c, t, bL, mL) # puhhh, now the likelihood... let ecc = recoEv.cluster[0].geometry.eccentricity let ldiv = recoEv.cluster[0].geometry.lengthDivRmsTrans let frin = recoEv.cluster[0].geometry.fractionInTransverseRms let logL = calcLikelihoodForEvent(energy, ecc, ldiv, frin, refSetTuple) # finally done energies.add energy logLs.add logL rmss.add recoEv.cluster[0].geometry.rmsTransverse eccs.add ecc ldivs.add ldiv frins.add frin cxxs.add recoEv.cluster[0].centerX cyys.add recoEv.cluster[0].centerY lengths.add recoEv.cluster[0].geometry.length inc count let df = toDf({ "energyFromCharge" : energies, "likelihood" : logLs, "runNumber" : num, "rmsTransverse" : rmss, "eccentricity" : eccs, "lengthDivRmsTrans" : ldivs, "centerX" : cxxs, "centerY" : cyys, "length" : lengths, "fractionInTransverseRms" : frins }) result.add df proc applyLogLCut(df: DataFrame, cutTab: CutValueInterpolator): DataFrame = result = df.mutate(f{float: "passLogL?" ~ (block: #echo "Cut value: ", cutTab[idx(igEnergyFromCharge.toDset())], " at dset ", toRefDset(idx(igEnergyFromCharge.toDset())), " at energy ", idx(igEnergyFromCharge.toDset()) idx(igLikelihood.toDset()) < cutTab[idx(igEnergyFromCharge.toDset())])}) proc readRunData(h5f: H5File): DataFrame = result = h5f.readDsets(chipDsets = some((chip: 3, dsets: @[igEnergyFromCharge.toDset(), igRmsTransverse.toDset(), igLengthDivRmsTrans.toDset(), igFractionInTransverseRms.toDset(), igEccentricity.toDset(), igCenterX.toDset(), igCenterY.toDset(), igLength.toDset(), igLikelihood.toDset()]))) proc filterEvents(df: DataFrame, energy: float = Inf): DataFrame = let xrayCutsTab {.global.} = getXrayCleaningCuts() template applyFilters(dfI: untyped): untyped {.dirty.} = let minRms = xrayCuts.minRms let maxRms = xrayCuts.maxRms let maxLen = xrayCuts.maxLength let maxEcc = xrayCuts.maxEccentricity dfI.filter(f{float -> bool: idx(igRmsTransverse.toDset()) < RmsCleaningCut and inRegion(idx("centerX"), idx("centerY"), crSilver) and idx("rmsTransverse") >= minRms and idx("rmsTransverse") <= maxRms and idx("length") <= maxLen and idx("eccentricity") <= maxEcc }) if "Peak" in df: doAssert classify(energy) == fcInf result = newDataFrame() for (tup, subDf) in groups(df.group_by("Peak")): case tup[0][1].toStr of "Escapepeak": let dset = 5.9.toRefDset() let xrayCuts = xrayCutsTab[dset] result.add applyFilters(df) of "Photopeak": let dset = 2.9.toRefDset() let xrayCuts = xrayCutsTab[dset] result.add applyFilters(df) else: doAssert false, "Invalid name" else: doAssert classify(energy) != fcInf let dset = energy.toRefDset() let xrayCuts = xrayCutsTab[dset] result = applyFilters(df) proc splitPeaks(df: DataFrame): DataFrame = let eD = igEnergyFromCharge.toDset() result = df.mutate(f{float -> string: "Peak" ~ ( if idx(eD) < 3.5 and idx(eD) > 2.5: "Escapepeak" elif idx(eD) > 4.5 and idx(eD) < 7.5: "Photopeak" else: "None")}) .filter(f{`Peak` != "None"}) proc handleFile(fname: string, cutTab: CutValueInterpolator): DataFrame = ## Given a single input file, performs application of the likelihood cut for all ## runs in it, split by photo & escape peak. Returns a DF with column indicating ## the peak, energy of each event & a column whether it passed the likelihood cut. ## Only events that are pass the input cuts are stored. let h5f = H5open(fname, "r") randomize(423) result = newDataFrame() let data = h5f.readRunData() .splitPeaks() .filterEvents() .applyLogLCut(cutTab) result.add data when false: ggplot(result, aes("energyFromCharge")) + geom_histogram(bins = 200) + ggsave("/tmp/ugl.pdf") discard h5f.close() proc handleFakeData(fname: string, energy: float, cutTab: CutValueInterpolator): DataFrame = let h5f = H5open(fname, "r") var data = generateFakeData(h5f, 5000, energy = energy) .filterEvents(energy) .applyLogLCut(cutTab) result = data discard h5f.close() proc getIndices(dset: string): seq[int] = result = newSeq[int]() applyLogLFilterCuts(CdlFile, RefFile, dset, yr2018, igEnergyFromCharge): result.add i proc plotRefHistos(df: DataFrame, energy: float, cutTab: CutValueInterpolator, dfAdditions: seq[tuple[name: string, df: DataFrame]] = @[]) = # map input fake energy to reference dataset let grp = energy.toRefDset() let passedInds = getIndices(grp) let h5f = H5open(RefFile, "r") let h5fC = H5open(CdlFile, "r") const xray_ref = getXrayRefTable() #for (i, grp) in pairs(xray_ref): var dfR = newDataFrame() for dset in IngridDsetKind: try: let d = dset.toDset() if d notin df: continue # skip things not in input ## first read data from CDL file (exists for sure) ## extract all CDL data that passes the cuts used to generate the logL histograms var cdlFiltered = newSeq[float](passedInds.len) let cdlRaw = h5fC[cdlGroupName(grp, "2019", d), float] for i, idx in passedInds: cdlFiltered[i] = cdlRaw[idx] echo "Total number of elements ", cdlRaw.len, " filtered to ", passedInds.len dfR[d] = cdlFiltered ## now read histograms from RefFile, if they exist (not all datasets do) if grp / d in h5f: let dsetH5 = h5f[(grp / d).dset_str] let (bins, data) = dsetH5[float].reshape2D(dsetH5.shape).split(Seq2Col) let fname = &"/tmp/{grp}_{d}_energy_{energy:.1f}.pdf" echo "Storing histogram in : ", fname # now add fake data let dataSum = simpson(data, bins) let refDf = toDf({"bins" : bins, "data" : data}) .mutate(f{"data" ~ `data` / dataSum}) let df = df.filter(f{float: idx(d) <= bins[^1]}) ggplot(refDf, aes("bins", "data")) + geom_histogram(stat = "identity", hdKind = hdOutline, alpha = 0.5) + geom_histogram(data = df, aes = aes(d), bins = 200, alpha = 0.5, fillColor = "orange", density = true, hdKind = hdOutline) + ggtitle(&"{d}. Orange: fake data from 'reducing' 5.9 keV data @ {energy:.1f}. Black: CDL ref {grp}") + ggsave(fname, width = 1000, height = 600) except AssertionError: continue # get effect of logL cut on CDL data dfR = dfR.applyLogLCut(cutTab) var dfs = @[("Fake", df), ("Real", dfR)] if dfAdditions.len > 0: dfs = concat(dfs, dfAdditions) var dfPlot = bind_rows(dfs, "Type") echo "Rough filter removes: ", dfPlot.len dfPlot = dfPlot.filter(f{`lengthDivRmsTrans` <= 50.0 and `eccentricity` <= 5.0}) echo "To ", dfPlot.len, " elements" ggplot(dfPlot, aes("lengthDivRmsTrans", "fractionInTransverseRms", color = "eccentricity")) + facet_wrap("Type") + geom_point(size = 1.0, alpha = 0.5) + ggtitle(&"Fake energy: {energy:.2f}, CDL dataset: {grp}") + ggsave(&"/tmp/scatter_colored_fake_energy_{energy:.2f}.png", width = 1200, height = 800) # plot likelihood histos ggplot(dfPlot, aes("likelihood", fill = "Type")) + geom_histogram(bins = 200, alpha = 0.5, hdKind = hdOutline) + ggtitle(&"Fake energy: {energy:.2f}, CDL dataset: {grp}") + ggsave(&"/tmp/histogram_fake_energy_{energy:.2f}.pdf", width = 800, height = 600) discard h5f.close() discard h5fC.close() echo "DATASET : ", grp, "--------------------------------------------------------------------------------" echo "Efficiency of logL cut on filtered CDL data (should be 80%!) = ", dfR.filter(f{idx("passLogL?") == true}).len.float / dfR.len.float echo "Elements passing using `passLogL?` ", dfR.filter(f{idx("passLogL?") == true}).len, " vs total ", dfR.len let (hist, bins) = histogram(dfR["likelihood", float].toRawSeq, 200, (0.0, 30.0)) ggplot(toDf({"Bins" : bins, "Hist" : hist}), aes("Bins", "Hist")) + geom_histogram(stat = "identity") + ggsave("/tmp/usage_histo_" & $grp & ".pdf") let cutval = determineCutValue(hist, eff = 0.8) echo "Effficiency from `determineCutValue? ", bins[cutVal] proc main(files: seq[string], fake = false, real = false, refPlots = false, energies: seq[float] = @[]) = ## given the input files of calibration runs, walks all files to determine the ## 'real' software efficiency for them & generates a plot let cutTab = calcCutValueTab(CdlFile, RefFile, yr2018, igEnergyFromCharge) var df = newDataFrame() if real and not fake: for f in files: df.add handleFile(f, cutTab) var effEsc = newSeq[float]() var effPho = newSeq[float]() var nums = newSeq[int]() for (tup, subDf) in groups(df.group_by(@["runNumber", "Peak"])): echo "------------------" echo tup #echo subDf let eff = subDf.filter(f{idx("passLogL?") == true}).len.float / subDf.len.float echo "Software efficiency: ", eff if tup[1][1].toStr == "Escapepeak": effEsc.add eff elif tup[1][1].toStr == "Photopeak": effPho.add eff # only add in one branch nums.add tup[0][1].toInt echo "------------------" let dfEff = toDf({"Escapepeak" : effEsc, "Photopeak" : effPho, "RunNumber" : nums}) echo dfEff.pretty(-1) let stdEsc = effEsc.standardDeviationS let stdPho = effPho.standardDeviationS let meanEsc = effEsc.mean let meanPho = effPho.mean echo "Std Escape = ", stdEsc echo "Std Photo = ", stdPho echo "Mean Escape = ", meanEsc echo "Mean Photo = ", meanPho ggplot(dfEff.gather(["Escapepeak", "Photopeak"], "Type", "Value"), aes("Value", fill = "Type")) + geom_histogram(bins = 20, hdKind = hdOutline, alpha = 0.5) + ggtitle(&"σ_escape = {stdEsc:.4f}, μ_escape = {meanEsc:.4f}, σ_photo = {stdPho:.4f}, μ_photo = {meanPho:.4f}") + ggsave("/tmp/software_efficiencies_cast_escape_photo.pdf", width = 800, height = 600) for (tup, subDf) in groups(df.group_by("Peak")): case tup[0][1].toStr of "Escapepeak": plotRefHistos(df, 2.9, cutTab) of "Photopeak": plotRefHistos(df, 5.9, cutTab) else: doAssert false, "Invalid data: " & $tup[0][1].toStr if fake and not real: var effs = newSeq[float]() for e in energies: if e > 5.9: echo "Warning: energy above 5.9 keV not allowed!" return df = newDataFrame() for f in files: df.add handleFakeData(f, e, cutTab) plotRefHistos(df, e, cutTab) echo "Done generating for energy ", e effs.add(df.filter(f{idx("passLogL?") == true}).len.float / df.len.float) let dfL = toDf({"Energy" : energies, "Efficiency" : effs}) echo dfL ggplot(dfL, aes("Energy", "Efficiency")) + geom_point() + ggtitle("Software efficiency from 'fake' events") + ggsave("/tmp/fake_software_effs.pdf") if fake and real: doAssert files.len == 1, "Not more than 1 file supported!" let f = files[0] let dfCast = handleFile(f, cutTab) for (tup, subDf) in groups(dfCast.group_by("Peak")): case tup[0][1].toStr of "Escapepeak": plotRefHistos(handleFakeData(f, 2.9, cutTab), 2.9, cutTab, @[("CAST", subDf)]) of "Photopeak": plotRefHistos(handleFakeData(f, 5.9, cutTab), 5.9, cutTab, @[("CAST", subDf)]) else: doAssert false, "Invalid data: " & $tup[0][1].toStr #if refPlots: # plotRefHistos() when isMainModule: dispatch main
UPDATE : The discussion about the results of the above code here is limited to the results relevant for the systematic of the software efficiency. For the debugging of the unexpected software efficiencies computed for the calibration photo & escape peaks, see section 14.7.
After the debugging session trying to figure out why the hell the software efficiency is so different, here are finally the results of this study.
The software efficiencies for the escape & photo peak energies from the calibration data at CAST are determined as follows:
- filter to events with
rmsTransverse<= 1.5 - filter to events within the silver region
- filter to events passing the 'X-ray cuts'
- for escape & photo peak each filter to energies of 1 & 1.5 keV around the peak
The remaining events are then used as the "basis" for the evaluation. From here the likelihood cut method is applied to all clusters. In the final step the ratio of clusters passing the logL cut over all clusters is computed, which gives the effective software efficiency for the data.
For all 2017 and 2018 runs this gives:
Dataframe with 3 columns and 67 rows: Idx Escapepeak Photopeak RunNumber dtype: float float int 0 0.6886 0.756 83 1 0.6845 0.794 88 2 0.6789 0.7722 93 3 0.7748 0.7585 96 4 0.8111 0.769 102 5 0.7979 0.765 108 6 0.7346 0.7736 110 7 0.7682 0.7736 116 8 0.7593 0.775 118 9 0.7717 0.7754 120 10 0.7628 0.7714 122 11 0.7616 0.7675 126 12 0.7757 0.7659 128 13 0.8274 0.7889 145 14 0.7974 0.7908 147 15 0.7969 0.7846 149 16 0.7919 0.7853 151 17 0.7574 0.7913 153 18 0.835 0.7887 155 19 0.8119 0.7755 157 20 0.7738 0.7763 159 21 0.7937 0.7736 161 22 0.7801 0.769 163 23 0.8 0.7801 165 24 0.8014 0.785 167 25 0.7922 0.787 169 26 0.8237 0.7945 171 27 0.8392 0.781 173 28 0.8092 0.7756 175 29 0.8124 0.7864 177 30 0.803 0.7818 179 31 0.7727 0.7742 181 32 0.7758 0.7676 183 33 0.7993 0.7817 185 34 0.8201 0.7757 187 35 0.824 0.8269 239 36 0.8369 0.8186 241 37 0.7953 0.8097 243 38 0.8205 0.8145 245 39 0.775 0.8117 247 40 0.8368 0.8264 249 41 0.8405 0.8105 251 42 0.7804 0.803 253 43 0.8177 0.7907 255 44 0.801 0.7868 257 45 0.832 0.8168 259 46 0.8182 0.8074 260 47 0.7928 0.7995 262 48 0.7906 0.8185 264 49 0.7933 0.8039 266 50 0.8026 0.811 269 51 0.8328 0.8086 271 52 0.8024 0.7989 273 53 0.8065 0.7911 275 54 0.807 0.8006 277 55 0.7895 0.7963 280 56 0.8133 0.7918 282 57 0.7939 0.8037 284 58 0.7963 0.8066 286 59 0.8104 0.8181 288 60 0.8056 0.809 290 61 0.762 0.7999 292 62 0.7659 0.8021 294 63 0.7648 0.79 296 64 0.7868 0.7952 300 65 0.7815 0.8036 302 66 0.8276 0.8078 304with the following statistical summaries:
Std Escape = 0.03320160467567293 Std Photo = 0.01727763707839311 Mean Escape = 0.7923601424260915 Mean Photo = 0.7909126317171645
(where
Stdreally is the standard deviation. For the escape data this is skewed due to the first 3 runs as visible in the DF output above).The data as a histogram:
Figure 359: Histogram of the effective software efficiencies for escape and photopeak data at CAST for all 2017/18 calibration runs. The low efficiency outliers are the first 3 calibration runs in 2017. Further, we can also ask for the behavior of fake data now. Let's generate a set and look at the effective efficiency of fake data.
Figure 360: Fake effective software efficiencies at different energies. Clusters are generated from valid 5.9 keV Photopeak clusters (that pass the required cuts) by randomly removing a certain number of pixels until the desired energy is reached. Given the approach, the achieved efficiencies seem fine. Figure 361: Histograms showing the different distributions of the properties for the generated fake data compared to the real reference data from the CDL. At the lowest energies the properties start to diverge quite a bit, likely explaining the lower efficiency there. Figure 362: Scatter plots of the different parameters going into the logL cut method comparing the CDL reference data & the fake generated data. The cuts (X-ray for fake & X-ray + reference for CDL) are applied. NOTE: One big TODO is the following:
[ ]Currently the cut values for the LogL are computed using a histogram of 200 bins, resulting in significant variance already in the CDL data of around 1%. By increasing the number of bins this variance goes to 0 (eventually it depends on the number of data points). In theory I don't see why we can't compute the cut value purely based on the unbinned data. Investigate / do this![ ]Choose the final uncertainty for this variable that we want to use.
- (While generating fake data) Events with large energy, but few pixels
While developing some fake data using existing events in the photo peak & filtering out pixels to end up at ~3 keV, I noticed the prevalence of events with <150 pixels & ~6 keV energy.
Code produced by splicing in the following code into the body of
generateFakeData.for i in 0 ..< xs.len: if xs[i].len < 150 and energyInput[i] > 5.5: # recompute from data let pp = toSeq(0 ..< xs[i].len).mapIt((x: xs[i][it], y: ys[i][it], ch: ts[i][it])) let newEnergy = h5f.computeEnergy(pp, group, a, b, c, t, bL, mL) echo "Length ", xs[i].len , " w/ energy ", energyInput[i], " recomp ", newEnergy let df = toDf({"x" : pp.mapIt(it.x.int), "y" : pp.mapIt(it.y.int), "ch" : pp.mapIt(it.ch.int)}) ggplot(df, aes("x", "y", color = "ch")) + geom_point() + ggtitle("funny its real") + ggsave("/tmp/fake_event.pdf") sleep(200) if true: quit()
This gives about 100 events that fit the criteria out of a total of O(20000). A ratio of 1/200 seems probably reasonable for absorption of X-rays at 5.9 keV.
While plotting them I noticed that they all share that they are incredibly dense, like:
These events must be events where the X-ray to photoelectron conversion happens very close to the grid! This is one argument "in favor" of using ToT instead of ToA on the Timepix1 and more importantly a good reason to keep using the ToT values instead of pure pixel counting for at least some events!
[ ]We should look at number of pixels vs. energy as a scatter plot to see
what this gives us.
24.2. Implementation
The basic gist is that essentially every uncertainty listed above either has an effect on the S or B term (or maybe both).
That means, if we understand what the impact of each of these uncertainties is on either S or B, we can combine all possible uncertainties on S and B quadratically to get a combined uncertainty on these values.
We can assume that our used value is the most likely value, i.e. we can model the uncertainty as a gaussian fluctuation around our default parameter.
So, going back to the derivation of our used likelihood function from the product of the ratios of two Poissons, we should be able to derive a modified form of it, such that we (likely) get some penalty term in the final likelihood.
Our starting point for this would be not a regular Poisson, but a Poisson, where the mean value is actually a gaussian with λ = our parameter and a σ = our deduced uncertainty on S and B each.
24.2.1. Klaus discussion w/ Philip Bechtle
Klaus discussed this w/ Philip.
Summary as a figure:

what Klaus wrote to me:
Um die "marginalisierte Likelihood" zu bekommen, müssen wir nur für jeden Wert von gag über den/die Nuisance-Parameter integieren. also LM (gag) = int -infty ^+infty ( L(gag, theta) dtheta und dann das 95% CL bei LM ablesehen anstatt bei L Den Constraint durch einen Nuisance Parameter schreibt man in log L einfach als (theta-theta0)2/sigmatheta2, bzw. wenn man theta0 = 0 wählt noch einfacher. Die NP-Abhängigkeit von s und b muss man dann entsprechend hinschreiben, also z.B. für einen systematischen Fehler auf den Background: b(theta) = b(theta=0) * (1 + theta) sigmatheta wäre dann die von uns geschätzte/bestimmte relative Unsicherheit auf den Untergrund
Two things:
- what he writes, the
(Θ - Θ₀)²term should be negative, as it comes from multiplying a Gaussian to the initial "Q" (from which we derive our ln L, see eq. [BROKEN LINK: eq:likelihood_1_plus_s_over_b_form]) - the implementation of this hinges on whether the changed L can be integrated analytically or not.
Reg 1: First let's look at how we actually get to our likelihood function that includes nuisance parameters.
To derive the full likelihood function (not log likelihood) we just take the exponential of the likelihood. In order to include a nuisance parameter that is normally distributed around our hypothesis value (b or s), we extend the initial description of the term that we use to derive the likelihood function.
Remember that we derived our ln L from a ratio of probabilities, done in equation [BROKEN LINK: eq:likelihood_1_plus_s_over_b_form]. A gaussian nuisance parameter then is just another probability multiplied to the ratio, which is 1 for our hypothesized values and decreases exponentially from there:
\[ \mathcal{Q}' = \left(\prod_i \frac{P_{\text{pois}}(n_i; s_i + b_i)}{P_{\text{pois}}(n_i; b_i)}\right) \cdot \mathcal{N}(θ_s, σ_s) \cdot \mathcal{N}(θ_b, σ_b) \] where \(\mathcal{N}(θ, σ)\) refers to a normal distribution with standard deviation \(σ\), mean zero and \(θ\) the parameter.
In this form we add two nuisance parameters \(θ_s\) and \(θ_b\) for signal and background respectively.
It is relatively straightforward to see that by taking the log of the expression, we get back our initial ln L form, with two additional terms, a \(\left(\frac{θ}{σ}\right)²\) for each nuisance parameter (this would be a \(\left(\frac{x - θ}{σ}\right)²\) term had we included a mean unequal zero, harking back to the term written in the image by Klaus above).
Question: where does the arise from to modify the terms s_i and
b_i by s_i * (1 + θ_s) and b_i * (1 + θ_b) respectively? It
makes sense intuitively, but I don't quite understand it. Do these
actually appear if we exercise through the calculation? I don't think so.
Check that!
So using this, we can deduce the likelihood function we need to
integrate over. Starting from our ln L:
\[
\ln \mathcal{L} = -s + Σ_i \ln \left( 1 + \frac{s_i}{b_i} \right)
\]
we add the two nuisance terms & replace the s, s_i and b_i by
their respective versions:
\[
x' = x \cdot (1 + θ)
\]
to get:
\[
\ln \mathcal{L}' = -s' + Σ_i \ln \left( 1 + \frac{s_i'}{b_i'} \right) -
\left(\frac{θ_s}{σ_s}\right)² - \left(\frac{θ_b}{σ_b}\right)²
\]
Now just take the exponential of the expression
\begin{align*} \mathcal{L}' &= \exp\left[\ln \mathcal{L}'\right] \\ &= \exp\left[-s' + Σ_i \ln \left(1 + \frac{s_i'}{b_i'}\right) - \left(\frac{θ_s}{σ_s}\right)² - \left(\frac{θ_b}{σ_b}\right)²\right] \\ &= \exp[-s'] \cdot Π_i \left(1 + \frac{s_i'}{b_i'}\right) \cdot \exp\left[-\left(\frac{θ_s}{σ_s}\right)²\right] \cdot \exp\left[-\left(\frac{θ_b}{σ_b}\right)²\right] \\ \end{align*}What we thus use as our actual likelihood function to compute the limit is:
\[ \ln \mathcal{L}_M = \ln ∫_{-∞}^∞∫_{-∞}^∞ \mathcal{L(θ_s, θ_b)}' \, \mathrm{d}\,θ_s \mathrm{d}\,θ_b \] which then only depends on \(g_{ae}\) again.
The big problem with the expression in general is the singularity provided by \(b_i'\). It is visible, once we insert the definition of \(b_i'\):
\[ \ln \mathcal{L}_M = \ln ∫_{-∞}^∞ ∫_{-∞}^∞ \exp[-s'] \cdot Π_i \left(1 + \frac{s_i'}{b_i(1 + θ_b)}\right) \cdot \exp\left[-\left(\frac{θ_s}{σ_s}\right)²\right] \cdot \exp\left[-\left(\frac{θ_b}{σ_b}\right)²\right] \, \mathrm{d}\,θ_s \mathrm{d}\,θ_b \]
Reg 2: Using sagemath
θ = var('θ')
s = var('s')
s_i = var('s_i')
b_i = var('b_i')
σ = var('σ')
Give sage our assumptions:
assume(s > 0) assume(s_i > 0) assume(b_i > 0) assume(σ > 0)
First start with the signal only modification:
L_s(s, s_i, b_i, σ, θ) = exp(- s * (1 + θ)) * (1 + s_i * (1 + θ) / b_i) * exp(-(θ / σ)^2 )
And integrate it from -∞ to ∞:
from sage.symbolic.integration.integral import definite_integral definite_integral(L_s(s, s_i, b_i, σ, θ), θ, -oo, oo)
We can see that the term including a nuisance parameter modifying our
signal s can be integrated analytically with the above expression.
This is good news, as it implies we can simply use that result in
place of the \(∫_{-∞}^∞ \mathcal{L} dθ\) (assuming this \(θ\) refers to
the \(θ\) used for s.
One question is: do we get the same if we integrate the ln L version
and taking exp of the result?
lnL_s(s, s_i, b_i, σ, θ) = -s * (1 + θ) + ln(1 + s_i * (1 + θ) / b_i) - (θ / σ)^2 definite_integral(lnL_s(s, s_i, b_i, σ, θ), θ, -oo, oo)
Which diverges (also if we choose 0 as lower bound). So the answer is
clearly: ln and integration do not commute here!
Another question: How does the above change if we don't have 1, but
multiple s_i? Compare the different cases with 1, 2 and 3 s_i:
s_i2 = var('s_i2')
s_i3 = var('s_i3')
L_s2(s, s_i, s_i2, b_i, σ, θ) = exp(- s * (1 + θ)) * (1 + s_i * (1 + θ) / b_i) * (1 + s_i2 * (1 + θ) / b_i) * exp(-(θ / σ)^2 )
L_s3(s, s_i, s_i2, s_i3, b_i, σ, θ) = exp(- s * (1 + θ)) * (1 + s_i * (1 + θ) / b_i) * (1 + s_i2 * (1 + θ) / b_i) * (1 + s_i3 * (1 + θ) / b_i) * exp(-(θ / σ)^2 )
latex(definite_integral(L_s(s, s_i, b_i, σ, θ), θ, -oo, oo))
latex(definite_integral(L_s2(s, s_i, s_i2, b_i, σ, θ), θ, -oo, oo))
latex(definite_integral(L_s3(s, s_i, s_i2, s_i3, b_i, σ, θ), θ, -oo, oo))
\[ -\frac{\sqrt{\pi} {\left(s s_{i} σ^{2} - 2 \, b_{i} - 2 \, s_{i}\right)} σ e^{\left(\frac{1}{4} \, s^{2} σ^{2} - s\right)}}{2 \, b_{i}} \] \[ \frac{{\left(2 \, \sqrt{\pi} s_{i} s_{i_{2}} σ^{3} + \sqrt{\pi} {\left(s^{2} s_{i} s_{i_{2}} σ^{4} - 2 \, {\left(b_{i} s s_{i} + {\left(b_{i} s + 2 \, s s_{i}\right)} s_{i_{2}}\right)} σ^{2} + 4 \, b_{i}^{2} + 4 \, b_{i} s_{i} + 4 \, {\left(b_{i} + s_{i}\right)} s_{i_{2}}\right)} σ\right)} e^{\left(\frac{1}{4} \, s^{2} σ^{2} - s\right)}}{4 \, b_{i}^{2}} \] \[ -\frac{{\left(2 \, \sqrt{\pi} {\left(3 \, s s_{i} s_{i_{2}} s_{i_{3}} σ^{2} - 2 \, b_{i} s_{i} s_{i_{2}} - 2 \, {\left(b_{i} s_{i} + {\left(b_{i} + 3 \, s_{i}\right)} s_{i_{2}}\right)} s_{i_{3}}\right)} σ^{3} + \sqrt{\pi} {\left(s^{3} s_{i} s_{i_{2}} s_{i_{3}} σ^{6} - 2 \, {\left(b_{i} s^{2} s_{i} s_{i_{2}} + {\left(b_{i} s^{2} s_{i} + {\left(b_{i} s^{2} + 3 \, s^{2} s_{i}\right)} s_{i_{2}}\right)} s_{i_{3}}\right)} σ^{4} - 8 \, b_{i}^{3} - 8 \, b_{i}^{2} s_{i} + 4 \, {\left(b_{i}^{2} s s_{i} + {\left(b_{i}^{2} s + 2 \, b_{i} s s_{i}\right)} s_{i_{2}} + {\left(b_{i}^{2} s + 2 \, b_{i} s s_{i} + {\left(2 \, b_{i} s + 3 \, s s_{i}\right)} s_{i_{2}}\right)} s_{i_{3}}\right)} σ^{2} - 8 \, {\left(b_{i}^{2} + b_{i} s_{i}\right)} s_{i_{2}} - 8 \, {\left(b_{i}^{2} + b_{i} s_{i} + {\left(b_{i} + s_{i}\right)} s_{i_{2}}\right)} s_{i_{3}}\right)} σ\right)} e^{\left(\frac{1}{4} \, s^{2} σ^{2} - s\right)}}{8 \, b_{i}^{3}} \]
Crap. While the result seems to be "regular" in a sense, it doesn't seem like there's an easy way to generalize the expression to some product again. Regular things to note:
- the exponential term remains unchanged
- the denominator is \(2^n b_i^n\) where \(n\) is the number of elements in the product
- the remaining parts (products of s, si, …) seem to be some pascal triangle thing, but not quite.
res = -1/8*(2*sqrt(pi)*(3*s*s_i*s_i2*s_i3*σ^2 - 2*b_i*s_i*s_i2 - 2*(b_i*s_i + (b_i + 3*s_i)*s_i2)*s_i3)*σ^3 + sqrt(pi)*(s^3*s_i*s_i2*s_i3*σ^6 - 2*(b_i*s^2*s_i*s_i2 + (b_i*s^2*s_i + (b_i*s^2 + 3*s^2*s_i)*s_i2)*s_i3)*σ^4 - 8*b_i^3 - 8*b_i^2*s_i + 4*(b_i^2*s*s_i + (b_i^2*s + 2*b_i*s*s_i)*s_i2 + (b_i^2*s + 2*b_i*s*s_i + (2*b_i*s + 3*s*s_i)*s_i2)*s_i3)*σ^2 - 8*(b_i^2 + b_i*s_i)*s_i2 - 8*(b_i^2 + b_i*s_i + (b_i + s_i)*s_i2)*s_i3)*σ)*e^(1/4*s^2*σ^2 - s)/b_i^3 res.full_simplify()
Can we express the product as a symbolic product?
L_sP(s, s_i, b_i, σ, θ) = exp(- s * (1 + θ)) * product(1 + s_i * (1 + θ) / b_i, s_i) * exp(-(θ / σ)^2 ) definite_integral(L_sP(s, s_i, b_i, σ, θ), θ, -oo, oo)
Not really, as this cannot be integrated…
With all the above, it seems like analytical integration is out of the question for the problem in general. I suppose we'll just live with the fact that we need numerical integration.
Let's look at the background nuisance parameter.
First we define the function:
L_b(s, s_i, b_i, σ, θ) = exp(- s) * (1 + s_i / (b_i * (1 + θ))) * exp(-(θ / σ)^2 )
now compute the integral:
definite_integral(L_b(s, s_i, b_i, σ, θ), θ, -oo, oo)
which means integration is not possible due to the singularity (though: I don't understand the error message really!)
Let's look at the integral from 0 to ∞:
assume(θ > 0) definite_integral(L_b(s, s_i, b_i, σ, θ), θ, 0, oo)
which already gives us a not very helpful result, i.e. the integral rewritten slightly more compactly.
We can also ask it for the indefinite integral:
from sage.symbolic.integration.integral import indefinite_integral indefinite_integral(L_b(s, s_i, b_i, σ, θ), θ)
which is about as helpful as before…
assume(θ > -1) L2(s, s_i, b_i, σ, θ) = exp(- s) * (1 + s_i / (b_i * (1 + θ))) * exp(-(θ / σ)^2 ) definite_integral(L2(s, s_i, b_i, σ, θ), θ, -0.99, oo)
This implies we won't be able to work around using numerical integration. Let's look at the result using numerical integration:
from sage.calculus.integration import numerical_integral L_b(θ, s, s_i, b_i, σ) = exp(- s) * (1 + s_i / (b_i * (1 + θ))) * exp(-(θ / σ)^2 ) sv = 50.0 s_iv = 2.5 b_iv = 3.5 σv = 0.2 L_bθ(θ) = L_b(θ, sv, s_iv, b_iv, σv) #L_bInt(s, s_i, b_i, σ, θ) = lambda x: L_b(s, s_i, b_i, σ, x) #numerical_integral(L_b, -oo, oo, params = [70.0, 4.0, 4.0, 0.2]) numerical_integral(L_bθ, -oo, oo)
Let's see if we can reproduce this result (the numerical background) with numericalnim:
import numericalnim, math proc L_b(θ, s, s_i, b_i, σ: float, nc: NumContext[float, float]): float = exp(-s) * (1 + s_i / (b_i * (1 + θ))) * exp(-pow(θ / σ, 2) ) proc L_bθ(θ: float, nc: NumContext[float, float]): float = let sv = 50.0 let s_iv = 4.0 let b_iv = 3.5 let σv = 0.2 #if abs(θ + 1.0) < 1e-5: result = 0 #else: result = L_b(θ, sv, s_iv, b_iv, σv, nc = nc) echo adaptiveGauss(L_bθ, -1, Inf)
1.514138315562322e-22
Hmm, so with all this playing around here, we notice two things:
- the singularity is an actual problem, if we have it within our integration bounds & get unlucky and the algorithm evaluates at -1. Problem is the function is essentially a 1/x, which is highly non-polynomial near 0 (or in our case -1).
- the contribution to the integral of the area around -1 is tiny,
as expected, because we're already at the tail of the exponential
term. This should imply that from a physical standpoint the
integral around that area shouldn't matter, I would say. A theta of
-1 would anyhow mean that our value
b_0(our background hypothesis w/o uncertainty) is completely wrong, as we would end up at an effective background of 0 / and or multiple sigmas away from our hypothesis.
- TODO Understand values of θ and σ better
I'm confused about this: The θ term appears with
b_i.b_iis the background value for a specific candidate, i.e. at a specific location and energy. If we assign an uncertainty of σ to the background (around ourb_i,0values) the σ will be considered "absolute" i.e. 5%, 20% etc. of the full background hypothesis.Given that individual
b_ithemselves can already be essentially 0 in our computation, what does this imply for the likelihood of having a valid θ = -1 term? It shouldn't matter, right? - TODO add section explaining origin of
s_i'andb_i'terms
They come from substituting the normal distribution such that it is centered around θ = 0 with a relative σ, if one replaces the
s_iandb_iterms by those that enter the normal distribution initially. - TODO Understand integrated L for σ = 0
Intuitively we would think that if we take the result for the function integrated over the nuisance parameter and set σ to 0, we should again obtain the result of the regular L.
Consider
L_sfrom the sagemath session above:L_s(s, s_i, b_i, σ, θ) = exp(- s * (1 + θ)) * (1 + s_i * (1 + θ) / b_i) * exp(-(θ / σ)^2 )
which results in:
-1/2*sqrt(pi)*(s*s_i*σ^2 - 2*b_i - 2*s_i)*σ*e^(1/4*s^2*σ^2 - s)/b_i
after the definite integral.
This term for \(σ = 0\) is 0, no?
If we take the log of this expression of course the
ln σterms are separate terms. While we can just "drop" them, that's still wrong, as these terms end up asln 0, i.e. ∞. So dropping them from the sum isn't valid (they are not constant, which is the only time we can reasonably drop something under the pretense that we only care about the change in L for our limit).One thing to keep in mind (which may be unrelated) is that the
L_swe implemented only has a single term for thes_i,b_irelated parts. In practice this is still a sum / product of these terms. Does that change anything?However: If we simply take the \(\ln\) of the integrated out result, split off the terms that are constant or set those to 0 that have a \(σ\), we do indeed get back the result of our original \(\mathcal{L}\). This is pretty easy to see:
- absorb
-1/2into the parenthesis containing2 s_i + 2 b_i. - drop the
ln(√π)term - drop the
ln(σ)term - drop the
1/4 s² σ²term - division by
b_ibecomes-ln b_iterm
And we're back where we started.
I take this as good enough reason to simply use this expression for the \(θ_s\) solution. Then integrate that result for the \(θ_b\) nuisance parameter.
- absorb
- \(s'\) is equivalent to \(s_i'\) ?
\begin{align*} s &= Σ_i s_i \\ s_i' &= s_i (1 + θ_s) \\ s' &= Σ_i s_i' \\ &= Σ_i s_i (1 + θ_s) \\ &\text{as }(1 + θ_s)\text{ is constant} \\ &= (1 + θ_s) Σ_i s_i \\ &= (1 + θ_s) s \\ s' &= s (1 + θ_s) \\ \end{align*}so indeed, this is perfectly valid.
- ln L definitions for each uncertainty case
Before we look at explicit cases, we need to write down the full likelihood function with both nuisance parameters as reference and as a starting point:
\[ \mathcal{L}' = \exp[-s'] \cdot Π_i \left(1 + \frac{s_i'}{b_i'}\right) \cdot \exp\left[-\left(\frac{θ_s}{σ_s}\right)²\right] \cdot \exp\left[-\left(\frac{θ_b}{σ_b}\right)²\right] \]
Or as a
exp(log()):\[ \mathcal{L}' = \exp\left[ -s' + Σ_i \ln \left(1 + \frac{s_i'}{b_i'}\right) - \left(\frac{θ_s}{σ_s}\right)² - \left(\frac{θ_b}{σ_b}\right)² \right] \]
We should implement this function as a base (except for the no uncertainty case) and either input \(s_i\) or \(s_i'\) (and setting \(θ_s\) to 0 in case of \(s_i\)) depending on which thing we're integrating over.
Case 1: No uncertainties
\[ \ln \mathcal{L} = -s_{\text{tot}} + Σ_i \ln\left( 1 + \frac{s_i}{b_i}\right) \]
(if \(s_i\) or \(b_i\) is 0, the result is set to 1)
Case 2: Uncertainty on signal
Start from
sagemathintegration ofL_s(s, s_i, b_i, σ, θ) = exp(- s * (1 + θ)) * (1 + s_i * (1 + θ) / b_i) * exp(-(θ / σ)^2 )
i.e.:
-1/2*sqrt(pi)*(s*s_i*σ^2 - 2*b_i - 2*s_i)*σ*e^(1/4*s^2*σ^2 - s)/b_i
we simply take the
lnto arrive at: \[ \ln \mathcal{L}_{SM} = -s_{\text{tot}} + \frac{s_{\text{tot}}²σ_s²}{4} + \ln(σ_s\sqrt π)+ Σ_i \ln\left[ 1 + \frac{s_i}{b_i} \cdot (1 - 2 s_{\text{tot}} σ_s²) \right] \] (the subscript \(SM\) refers to "signal" and "marginalized" as the signal related nuisance parameter has been integrated out)Note that the product of the s/b ratio with the
2 s σ²doesn't make sense from a perspective of the units that we currently use to compute the individual \(s_i\) or \(b_i\) terms. We can "fix" that by assigning a unit to \(σ\)?Further: this is only valid for a single candidate! See the discussion in the parent section that this does not generalize (at least not easily) for N candidates and that therefore we need to utilize numerical integration also for the signal case.
Thus:
\begin{align*} \mathcal{L}_{SM} &= ∫_{-∞}^∞ \exp(-s'_{\text{tot}}) \cdot Π_i \left(1 + \frac{s_i'}{b_i}\right) \cdot \exp\left[-\frac{θ_s²}{σ_s²}\right] \, \mathrm{d}\,θ_s \\ &= ∫_{-∞}^∞ \exp(-s_{\text{tot}} (1 + θ_s)) \cdot Π_i \left(1 + \frac{s_i (1 + θ_s)}{b_i}\right) \cdot \exp\left[-\frac{θ_s²}{σ_s²}\right] \, \mathrm{d}\,θ_s \end{align*}For the \(\log\mathcal{L}\) we simply take the log of the result.
Case 3: Uncertainty on background
For the case of pure uncertainty on the background, we simply integrate over the modified 'Case 1' version, after taking the
\begin{align*} \ln \mathcal{L}_{BM} &= -s_{\text{tot}} + Σ_i \ln\left( 1 + \frac{s_i}{b_i(1 + θ_b)}\right) - \frac{θ_b²}{σ²} \\ \mathcal{L}_{BM} &= \exp\left[ -s_{\text{tot}} + Σ_i \ln\left( 1 + \frac{s_i}{b_i(1 + θ_b)}\right) - \frac{θ_b²}{σ²}\right] \\ \end{align*} \begin{align*} \mathcal{L}_{BM} &= ∫_{-∞}^∞ \exp(-s_{\text{tot}}) \cdot Π_i \left(1 + \frac{s_i}{b_i'}\right) \cdot \exp\left[-\frac{θ_b²}{σ_b²}\right] \, \mathrm{d}\,θ_b \\ &= ∫_{-∞}^∞ \exp(-s_{\text{tot}}) \cdot Π_i \left(1 + \frac{s_i}{b_i (1 + θ_b)}\right) \cdot \exp\left[-\frac{θ_b²}{σ_b²}\right] \, \mathrm{d}\,θ_b \end{align*}expof it:where the integration is performed similar to 1.
Case 4: Uncertainty on both
This is simply a combination of 3 and 4. Instead of integrating over 'Case 1', we integrate over the corresponding 'Case 2' version:
\[ \ln \mathcal{L}_{SBM} = ∫_{-∞}^∞ -s_{\text{tot}} + \frac{s_{\text{tot}}²σ_s²}{4} + \ln(σ_s\sqrt π)+ Σ_i \ln\left[ 1 + \frac{s_i}{b_i(1 + θ_b)} \cdot (1 - 2 s_{\text{tot}} σ_s²) \right] - \frac{θ_b²}{σ_b²} \, \mathrm{d}\,θ_b \]
\begin{align*} \mathcal{L}_{SBM} &= ∫_{-∞}^∞∫_{-∞}^∞ \exp(-s'_{\text{tot}}) \cdot Π_i \left(1 + \frac{s_i'}{b_i'}\right) \cdot \exp\left[-\frac{θ_b²}{σ_b²}\right] \cdot \exp\left[-\frac{θ_s²}{σ_s²}\right] \, \mathrm{d}\,θ_b \mathrm{d}\,θ_s \\ &= ∫_{-∞}^∞ ∫_{-∞}^∞ \exp(-s_{\text{tot}}(1 + θ_s)) \cdot Π_i \left(1 + \frac{s_i(1 + θ_s)}{b_i (1 + θ_b)}\right) \cdot \exp\left[-\frac{θ_b²}{σ_b²}\right] \cdot \exp\left[-\frac{θ_s²}{σ_s²}\right] \, \mathrm{d}\,θ_b \mathrm{d}\,θ_s \end{align*}- Note about integration style
Note that we use products of exponentials instead of an exponential of the sum of the arguments to avoid numerical issues with the arguments.
The issue when writing it as an exponential of an effective "log" like argument, is that we have a sum of the \(\ln(1 + \frac{s_i'}{b_i'}\) terms. If any \(s_i'\) or \(b_i'\) becomes negative we're in trouble as the logarithm isn't defined in that domain. Instead of restricting these values artificially, we get around problems by simply using the product of regular numbers. While this might have issues with the interpretation (effectively a "negative" probability) it is at least numerically stable.
- Note about integration style
- Debugging "infinite" loop in double integral
While implementing the numerical integrals described in the previous section, we encountered a case where the integral apparently did not converge. Or rather the MC step at index 439 simply never finished.
Digging into this by using
flattyand storing the candidates with their position and energies in a file to be able to quickly reproduce the specific problematic case, shed light on it. Instead of the integration never finishing, the problem is actually that our algorithm to determine the 95% value of thelogLnever finishes, because the likelihood values computed do not decrease, but rather keep on increasing. So something in the set of candidates causes the logL to be an ever increasing likelihood function. Maybe?As it turned out the issue was that for this set of candidates the integration range for the background case, starting from
-0.99simply was too close to the singularity.Let's checks this in detail.
The candidates that are problematic are the following, stored in ./../resources/problematic_candidates_syst_uncert_limit.csv
The candidates in form of a plot are:
Figure 363: Set of problematic candidates, which cause the integration to take effectively forever. With these candidates separately, the limit calculation was debugged. It turned out to not be the actual integral (which was slow sometimes), but rather the behavior of
Lfor increasing coupling constants. Instead of decreasing, the values forLkept on increasing forever?[X]check what happens if stepping size is increased. Does the increase stop at some point?
An excerpt of the integration results:
Limit step 0 at curL 0.07068583486571436 at g_ae²: 0.0 Limit step 1 at curL 23.10461624419532 at g_ae²: 5e-21 Limit step 2 at curL 57088.5769711143 at g_ae²: 9.999999999999999e-21 Limit step 3 at curL 1459616.8705168 at g_ae²: 1.5e-20 Limit step 4 at curL 5007822.77502083 at g_ae²: 2e-20 Limit step 5 at curL 5797529.015339485 at g_ae²: 2.5e-20 Limit step 7 at curL 1407002.639815327 at g_ae²: 3.499999999999999e-20 Limit step 8 at curL 435658.5012481307 at g_ae²: 3.999999999999999e-20 Limit step 9 at curL 114540.0461727056 at g_ae²: 4.499999999999999e-20 Limit step 10 at curL 27313.76069930456 at g_ae²: 4.999999999999999e-20 Limit step 11 at curL 6188.470608073056 at g_ae²: 5.499999999999998e-20 Limit step 12 at curL 1377.326797388224 at g_ae²: 5.999999999999998e-20 Limit at = 3.499999999999
So indeed there is a maximum and at some point the log values start to decrease again!
But obviously the resulting values are everything but sane!
The reason this behavior appears (looking at the points being evaluated in the integration) is that the algorithm tries its best to estimate what the function looks like near
-0.99.The singularity simply becomes extremely dominant for this set of candidates.
Arguably the reason must be that the signal in this set of candidates is large such that values approaching
-0.99are already yielding very large values. The size of the nominator of course directly correlates with how close we need to approach the singularity for terms to grow large.We can further investigate what the likelihood functions actually look like in \(θ\) space, i.e. by plotting the likelihood value against \(θ\).
This is done using the
plotLikelihoodCurvesfunction in the code. We'll use the same candidates as a reference, as we know these cause significant issues.Figure 364: Likelihood function for signal uncertainty. Integrated numerically at each point in \(θ\) space. Figure 365: Likelihood function for background uncertainty. Integrated numerically at each point in \(θ\) space. Figure 366: Likelihood function for signal & background uncertainty. Integrated numerically for the background and then scanned for the signal as the "outer" integral. Figure 367: Likelihood function for signal & background uncertainty. Integrated numerically for the signal and then scanned for the background as the "outer" integral. In figures 365 and to a lesser extent in 367 it is visible that the part of the function towards
-1clearly dominates the space, but obviously we are interested in the "gaussian" part near 0.Interestingly, in all cases the maximum of the likelihood function is actually at negative values.
Given that the terms for the signal and background appear in the nominator and denominator respectively, it does seem to explain why one of the two yields an improvement in the limit and the other worsening of the limit.
- Investigate why signal uncertainty causes improvement in limits
[1/1]
In addition why the effect of a 15% systematic on the background has such a strong effect.
To investigate the former, let's see what happens if we set the \(σ_s\) to 0.
[X]does using theLtemplate with theσset to 0 and noθterms reproduce the same result as our regular limit calculation? -> yes it does. Implemented the "certain" logL function using the template with 0 arguments for σ and θ and it indeed reproduces the same result as the regular code.
- Replace nuisance parameter distribution from normal to log-normal
UPDATE: We have dropped looking into this further for now and instead decided to just cut off the gaussian at a specific value.
The behavior of using a normal distribution for the nuisance parameters has multiple problems.
As mentioned above, we need to get the 'marginalized likelihood' function by integrating out the nuisance parameter:
\[ \mathcal{L}_M = ∫_{-∞}^∞ \mathcal{L}(θ)\, \mathrm{d}θ \]
However, due to the structure of our likelihood function, essentially a
s/b, the modified \(b\) (\(b' = b(1+θ)\)) becomes 0 at a specific value (depending on the normalization, in this case \(θ = -1\)).One way to work around this is to choose a distribution that is not non-zero on whole ℝ, but rather one that is not defined where \(b' = 0\).
One option for that is a log-normal distribution.
Let's plot the log-normal for multiple parameters:
import ggplotnim, math proc logNormal(x, μ, σ: float): float = result = 1.0 / (x * σ * sqrt(2 * PI)) * exp(- pow(ln(x) - μ, 2) / (2 * σ*σ) ) var df = newDataFrame() for μ in @[0.0, 0.5, 1.0]: #, 3.0]: for σ in @[0.1, 0.25, 0.5, 1.0]: let x = linspace(1e-6, 5.0, 1000) # 30.0, 1000) df.add toDf({"x" : x, "y": x.map_inline(logNormal(x, μ, σ)), "σ" : σ, "μ" : μ}) ggplot(df, aes("x", "y", color = factor("σ"))) + facet_wrap("μ") + #, scales = "free") + #facetMargin(0.35) + geom_line() + ggsave("/tmp/log_normal.pdf") proc logNormalRenorm(b, θ, σ: float): float = let σ_p = σ / b let b_p = exp(b * (1 + θ)) result = 1.0 / (b_p * σ_p * b * sqrt(2.0 * PI)) * exp(-pow(θ / (sqrt(2.0) * σ_p), 2.0)) var dfR = newDataFrame() for b in @[0.1, 0.5, 1.0, 10.0]: for σ in @[0.1, 0.25, 0.5, 1.0]: let θ = linspace(-10.0, 10.0, 1000) dfR.add toDf({"x" : θ, "y": θ.map_inline(logNormalRenorm(b, x, σ)), "σ" : σ, "b" : b}) ggplot(dfR, aes("x", "y", color = factor("σ"))) + facet_wrap("b") + geom_line() + ggsave("/tmp/log_normal_renorm.pdf")
- Likelihood in \(θ_b\) space
As discussed in other parts (search for the
sagecode about integrating the likelihood function) the likelihood function including a nuisance parameter \(θ_b\) results in a singularity at \(θ_b = -1\) as it yields a \(b_i' = 0\) at that point (which is in the denominator).From a purely statistical point of view this represents somewhat of a problem, as the marginalized likelihood is defined by the integral over all \(θ_b\). Of course from an experimental standpoint we know that the singularity is unphysical. In addition to that the normal distribution that is added anyway limits the parts that contribute meaningfully to the integration. As long as the relative uncertainty is small enough, the likelihood will be small compared to the maximum before one approaches the singularity. That makes it easier to cut it off from an experimental standpoint. However, if the σ is too large, the "real" likelihood will still be non negligible when approaching the singularity, making it much more difficult to 1. integrate the parts that are important and 2. argue why a specific cutoff was chosen.
To get an idea:
shows the space of \(θ_b\) around 0 for three different σ for a fixed set of candidates.
We can see that for 0.15 and 0.2 the drop off is large enough that a cutoff at e.g. \(θ_b = -0.8\) is perfectly fine. However, at σ = 0.3 this isn't the case anymore. What should be done there? Or are we certain enough that our background model is more accurate than 30%?
- Behavior of expected limit for different uncertainties
With the uncertainties for signal and background implemented in the limit calculation code, it is time to look at whether the behavior for different uncertainties is actually as we would expect them.
That is, an increase in a systematic uncertainty should make the expected limit worse. To see whether that is the case, we first have to define what we mean by expected limit. Given a set of toy experiments, where \(l_i\) is the obtained limit for a single set of candidates. Then the expected limit is defined by
\[ l_{\text{expected}} = \text{median}( \{ l_i \} ) \]
i.e. simply the median of the set of all 'observed' toy limits.
By computing expected limits for different setups of \(σ_s\) and \(σ_b\) we can plot the behavior.
- Starting with the uncertainty on the background (where we integrate from \(θ_b = -0.8\) to avoid the singularity at \(θ_b = -1\)). The change in expected limits is shown in fig. 368. As can be seen, the expected limit does indeed increase the larger the uncertainty becomes.
- Second is the uncertainty on the signal. This is shown in fig. 370 where we can fortunately see the same behavior (even if the \(R_T = 0\) line might move a tiny bit to smaller values in presence of an uncertainty on the signal), i.e. an increase to larger values.
- Finally, the uncertainty on both the signal and background. By computing a grid of different combinations and drawing them with a color scale for the limit, we can see in fig. [BROKEN LINK: expected_limits_σ_s_and_σ_b] that also here there is a trend to larger values to further right / up one goes.
Figure 368: Behavior of the expected limits (median of toys) for the variation of the expected uncertainty on the background \(σ_b\) (integrated from \(θ_b = -0.8\)) given 1000 toy experiments each. As one might expect, the expected limit gets worse for increasing uncertainty. Figure 369: Behavior of the expected limits (median of toys) for the variation of the expected uncertainty on the signal \(σ_s\) given 1000 toy experiments each. As one might expect, the expected limit gets worse for increasing uncertainty. Figure 370: Grid of the behavior of the expected limits (median of toys) for the variation of the expected uncertainty on the signal \(σ_s\) and background together \(σ_b\) given 1000 toy experiments each. Visible in the color scale is that the expected limit gets worse the larger either of the two uncertainties gets & largest if both are large. The effect of the signal uncertainty seems to be larger. - Behavior of θx and θy nuisance parameters
The nuisance parameters for \(θ_x\) and \(θ_y\) account for the systematic uncertainty on the position of the axion signal.
Starting from the likelihood \(\mathcal{L}\)
\[ \mathcal{L} = \exp[-s] \cdot Π_i \left(1 + \frac{s_i}{b_i}\right) \]
Consider that \(s_i\) actually should be written as:
\[ s_i = f(E_i) \cdot P_{a \rightarrow γ} \cdot ε(E_i) \cdot r(x_i, y_i) \] where \(f\) returns the expected axion flux, \(ε\) encodes the detection efficiency of the detector and \(r\) encodes the X-ray optics obtained using the raytracing simulation (which is normally centered around \((x_i, y_i) = (7, 7)\), the center of the chip). With this we can introduce the nuisance parameters by replacing \(r\) by an \(r'\) such that \[ r' ↦ r(x_i - x'_i, y_i - y'_i) \] which effectively moves the center position by \((x'_i, y'_i)\).
In addition we need to add penalty terms for each of these introduced parameters:
\[ \mathcal{L}' = \exp[-s] \cdot Π_i \left(1 + \frac{s'_i}{b_i}\right) \cdot \exp\left[-\left(\frac{x_i - x'_i}{\sqrt{2}σ} \right)² \right] \cdot \exp\left[-\left(\frac{x_i - x'_i}{\sqrt{2}σ} \right)² \right] \] where \(s'_i\) is now the modification from above using \(r'\) instead of \(r\).
By performing the same substitution as we do for \(θ_b\) and \(θ_s\) we can arrive at: \[ \mathcal{L}' = \exp[-s] \cdot Π_i \left(1 + \frac{s'_i}{b_i}\right) \cdot \exp\left[-\left(\frac{θ_x}{\sqrt{2}σ_x} \right)² \right] \cdot \exp\left[-\left(\frac{θ_y}{\sqrt{2}σ_y} \right)² \right] \]
The substitution for \(r'\) means the following for the parameters: \[ r' = r\left(x (1 + θ_x), y (1 + θ_y)\right) \] where essentially a deviation of \(|θ| = 1\) means we move the spot to the edge of the chip.
Implementing this in code is more computationally expensive than the nuisance parameters for \(θ_s\) and \(θ_b\), because we need to compute the raytracing interpolation evaluation for each iteration. Therefore the (current) implementation computes everything possible once (conversion probability and detection efficiency of the detector) and only lookup the raytracing interpolation for each \((θ_x, θ_y)\) pair. As such we have:
let s_tot = expRate(ctx) var cands = newSeq[(float, float)](candidates.len) let SQRT2 = sqrt(2.0) for i, c in candidates: let sig = ctx.detectionEff(c.energy) * ctx.axionFlux(c.energy) * conversionProbability() cands[i] = (sig.float, ctx.background(c.energy, c.pos).float) let σ_p = ctx.σ_p proc likeX(θ_x: float, nc: NumContext[float, float]): float = ctx.θ_x = θ_x proc likeY(θ_y: float, nc: NumContext[float, float]): float = ctx.θ_y = θ_y result = exp(-s_tot) result *= exp(-pow(θ_x / (SQRT2 * σ_p), 2)) * exp(-pow(θ_y / (SQRT2 * σ_p), 2)) for i in 0 ..< cands.len: let (s_init, b_c) = cands[i] if b_c.float != 0.0: let s_c = (s_init * ctx.raytracing(candidates[i].pos)).float result *= (1 + s_c / b_c) result = simpson(likeY, -1.0, 1.0) let res = simpson(likeX, -1.0, 1.0) result = ln( res )
where we removed everything that is not relevant.
This approaches a ~reasonable runtime, yet is still quite slow (we're using
simpsonhere as it's quite a bit faster than adaptive gauss).However, while the scan of the θ space looks somewhat reasonable, we have an issue with the scan of \(\mathcal{L}\) for increasing \(g_{ae}²\), because the likelihood seems to increase exponentially.
In any case, let's look at what the phase space of \(θ\) look like. First separately and then as a heatmap.
Similar to the scan of \(\mathcal{L}\) for combined \(θ_s\) and \(θ_b\) we look at one by integrating out the other first.
Thus, fig. 372 shows the scan of \(θ_x\) and fig. [BROKEN LINK: fig:likelihood_theta_y] the equivalent for \(θ_y\).
It is clearly visible that both are somewhat symmetric around \(θ = 0\), but clear peaks around other points are visible.
Figure 371: Likelihood scan of \(θ_x\) after integrating out \(θ_y\) for a \(σ = 0.1\). While somewhat symmetric around \(θ_x = 0\) as expected due to the penalty term, there is a clear bias to a value around \(θ_x = -0.15\). Further another set of peaks is visible further away. Figure 372: Likelihood scan of \(θ_y\) after integrating out \(θ_x\) for a \(σ = 0.1\). Also mostly symmetric around \(θ_y = 0\) as expected due to the penalty term, there is also a (smaller) bias to a value around \(θ_y = 0.15\). Looking at the full phase space around both at the same time, we see that we have a clear tendency to a point away from the center, as shown in fig. 373.
Figure 373: Scan of the \((θ_x, θ_y)\) phase space, showing a clear maximum at the positions that roughly peak for the individual scans. Note that \(|θ| = 1\) corresponds to the signal center being at the edge of the chip, thus it more or less directly directly maps to the chip position. This is for the candidates shown in fig. 374.
Figure 374: Candidates and their energies for the plots shown in the likelihood scans above. It begs the question whether changing the parameters has a strong effect on the position of the peak in the x/y scan or not. We'll redraw and check again.
Further investigation using different candidate sets & different \(σ\) yielded the following:
- different sets of candidates have a very significant impact on the theta x/y space!
- changing \(σ\) has a drastic impact on the distance of the largest possible deviation from the center of the chip & the absolute value found. From the 1e-3 range it easily exceeds 6 (!) in absolute numbers for one set of candidates going from σ = 0.01 to 0.1. At the same time the position moves further towards one corner where there are denser populations of low energy clusters.
If this was all, that would be fine maybe. But beyond that, we still suffer from problems computing the actual limit, because the likelihood increases with increasing coupling constant instead of decreasing. Further investigation into that however seems to indicate that this increase is due to too large steps in \(g_{ae}²\). By using smaller steps there is a decrease at first before it starts rising exponentially. Unfortunately, the range in which it decreases before rising is rather small (16e-3 to 14e-3 in one case). NOTE: see about increase in L in next section
What to do?
Plots:
alternative candidates:
smaller sigma
even smaller sigma
- DONE Understand increase in \(\mathcal{L}\) for increasing \(g_{ae}\)
In the previous section 24.2.1.11 we noticed that (something we had seen previously at some point) the likelihood seemed to increase for increasing \(g_{ae}\) at some point again. There was a small dip before rising again. This is the same reason we initially added the "break from loop if increase again" logic to the while limit loop.
The behavior is shown in fig. 375.
We see that first it goes down as expected, but suddenly rises significantly again. This is due to our choice of candidates here. This is for the old case of "no candidates in signal sensitive region", which in the past meant we push candidates to \((x, y) = (14, 14)\), i.e. the corner. However, now we marginalize over the signal position, this causes these problems. Solution: simply draw no candidates for this case (or place them outside the chip, which requires a) removing the
clampcall in theraytracingproc { done } and b) avoid theChipCoordcheck { done by compiling with-d:danger, but not a good solution as unsafe code!!. Used as cross check though and does indeed give the same results } ).Figure 375: Behavior of different parts of likelihood terms (integrated separately over \(θ_x\) and \(θ_y\) so cannot be multiplied!). The main one of interest is the pure L term that is simply the marginalized L over \(θ_x\) and \(θ_y\) for \(σ = 0.05\). We see that first it goes down as expected, but suddenly rises significantly again. This is due to our choice of candidates here. This is for the old case of "no candidates in signal sensitive region", which in the past meant we push candidates to \((x, y) = (14, 14)\), i.e. the corner. However, now we marginalize over the signal position, this causes these problems. Solution: simply draw no candidates for this case. - DONE Understand limits being smaller w/ uncertainty than without
Further we saw that the limits seemed to be much smaller if uncertainties were used than if they are not used. This turned out to be a side-effect of the logic we used in the
bayesLimitprocedure.We still used a hardcoded cutoff for L to determine when we scanned "enough" of L to be able to accurately determine the 95% cutoff. Due to the added penalty terms from the nuisance parameters though, the absolute values of L were smaller than for the original no nuisance parameter case. This meant that our hardcoded cutoff was too large. L often only starts at ~0.015 and previously we had 5e-3 = 0.005 as a cutoff. This meant that we scanned only a small portion of the range and thus our limit was artificially lowered.
We currently fixed it by setting it to L@g = 0 / 250, but that is not great either. We need an adaptive solution (something like a binary search in log space? A newton method? LM fit to a set of points? Something…
- Improvements to limit algorithm
While dealing with the slowness of the \(θ_x\) and \(θ_y\) algorithm, we made some smaller and some larger changes to the way we compute the limit.
First of all we changed the integration routine used for the marginalized likelihood, integrating out the \(θ\) parameters. Instead of using an adaptive Gauss-Kronrod quadrature, we use
rombergwith a slightly lower depth 6 instead of the default 8. This massively improves runtime and makes it possible in the first place.However, even with that approach the method is rather slow, if we want a fine binning for the limits (i.e. making small steps in the coupling constant). That's why we changed the way we compute the 95% point in the first place.
The old algorithm in pseudo code was just:
let L_start = eval_L_at(g_ae² = 0) let g_ae²_step = 5e-23 Ls = [] while L < L_start / 500.0: g_ae² += g_ae²_step L = eval_L_at(g_ae²) Ls.add L limit = Ls.toCdf().lowerBound(0.95)
i.e. a linear scan from \(g_{ae}² = 0\) to a value that is sufficiently small (e.g. starting L divided by reasonably large number) and then treat that as all contributing terms to the integral. From there compute the cumulative distribution function and extract the point at which the value is 0.95.
The step distance had to be made rather large to make the code run fast enough, which is not ideal. Therefore, an approach that changes the spacing automatically depending on certain factors was implemented. The main idea is that we start from a rough scan of 10 points from \(g_{ae}² = 0\) to a large coupling constant and then add more points in between wherever the piecewise function describing the CDF based on the individual lines between data points is not smooth enough.
An example plot of the one created now is 376. We can see the resulting grid as points along the line.
Figure 376: Example of a MC toy in which we use the adaptive grid for the limit calculation which mainly depends on the difference in y (CDF) & the slope between lines between points until the difference between a point and the 95% point is smaller than some ε. - Effect of increasing tracking time to 2x
The plots in ./../Figs/statusAndProgress/limitCalculation/artificialTrackingTime/, namely
show the impact of doubling the tracking time from 180h to 360h, but keeping everything else the same (same background rate etc).
It improves the expected limit from
g_ae² = <5e-21tog_ae² = 2.8e-21, which converts to:- \(g_{ae} g_{aγ} = \SI{7.07}{\per\Giga\electronvolt}\)
- \(g_{ae} g_{aγ} = \SI{5.29}{\per\Giga\electronvolt}\)
so a significant improvement, but a limit. Real improvement will be less than that.
- TODO Investigate software efficiency using calibration runs
Take all calibration runs, filter out some very background like events using (rmsTrans & eccentricity), then apply likelihood method and see what it yields as a 'software' efficiency, i.e. fraction of events left compared to input.
- Combined likelihood with all nuisance parameters
\[ \mathcal{L}_{SBM} = ∫_{-∞}^∞∫_{-∞}^∞∫_{-∞}^∞∫_{-∞}^∞ \exp(-s'_{\text{tot}}) \cdot \prod_i \left(1 + \frac{s_i''}{b_i'}\right) \cdot \exp\left[-\frac{θ_b²}{2 σ_b²} - \frac{θ_s²}{2 σ_s²} - \frac{θ_x²}{2 σ_x²} - \frac{θ_y²}{2 σ_y²} \right] \, \mathrm{d}\,θ_b \mathrm{d}\,θ_s \mathrm{d}\,θ_x \mathrm{d}\,θ_y \]
24.2.2. TODO Things to do for final limit calculation [/]
For the final code that is going to be used for the real expected & real limits, we need to change a few things in the code up there.
[ ]extract the code and make it standalone, part of TPA[ ]make sure we have the latest raytracing image that matches what we expect, including rotation etc.[ ]make sure to have the correct amount of solar tracking time & background times. In particular also the background hypothesis should not include the tracking data!
25. STARTED Check all sampling via CDF code whether it uses normalized input!!
I just realized there was a bug in the sampling code of the muon flux. I was sampling from a wrong CDF. One that wasn't normalized to one!
I know I do sampling inn the ray tracing code as well. Check that I do it correctly there too!
UPDATE: A short look into raytracer2018.nim shows that there
indeed I correctly normalize by the last element.
26. General things done / Notes
26.1. Spark filtering
Event with a typical spark: Run 89, event 20933
26.2. Hough transformation for cluster finding
I started a Hough trafo playground in ./../../CastData/ExternCode/TimepixAnalysis/Tools/houghTrafoPlayground/houghTrafoPlayground.nim.
Reading up on Hough transformations is a bit confusing, but what we are doing for the moment:
- compute connecting lines between each point pair in a septem event (so for N hits, that's N² lines)
- for each line compute the slope and intersect
From this information we can look at different things:
- the plots of all lines. Very messy, but gives an idea if the lines are correct.
- a histogram of all found slopes
- a histogram of all found intersects
- a scatter plot of slopes vs. intersects
The "algorithm" to compute the Hough transformation is pretty dumb at the moment:
var xs = newSeqOfCap[int](x.len * x.len) var ys = newSeqOfCap[int](x.len * x.len) var ids = newSeqOfCap[string](x.len * x.len) var slopes = newSeqOfCap[float](x.len * x.len) var intersects = newSeqOfCap[float](x.len * x.len) echo x for i in 0 ..< x.len: for j in 0 ..< x.len: if i != j: # don't look at same point xs.add x[j] ys.add y[j] xs.add x[i] ys.add y[i] ids.add $i & "/" & $j ids.add $i & "/" & $j if xs[^1] - xs[^2] > 0: # if same x, slope is inf let slope = (ys[^1] - ys[^2]).float / (xs[^1] - xs[^2]).float slopes.add slope # make sure both points yield same intercept doAssert abs( (y[j].float - slope * x[j].float) - (y[i].float - slope * x[i].float) ) < 1e-4 intersects.add (y[j].float - slope * x[j].float)
Let's look at a couple of examples:
26.2.1. Example 0 septemEvent_run_272_event_95288.csv

26.2.2. Example 1 septemEvent_run_265_event_1662.csv


26.2.3. Example 2 septemEvent_run_261_event_809.csv


26.2.4. Example 3 septemEvent_run_291_event_31480.csv


26.2.5. Example 4 septemEvent_run_306_event_4340.csv


26.2.6. Conclusion
The hough transformation produces too much data that is too hard to interpret in the context of our goal. It doesn't actually help us a lot here, so we'll drop the pursuit of that.
26.3. TODO Reconstruct the CDL data
Given that we changed the gas gain computation to use slices and filter on the cluster size, the energy and gas gain values used in the CDL data are also outdated and need to be recomputed.
26.4. TODO insert following images
- all: ~/org/Figs/statusAndProgress/binnedvstime/energyFromCharge*.pdf
26.5.
Implemented: https://github.com/Vindaar/TimepixAnalysis/issues/44.
Every reconstruction file now creates its own unique directory, in which all plots are placed. Makes it much nicer to work with reconstruction.
Also added an overview facet plot for all datasets going into the Fe spectra cuts.
26.6. Meeting Klaus about TPA analysis
- flow chart of the whole data analysis, calibration, CDL, likelihood pipeline
- should include all ingredients. That is things like (in name, not value
of course, so that one sees where each aspect comes into play. At least not
within the flow chart):
- parameters, e.g. cluster size & cutoff
- data files, e.g. raw data, Timepix calibration files…
- algorithms, e.g. nlopt optimization for eccentricity, linear fit to gas gain vs. fit calibration…
- …
- should include all ingredients. That is things like (in name, not value
of course, so that one sees where each aspect comes into play. At least not
within the flow chart):
- total charge for background binned over N minute intervals
- relatively easy to do. Just read all data. Can't do it conveniently using
dataframes though I think. In any case, just walk timestamps, until
ΔT = x min, calc average of those events.- do same, but applying a filter, e.g.
totalCharge > 1e5or whatever - do same, but don't do any averaging, just sum up
- do same, but applying a filter, e.g.
- see sec. 14.6
- relatively easy to do. Just read all data. Can't do it conveniently using
dataframes though I think. In any case, just walk timestamps, until
- compare calibration Fe data to Mn target CDL data
- plot histograms for each Fe spectrum
- calculate some "agreement" value between each two histograms and plot result as some kind of scatter plot or so
- morph CDL spectra between two energies.
- allow interpolation between the shape of two neighboring reference datasets. Should hopefully have the effect that the steps visble in the background rate disappear
- talk to Phips
- read up on morphing of distributions in other contexts
- send Klaus reference datasets
26.7. Meeting Klaus about TPA analysis
Notes taken during meeting below, expanded afterwards.
- make mean charge plots split by time intervals
- This refers to the plots I sent to Klaus on in
the mail with title "Update zu Limits", namely the file
- In it we can clearly see that the background data shown has 5 "periods" of data taking.
- split these plots into 5 individual plots (at least for the mean charge versions).
- add the mean charge values for the calibration runs as well.
- for this possibly perform additional normalization not only by number of clusters but by number of pixel in each cluster to normalize out effect of different energy spectra
- This refers to the plots I sent to Klaus on in
the mail with title "Update zu Limits", namely the file
- all calbration plots into one plot
- just huge grid of all Fe55 spectra. Both charge and pixel. Use facetwrap and be happy. Possibly split into 2, one for Run 2 and one for Run 3
- add CDL Mn Feb 2019 spectra as well as comparison
- ridgeline of CDL data:
- This refers to the plot
command used to generate the final PDF:
: 1601573472:0;pdfunite eccentricity_ridgeline_XrayReferenceDataSet.h5_2014.pdf fracRmsTrans_ridgeline_XrayReferenceDataSet.h5_2014.pdf \ lengthDivRmsTrans_ridgeline_XrayReferenceDataSet.h5_2014.pdf eccentricity_ridgeline_XrayReferenceFile2018.h5_2018.pdf \ fracRmsTrans_ridgeline_XrayReferenceFile2018.h5_2018.pdf lengthDivRmsTrans_ridgeline_XrayReferenceFile2018.h5_2018.pdf \ /tmp/CDL_reference_distributions_2014_2018.pdf
The fact that the 2014 PDFs contain the string:
XrayReferenceDataSet.h5_2014means that the reference files used are indeed the 2014 Marlin files. That can be seen, because theggsavecall inlikelikood.nimcreates the following filename:ggsave(&"out/lengthDivRmsTrans_{refFile.extractFilename}_{yearKind}.pdf",
namely contains the full filename of the reference file plus an underscore and the year. The CDL 2014 Marlin file is: ./../../../../mnt/1TB/CAST/CDL-reference/XrayReferenceDataSet.h5 and the CDL 2014 TPA file: ./../../../../mnt/1TB/CAST/2014_15/CDL_Runs_raw/XrayReferenceFile2014.h5 which means it was the 2014 Marlin file.
To be absolutely sure compare the C Kalpha files for the fraction of pixels within transverse RMS:
They look considerably different, but the TPA one is definitely smoother. The reason for the difference: check again the studies of the CDL differences in this file!
- add background to ridges
- Means we want a comparison of a background distribution for each energy bin in all properties. Essentially just read background data as well and create histogram from it binned by the corresponding energy ranges and normalized to the same height. Question is both whether the distributions look sufficiently different for a full background dataset and also whether there are fluctuations in time of the properties. Even if we have fluctuations e.g. in the gas gain it's more important whether there are fluctuations of the properties that actually go into the likelihood calculation in time. If that's the case there are systematic effects we might have to correct for.
- bin background by 10min/60min, check mean value of these over time
- Related to the time effects mentioned in the previous note. One could calculate the distributions of the logL properties for background binned by time in 10/60 min. Then for each energy bin calculate a few statistical moments (mean, RMS etc) of each distribution and plot these values against time. That should give us a good understanding of how stable the background properties are agains time. Another thing in this direction one might look at is what the same looks like for calibration data. Since that is only 2 different kinds of events (photo + escape peak) we should clearly see how photons are affected in these properties when the things we know change do change (gain).
- This refers to the plot
26.8. Meeting Klaus about TPA analysis
Discussed:
Search for events with fracRmsTrans events with ~ 0 and plot them as events.
Whats min number pixel we allow for CDL? Check here in doc.
result.lengthDivRmsTrans = result.length / result.rmsTransverse result.fractionInTransverseRms = ( filterIt(zip(xRot, yRot), distance(it[0], it[1]) <= result.rmsTransverse).len ).float / float(npix)
frac in RMS trans is sphere around rotated coordinates!!
TODO:
Cut away fracRmsTrans 0 events? Plot ecc for all fracRmsTrans events = 0?
26.8.1. DONE Energy per cluster binned plot against time (see added code, but
sefgaults) So the same as the existing plot, just the energy.
26.8.2. DONE Fe55 spectra for charge and not only pixels!
26.8.3. TODO scatter plot fe 55 peak pos against mean charge val of new plot
so take the charge position of the photopeak for each Fe55 run. Then take the corresponding values from the mean charge binned by 100min or similar (read kinda gas gain) and create a scatter plot of the two!
26.9. Meeting Klaus about TPA analysis
- done all of the above TODOs
- we looked at mainly 3 (to an extent 4) different plots
26.9.1. Energy vs time
The median energy of clusters in time bins of 100 minutes. Shows a variation in certain areas of the plot. Beginning of data taking and at later times as well. A perfect energy calibration should result in perfectly flat lines for both the background as well as the calibration data. The background of cosmics can be assumed to be a flat spectrum with a well defined mean or median given a large enough binning. Possibly 2 major reasons:
- TODO Bin background by time for more gas gain values
- each run is long enough to suffer from the time variation in the
charge as seen in
from the last meeting. This means that the gas gain varies too much to assign a single value for the gas gain for a whole run, resulting in a definite problem for the variation. Possible solution: change the way the gas gain is calculated in the reconstruction. Instead of calculating the polya for each run, bin it also by time (have to look at different binning times to find the shortest possible time which still gives us good enough statistics!) and then calibrate each of these intervals individually based on the energy calibtration function.
- each run is long enough to suffer from the time variation in the
charge as seen in
- TODO Change energy calib to use closest two calibration runs
- Change the energy calibtration to not use all runs and perform the "gas gain vs energy calibration slope fit". Instead only look at the weighted mean of the energy calibrations of the two closest calibtration runs, i.e. linear interpolation. Then the gas gain won't be needed at all anymore.
26.9.2. Eccentricity binned vs time
file:///home/basti/org/Figs/statusAndProgress/binned_vs_time/background_mean_eccentricity_binned_100.0_min_filtered.pdf Here the variation is still visible. This is important, because the energy calibtration does not enter the calculation in any way! Need to understand this behavior. Why does it fluctuate? How does it fluctuate in time? This should be as flat as possible. Variations in gas gain seem to play a role. Why? Either means noisy pixels are active sometimes that distort the geometry or we have more multi hits which affect the calculations. NOTE: maybe this could be visible if we did take into account the charge that each pixel sees. Currently we just treat each pixel with the same weight. In principle each computation could be weighted by its charge value. Problematic of course, due to influence of gaussian statistics of gas gain!
26.9.3. Eccentricity ridgeline: background vs. CDL
In comparison to the above. The question is: how does this behavior
change in time? The above plot shows the median value of this
variable against time, but doesn't tell us anything about the
behavior in different energy bins of the CDL intervals.
The problem combining the two by for instance classifying all
clusters based on their energy into the corresponding CDL bin will
fold the energy calibtration into the equation here.
- TODO Calculate distributions match statistically w/ time against full background dist.
Thus: it is important to first get the energy calibration right to give us a mostly flat plot in the above. Once that is done we can be confident enough about the classification to look at it. Then we can calculate these distributions for background also binned according to time. Using a test to compare each time sliced distribution to the "true" full background spectrum (possibly separate for the separate Run 2 and 3?):
- compare using \(\chi^2\) test, but problematic because depends strongly on individual bin differences
- better maybe: Kolmogorov-Smirnov test Klaus worry about it (he's probably right): I'd probably implement that myself. Is it hard to implement? Figure out and maybe do or use nimpy+scipy: https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.kstest.html also see:
26.9.4. Fe charge spectra
- talked about
but didn't open it and discuss it in detail. Take away: differences clearly visible though.
26.10. Meeting Klaus about TPA analysis
26.10.1. DONE make median energy cluster plot w/ 30 min
26.10.2. TODO only use clusters in calibration data above a certain energy value
to cut away escape peak photons
26.10.3. TODO make time dependent plot only of new 30 min binned gas gain
26.10.4. TODO plot occupancy of an Fe 55 run
26.10.5. TODO make cuts to gold region and reproduce median energy plot to see if
variation disappears. Is due to change in Fe55 run geometry?
26.10.6. TODO look at ratio of amplitudes in Fe55 run of escape peak to photo peak
26.11. Meeting Klaus about TPA analysis
To discuss:
- fig. 234
- fig. 233
- figs in ./../Figs/statusAndProgress/binned_vs_time/gas_gain_investigation/
- figs of occupancy:
- fig. 193
26.12. Meeting Klaus about TPA analysis
- create histograms of median of binned energy vs time plots
-> just make a
geom_histogramclassified by the run period -> add to plotTotalChargeVsTime script? -> DONE TODO: make the run periods in the sense used by gas gain vs time business (separated by more than 5 days) something more supported in TPA? Have something like an "auto run period detector" that does it?
- Could go beyond the run period stored in ingrid database. Have "major" run period stored in DB and "minor" based on time split
In principle can be determined straight from run numbers, but that might not be the nicest. Have attribute in run group to store that? In practice hard, because before and after not well defined concept!
- create the new gas gain vs time plot with hits < 500 filter -> DONE
create the new median energy vs time plot
- NOTE have to redo the gas gain vs energy calibration fits first, because one of them is broken
-> DONE
change calculation of gas gain vs energy calibration fitting
- replace by fit + gas gain per time bin
- leave as is and use mean of gas gain bins and one fit DONE
- add option to only use the nearest neighbors in linear interpolation as base for a gas gain for a backgroud run (no, for each cluster in a background run calculate new interpolation between points) -> need to read all gas gains + fit parameters + timestamps from calibration and then have these in memory to compute values on the fly
this option needs to be selectable from the config.toml file!
create full histograms of energy. without any binning, create for each run period a histogram of:
- energy clusters in bronze + silver + gold DONE
- silver + gold
- gold
without any logL cuts applied or anything like this. Should be doable today using plotData.
26.13. Meeting Klaus about TPA analysis
The main result from this meeting was the observation that the results as discussed in
- ./../Mails/KlausUpdates/klaus_update_15_12_20.html
- file:///home/basti/org/Mails/KlausUpdates/klaus_update_15_12_20_num2.html
look really good. The behaviors are all explainable and the main takeaway before checking off the detector calibration is to find the perfect binning for the length of the time interval.
The current 30 min interval is an arbitrary value.
If possible the idea would be to:
Perform an optimization on the whole pipleline of:
- gas gain computation in interval of length N min
- compute energies using these slices
- make the median energy per cluster plot
- have some measure on the:
- visibility of the time dependence (none should be visible)
- variation (sigma of histogram) should be as small as possible
and have an optimizer (NLopt?) optimize this.
The difficulty in that is that optimizing from such a high level is
probably going to fail. Need a proc, which makes "one iteration", that
internally performs multiple shell calls to modifiy the files and
finally compute some numbers and return. Each iteration will take up
to 2 h or so.
A thing to use as the loss function / value to be optimized:
- compute the histogram for each run period of the background data based on all median energy values
- minize something that takes into account both:
- the χ²/dof to make sure the data is still gaussian distributed (it won't be once time dependent behaviors show up! should be ~1)
- the sigma of the fitted gaussian (should be as small as possible)
- ⇒ some cross entropy loss or something? Essentially find the smallest sigma such that the χ² is still more or less compatible with 1.
26.14. Meeting Klaus about TPA analysis
We discussed the plots contained in:
These still have to be moved from the optimizeGasGainSliceTime
(created in /tmp) to the figs directory.
From the final ones in
/tmp/medianEnergy_vs_time_ind.pdf
we determined that the best possible value is probaly about 90 minutes. There may be some very slight time dependence in the first 2 bins of 2017 visible, but it looks fine otherwise. Comparing the ridgeline plots we can see that longer than 90 min the FWHM of the distributions doesn't really get smaller anyway, but time dependencies appear.
We have the options to
- remove the first two bins in background of 2017 if we find excess signal from these times
- can re run the background rate / limit calc also with 30 and 300 mins to compare the effect on the limit. This gives us an idea on the systematic error we have!
This concludes the calibration / time dependent behavior studies, we're happy with the results.
Beyond that:
- TODO make plots for the 3 logL variables against time, not only for the
medians, but:
- variance, skewness, kurtosis (super easy)
- TODO make plot for Mn CDL data combined with the Fe55 data for each observable (of the distributions)! Perform simple cuts somewhat similar to the cuts on the CDL reference data (be more strict possibly and take 1 sigma around photo peak). Look at few ind. runs and all runs combined. Is distribution compatible?
- TODO try to find out what the generalization of statistical central moments to higher dimensions is!!
- TODO compute the efficiency of the logL cuts outside of the selected 3 sigma peak of the CDL data, by applying the same cut value to all pixels in the "blue" spectrum shown in the distributions at the beginning of this file, using them as "photons of different energies". Also easy.
- possibly will need to remove the frac in rms Trans variable, because it's not smooth but discrete at low energies -> what's effect on background if this variable is reomved?
26.15. Meeting Klaus about TPA analysis
Two points from last year:
- TODO make plot for Mn CDL data combined with the Fe55 data for each observable (of the distributions)! Perform simple cuts somewhat similar to the cuts on the CDL reference data (be more strict possibly and take 1 sigma around photo peak). Look at few ind. runs and all runs combined. Is distribution compatible?
- TODO compute the efficiency of the logL cuts outside of the selected 3 sigma peak of the CDL data, by applying the same cut value to all pixels in the "blue" spectrum shown in the distributions at the beginning of this file, using them as "photons of different energies".
New points:
create the background rate in the gold region with the current setup, i.e.:
- 90 min gas gain binning
- filtering on cluster size & position
as a benchmark to compare possible changes in the future to. Also: Maybe the background rate is already improved? In particular shift seen in parts of data 8 keV -> 9-10 keV
morphing of observables to hopefully remove discrete jumps from one energy range to another, due to sudden change in efficiency.
- talk to Phispi / Philip Bechtle / Hübbi
Related:
- in HEP: https://indico.cern.ch/event/507948/contributions/2028505/attachments/1262169/1866169/atlas-hcomb-morphwshop-intro-v1.pdf
- https://mathematica.stackexchange.com/questions/208990/morphing-between-two-functions
- https://mathematica.stackexchange.com/questions/209039/convert-symbolic-to-numeric-code-speed-up-morphing
Morphing especially problematic for essentially discrete distributions like the fraction in transverse RMS at lower energies in which there are discrete values. The morphed function needs to retain these features in the data.
26.16. Meeting Klaus about TPA analysis
Main takeaways were:
- why do something complex if simple also works? Try to use linear interpolation!
- TODO: compute all linearly interpolated pairs of CDL distributions
- TODO: compute the current background rate
- need to compute the gas gain 90 min for calibration data
TODO: compute the background rate using linearly interpolated CDL distributions
- either compute new interpolation for each cluster energy
- or bin the CDL distributions to about 1000 distributions
And for each bin / distribution compute the CDF to get 80% cut value
- TODO: compute 2D interpolated raster plot of CDL distributions
- also add the line that corresponds to 80% efficiency in each and see what that looks like
Optional:
- TODO: compute interpolation based on:
- spline
- KDE
- KD-tree in 2D fashion?
26.17. Meeting Klaus about TPA analysis
- much larger background rate in 2014/15 data due to Mylar window? C
Kα, O Kα lines? Mylar is a stretched form of PET, i.e.
(C10H8O4)n, so lots of C and O around! (ref: https://xdb.lbl.gov/Section1/Table_1-2.pdf) - check the average muon ionization energy per distance in Ar/Iso 97.7/2.3 to verify where the "8 keV" hump should really be in the background spectrum NOTE: take into account that a muon that isn't perfectly orthogonal to the readout plane will traverse a longer path through gas, yielding higher energy deposition. That means if the >8 keV hump is due to cosmics there should be a scaling of eccentricity to higher energies in the background rate of the "hump". -> create a plot of remaining clusters vs. their eccentricity
- in addition: compute the background rate as a fn of logL signal efficiency, 50 to 99 %, more steps for higher efficiency (higher efficiency means including more background)
- create a background rate plot to higher energies (15-20 keV?)
- DONE:
- DONE:
- perform KDE (bin-wise) instead of linear interpolation. To validate: compute KDE based on all targets except the one we wish to reproduce
- apply interpolations on the logL cut. Add interpolation option in
likelihood.nimwithconfig.tomlselector for the method:- none
- linear
- KDE
26.18. Meeting Klaus about TPA analysis
Things to talk about:
- muon energy deposition. Unable to compute the value using bethe formula? Value by PDG is ~7.8 keV (3 cm)
- did a lot of raytracing stuff. Code over 100x faster now. OTOH implementation using CGI ray tracing approach nearing end, cylinder, parabolics missing then port can happen.
- KDE interpolation doesn't work, cause data is pre binned of
course.. Leads to either too large bandwidth (bad prediction) or
valleys without data (good prediction but obv. bad in between)
- see kde plots
- spline interpolation:
- looks better in some respects and much worse in others.
- see spline plots
From last week:
- apply interpolations on the logL cut. Add interpolation option in
likelihood.nimwithconfig.tomlselector for the method:- none
- linear
- compute the eccentricity of the remaining clusters in the 8-10 keV hump
New insights:
- Muon energy loss / ~8 keV peak orthogonal muons?
- computation of muon energy loss using PDG formula simply wrong, due to missing conversion of I(Z) from eV to MeV.
- effect that should play a role: muons at CAST need to traverse lead shielding. Changes muon spectrum ⇒ changes γ of muons ⇒ changes mean energy loss
- Individual muon interaction Landau distributed, but ~8 keV has ~400 e⁻, thus probably reasonably gaussian
- KDE / Spline approach not a good idea. Keep it simple, use linear. Not wrong for sure.
- background rates for different logL efficiencies:
- curious that 8-10 keV peak barely changes for increasing efficiency. Implies that the clusters in that peak are so photonic that they are not removed even for very sharp cuts. In theory "more photonic" than real photons: check: what is the difference in number of clusters at 0.995 vs. 0.5? Should in theory be almost a factor of 2. Is certainly less, but how much less?
- 6 keV peak smaller at 50% eff. than 8-10 keV, but larger at 99.5% peak -> more / less photonic in former / latter
- barely any background at 5 keV even for 99.5% efficiency
- largest background rate at lowest energy somewhat makes sense, due to worst separation of signal / background at those energies
- interval boundaries of the different CDL distributions becomes more and more prevalent the higher the efficiency is.
- pixel density of orthogonal muon tracks should have a different drop off than X-ray, due to convolution of many different gaussians. An X-ray has one point of origin from which all electrons drift according to diffusion from height h, ⇒ gaussian profile. In muon each pixel has diffusion d(hi) where each electron has its own height. Could probably compute the expected distribution based on: mean distance between interactions = detector height / num electrons and combine D = Σ d(hi) or something like this? Ref my plot from my bachelor thesis… :) Is this described by kurtosis? Also in theory (but maybe not in practice due to integration time) the FADC signal should also have encoded that information (see section on FADC veto / SiPM)
New TODOs:
26.18.1. TODO Muons
- Investigate (and ask Jochen) about effect of lead shielding on muon spectrum.
- Research muon spectrum at sea level / at Geneva level, both energy as well as angular (energy at detector angles specifically). Maybe start here?: https://arxiv.org/pdf/1606.06907.pdf
- Compute -⟨dE/dx⟩ for different possible muon γs. How does it scale with γ? How does it scale with likely slightly different angles of entry due to larger distance in detector? Largest possible angle given width of detector & typical track width? Just simulate a bunch of possible different cases of muon spectra and angles.
26.18.2. Background rates (and muons)
- TODO combine background rate plots for different logL signal efficiencies
into a single plot. Possibly just use
geom_line(aes = aes(color = Efficiency))or histogram with outlines and no fill. - DONE make histogram of length of all remaining clusters. Cut at median
and produce a background rate for left and right side.
- TODO partly: Or possibly do the same with some other measure on the size of each cluster or rather a likely conversion origin in the detector (near the cathode vs. near anode)
- DONE compute other geometrical properties of the remaining clusters (c/f last week eccentricity, but also other observables)
- compute "signal" / √background of each bin. Since we don't have signal, use ε = efficiency. Plot all these ratios of all signal efficiencies in one plot. In theory we want the efficiency that produces the largest ε / √B. Is 80% actually a good value in that regard?
- TODO orthogonal muons: try to find other ways to investigate shape of muons vs. x-rays. Possibly plot kurtosis of 8-10 keV events?
26.19. Meeting Klaus about TPA analysis
Discussion of ./../Mails/KlausUpdates/klaus_update_16_02_21.html.
- explanation of CDL data and reference spectra: done for Klaus to have a resource to understand the details of the method
- implementation of morphing applied to both logL distributions as well as X-ray reference spectra (for each logL observable)
Background with morphing:
- effect pretty small
- at least change in those bins where we expect it
- gives us some confidence that a large number of the remaining clusters really is because they are X-ray like and not an artifact that happens due to a sharp drop in logL effectiveness near the boundary of energy bins
Comparison of all clusters passing logL cut vs. those in 8-10 keV:
- main thing to note: eccentricity and both RMS values slightly lower
for 8-10 keV than for all data.
- question: is this due to a higher median energy in the 8-10 keV sample than in the full sample?
- alternative hypothesis: orthogonal muons should have slightly higher density towards their center, due to having larger contribution of electrons generated closer to the grid and thus experiencing less diffusion.
- check: what does the same plot look like when comparing the 8-10 keV passing clusters with those of the 8 keV line from the CDL data? Possibly take those values that are remained after both sets of cuts are applied and are below the corresponding cut value in their respective energy.
- to check: is RMS value really in mm? Should be. Input to
statsis pixel converted to mm iirc and RMS doesn't do any normalization I think. Check.
Comparison of clusters passing logL cuts where properties are smaller / larger than median value in each property:
- curious that likelihood data shows 1 keV peak only in > than median
logL value. Possible reasons:
- there is an energy dependence of the logL distributions. Compare with logL distribution ridgeline plot. From that alone expect lower energy events to have larger logL values. Could account for this effect by itself? Can we try to correct for the the energy dependence of the logL variable somehow? Simplest idea is to fit function to energy behavior of logL distributions and correct logL values by that. But possibly too inaccurate. check
- another effect is that separation power decreases with decreasing
energy. This is strongly correlated with the previous point of
course. In this sense it can also just mean that lower energy clusters
will be closer to possible cut values (which does not take into
account that of course also the "cut line" scales in some way with
energy).
check: it would be interesting to visualize the cut position
against energy. I.e. plot
- energy vs. cut value directly
- in heatmap of interpolated data (logL vs. energy in this case) draw the line of cut value on top of raster
other variables also show strong dependencies on different areas of the background rate. Identifying 3 features:
- 1 keV peak
- 3 keV peak
- 8-10 keV peak
for many variables they are split quite stark between lower / upper median half. Too hard to make proper conclusions on its own though.
Thus the TODOs:
- check RMS calculation
- compare properties of 8-10 keV passing clusters to 8 keV CDL data
- possibly try to correct logL values for energy and compare lower / upper median logL plot again after correction?
- possibly visualize cut value
And from last week:
- continue study of muons from theoretical angle
- compute signal efficiency over √Background for the background rate plots for signal efficiencies in an attempt to try to find the best signal efficiencies depending on energies. In this context take a look at ROC curves and possible ways to quantify those via some signal purity or S/N value for best possible cut position (that's what they are for after all)
26.20. Meeting Klaus about TPA analysis
Mainly talked about ./../Mails/KlausUpdates/klaus_update_23_02_21.html.
Gives some credence to the hypothesis that the 8-10 keV hump is at least to a good part made up from muons.
Error to note:
- units for flux are wrong, missing a
GeV. It's due to the currently broken / not really implementedpowimpl. where I hack around by convertingGeVtofloat. That's what confuses the type checker. The equation in the paper of course is only correct for arbitraryndue to numerator and denominator both usingpowwith a difference of 1 independently ofn!powfor real numberednotherwise does not make sense unit wise of course. Work around this and impl. apowfor static exponents?
Things that should be done on top of existing work:
- compute the actual muon rate that we expect at CAST by integrating angles and gold region area
- compute the spectrum of muons we actually expect, not just the mean value
- fold the spectrum with the detector resolution to get a cross check whether the 8-10 keV hump somewhat matches the expected muon signals?
Further TODOs beyond muons:
- is it possible to compute an expected # of events for Cu X-ray fluorescence? See if we can find understand efficiency to excite a Cu atom and make some simplified assumptions.
related to optimizing signal efficiency over √Background: Perform an optimization of the expected limit for the current background rate. Essentially start with first energy bin (using CDL intervals) and simply compute multiple for multiple signal efficiencies. Possibly look at first 2 intervals. In theory one could automate this as an 8 dimensional problem (due to 8 energy intervals). Wrapping the whole likelihood cutting + limit computation in one program, which uses an optimizer sounds doable, but potentially would result in a very long runtime, because each iteration step is possibly pretty long. If done, we should see if we can work with:
- read all background clusters in memory (should fit just fine -
just means we need to take out some code from
likelihood.nimand merge intolimit_calculation.nimor a combined wrapper so that we don't depend on HDF5 files as an intermediate step). - prepare the interpolated logL distributions
- single procedure, which takes parameters (ε of each interval) and a pointer to an object storing all required data
- compute logL remainder + limit
- result of procedure is expected limit
- use (global?) optimizer to compute minimum of limit
Pretty speculative if this can work well.
- read all background clusters in memory (should fit just fine -
just means we need to take out some code from
26.21. Meeting Klaus about TPA analysis
Discussion of: ./../Mails/KlausUpdates/klaus_update_04_03_21.html
- ε / √Background:
- Energy ranges which have more statistics show smaller variaton between different signal efficiencies. Implies scaling of √background roughly in line with increase in signal efficiency. The case for 3 keV as well as 8-10 keV
- 80% only best in some bins
- bins at very low energy: tend to improve for stronger cuts / smaller ε
- bins at higher energy: tend to improve for weaker cuts / larger ε
- study using Kolmorogov tests or similar a bit problematic, because needs signal information
- TODO: compute the expected limit for:
- different signal efficiencies
- applying a 60% efficiency in the energy range up to ~1 keV
- TODO verify if computation was done using morphing or not
- muon study:
- there should be a hard limit on muon energies based on γ. If γ too small, muons will never reach surface! TODO: compute limit on γ ⇒ E
- only a simple approximation: thus compute the spectrum based on assumption muons do not see any material before detector (ignore concrete, steel etc). If too simplistic can still try to see what happens if taken into account.
26.22. Meeting Klaus about TPA analysis
Discussion of: ./../Mails/KlausUpdates/klaus_update_09_03_21.html
26.22.1. Limit calculation dep. signal efficiencies
- all in all different limits seem to make sense as well as scaling seems reasonable
- 60% the best, to an extent perhaps expected based on looking at the previous plots from
- in the lowest bins we expect signal but don't have any background!
TODOs:
- limit calculation currently is essentially 1 toy experiment due to drawing of a set of expected candidates once and optimizing for CLs instead of using mclimit's expected limit computation directly. Find out if this is possible / there are any bugs with that.
- cross check again the numbers we get from ROOT vs. our impl
- compute expected limits for single + few bins where we know analytically what we might expect
- make sure to recompute the numbers here with linear interpolation (morphing) of logL
26.22.2. Muons
- the peak at 12 keV is a bit confusing and at least somewhat unexpected
- why does a heavier muon result in less energy deposition? Shouldn't it be the other way round?
comparing flux under 88° with 0° (fig. 15, 16) and taking energy loss in atmosphere along ~260 km (for 88°) into account yields about an energy loss of ~50 GeV. At 50 GeV the flux at ϑ = 0° is about
echo "S@88° = ", h * distanceAtmosphere(88.0.degToRad.rad, d = 15.0.km) echo intBetheAtmosphere(200.GeV, ϑ = 88.0.degToRad.Radian).to(GeV)
S@88° = 268.4 KiloMeter total Loss 56.00 GigaElectronVolt At this energy in the plot for ϑ = 0° we are half way between 0.01 and 0.02 and in the "real" flux at more or less 0.04. At least the order of magnitude is correct, which is reassuring.
TODOs:
- figure out if 12 keV peak is sensible / does behavioral change between muon masses make sense?
- implement muon lifetime into integration through atmosphere (essentially compute γ at each step and compute probability it has decayed up to that point)
- figure out best way to find flux in atmosphere
- from a dataset
- from known proton + He flux and decay channels
- by inversely integrating known flux from surface to atmosphere. Means we don't have information about low energy muons that decay! That's only a problem if one wants to compute fluxes at higher altitudes than sea level (if starting from sea level to get atmospheric flux)
26.23. Meeting Klaus about TPA analysis
TODOs:
- send Klaus dataset of background, signal, candidates as CSV file
- consider again TLimit with an analytically understandable Poisson case. E.g. compute 1 - α for a given background case, e.g. N = 3.2 (background after normalization to tracking time), and compute based on Poisson: P(k; λ) = λk e(-λ) / k! where λ = 3.2 is our expectation value. Then: Σk = 0k = 2 P(k; 3.2) = α for the case N = 3 for example. (or is that 1 - α?) So that we can then find the N such that we get a 5% value. This corresponds the an expected ⟨CLb⟩ for b = 3.2 and N = 3. This way we can compute the poisson case with 1 bin. The same can be extended to include possibly 2 bins. If one uses two bins with the same background then it's essentially "1 bin split in 2". Can also check that. Finally, one could later make qualitative checks about the behavior of the results on gets from TLimit. E.g. have a background histogram with different entries. If one then increases the signal in a background bin with low background the impact on the CLs+b should be larger than if the bin has a large background.
- start playing around with TRexFitter.
- compile it locally
testdirectory contains:- config files to run it with
- inputs in form of ROOT histograms
26.24. Mail to Klaus
I just sent a mail to Klaus about my lack of understanding of
mclimit and statistics in general. It lives here:
./../Mails/KlausUpdates/klaus_update_25_03_21.html
26.25. Meeting Klaus about TPA analysis
Main talking point was: ./../Mails/KlausUpdates/klaus_update_04_06_21/klaus_update_04_06_21.html (and the related comments file) as well as: ./../Mails/KlausUpdates/klaus_update_07_06_21/klaus_update_07_06_21.html
Regarding the reproduction of the 2013 limit.
Klaus explained to me the idea of using the likelihood method. My lack of understanding was related to how the limit is computed based on the best fit. In addition I didn't understand the comments about the first mentioned file regarding the Δχ² numbers with the example calculation.
As it turns out: The χ² method allows for a (scale) independent way to compute a limit in the sense that the χ² distribution is computed and from there a Δχ² of ~1.96² is added. This describes the 95% coverage of a hypothetical underlying Gaussian distribution (as far as I understand).
So a limit is determined by: Let \(x\) be the best fit value. Then \(x'\) is the desired limit:
\[ x' = x + g(χ²|_{\text{at x}} + Δχ²) \]
where \(g\) is the inverse function that returns the \(x\) value for a given value on the χ² distribution.
In plain words: find the lowest point of the distribution (== best fit), add ~1.96² to the χ² value at that point, draw a horizontal line and see where it cuts the χ² distribution. The cut value at larger x is the limit.
In case of unphysical parameters in the phase space there are apparently methods to perform rescaling. Essentially only the physical part of the distribution is considered: χ² . . .. … … …. Note the "cut values" shown .. .. are obv. very wrong in terms . . . of the distribution shown. .. . .. . . .. cut value of full 95% .. . . . . . .. . .. . .. . . . .. . cut value of physical 95% .. . .. . . .. . .. .. . … . … . .. . …. . .. . . ….. .. . . …. … . . …… …. unphysical . physical . …………. ……………………………………………………………….. x . . . . . . . . . . . . "physicality bound"
This may be how in the paper they came up with a ~Δχ² = 5.5 or so (instead of the direct ~4).
Aside from this lack of understanding 2 main things were discussed:
- The absolute value of the χ² values. My computations result in a factor of 58 / 20 ~= 2.9 larger than in the paper. At first Klaus suggested it might be from the poisson calculation, so different computations were tried, e.g. using sums of logs. This was done in section 17.5.1.4 and ./../Mails/KlausUpdates/klaus_update_08_06_21/klaus_update_08_06_21.html. It was found that indeed my computation is not the issue (which I thought, given that the likelihood values are only of the order of 1e-16, see fig. 247.
- More troubling (given that the χ² method should be independent of the absolute scale) the shape and width of my distribution is quite different. Thinking about this and in particular where my χ² seems to become ∞ compared to what happens in fig. 6 of the paper we came to the conclusion that this could be explained if our code overestimated the flux that is actually used in the paper. Doing a rough calculation of the MPE telescope effective area (TODO INSERT PAPER) yielded a effective area of ~5.5 cm². Taking the ratio of the coldbore of 14.5 cm² / 5.5 cm² ~= 2.64. Inserting this into the flux scaling procedure in the code indeed yields a plot that is much closer to the shape of the paper. TODO: revert those changes in the code about the paper / add that somewhere including the plots
Discussion of the limit calculation lead us to the conclusion that our result is pretty optimistic now.
Things we want to do now:
- apply the likelihood method to our data
- start by using same flux assumptions as in the reproduction code
- need to do toy experiments using MC for the candidates (because candidates are required in the likelihood method) to perform the limit computation many times to get an expected limit
- then: compute using background / 3, background / 5 (imagine a lower background than we have essentially)
- introduce ray tracing flux code. Can then compare ray tracing results with simplified likelihood method.
- I think I forgot one thing here.
With mclimit:
- what result do we get for background / 3, background / 5?
- what limit do we get for 2013 data if we optimize for CLsb (should be much closer to what we see using likelihood method. In particular given that the observed CLs+b is already much lower than <CLs> in the output of the optimization, meaning coupling can be much smaller).
Independent:
- Compare flux we get from simple computation as done for reproduction and compare that with flux at same coupling constants as we get it from ray tracing. (almost possible with plots in limit computation we did in ./../Mails/KlausUpdates/klaus_update_07_06_21/klaus_update_07_06_21.html in comparison to 2013 reproduction flux code)
- reproduce the effective area from the paper about the telescope or the gaγ limit and fold it with the flux numbers (Energy dependent). Use to recompute the 2013 limit based on that addition.
Further:
Septem Veto. Given the septem veto plot here in fig. 53 we see that the background mainly improves for the lowest energies. I already found this weird when I wrote that part.
- extract those clusters that are removed by the veto and look at them before and after the "septem event" creation. What changes and makes them disappear?
26.26. Meeting Klaus about TPA analysis
Discussion about ./../Mails/KlausUpdates/klaus_update_11_06_21/klaus_update_11_06_21.html.
Main takeaways:
- limit calculation seems to make sense
- Klaus was wondering why I didn't include detector window + gas absorption yet: plain and simply to do it step by step in order to see the effect of each change
- toy MC should also be done with limit calculation using
mclimit(by using observed limit of that) - extend LLNL telescope efficiencies below 1 keV by taking ratio of LLNL/MPE @ 1 keV and then using that ratio to extend it down along the path of MPE
- TODO: compute distribution of χ² values at the minimum. This should be a proper χ² distribution (interesting).
26.27. Meeting Klaus about TPA analysis
Had a short ~55 min meeting with Klaus.
We discussed ./../Mails/KlausUpdates/klaus_update_22_06_21/klaus_update_22_06_21.html
The results all look pretty much as we would expect.
Things to do after this:
- compute likelihood based limit including detector window + gas
- compute likelihood based limit with our background scaled down by a factor ~5
- change the way we compute the actual limit.
3 requires more explanation:
Currently we deduce the limit by walking the χ² "distribution" we get from the minimum to the right until we're at χ²min + 5.5 as 5.5 is our rough guesstimate what the 2013 paper did due to rescaling the unphysical part.
The way this should actually work though:
.
. ..
.. .
. .
. | ..
. | ..
. | ..
. | ..
. | ..
.. | ..
. | | |. .
..v | |.
-----------------------–—+---–— χ² min + 1
| .. | .. |
| …. | …. |
…………. -------–— χ² min
----------------------------------------------------------------------------–—
minimum| ……… …. | .. .. | .. . .. . .. .. .. … .. .. . .. . .. .. .. .. … .. .. …. …. …. ….. … ………
draw gaussian with σ determined by width at χ² min + 1 (oh, might actually plus 0.5, look at barlow).
From there we can integrate that gaussian from:
- left to 0.95 to get the "normal" 95% value (and compute the Δχ² for that for reference)
- compute from the 0 line (0 in g²ae) to right until 0.95 of that. That's our "actual" limit (that should correspond to the ~5.5 we used, but of course only for a single drawing of candidates)
So we need to include that into our limit calculation.
Another thing I remembered: The likelihood based limit method currently ignores the software efficiency! That means of course our ε = 60% was significantly better!
Finally, once these 3 things are done:
- extract all clusters that pass gold region likelihood cut
- create septemevents for all of them and plot.
Last:
- combine 2014 & 2018 data. Compute lnL for each, add them.
26.28. Meeting Klaus about TPA analysis
Mainly discussed the notes in ./../Mails/KlausUpdates/klaus_update_29_06_21/klaus_update_29_06_21.html.
Limits look all good mainly.
One thing to do:
- look at combination of 2014 + 2018 data by concating the 2014 data
to the
monteCarloLimitsequences and running it. Need mylar, MPE efficiencies mainly
26.29. Meeting Klaus about TPA analysis
Discussion mainly of the ~500 septem events where the center clusters pass the logL cut.
Things to take away:
- a very large number of especially low energetic clusters are (often directly) connected to tracks on outer chips
- many high energy events in the samples
- also quite a few events where there are tracks that do not point to the cluster center
- some are just weird (why did they pass the cut in the first place?)
- energy of cases with > 1 cluster on center chip sometimes choose the wrong energy: energy index access is wrong!
Things to do:
- DONE cut to energy (less than 5 keV or so) and only look at those plots
- DONE fix energy access
- DONE fix plots
- DONE fix passing of logL cut, it seems broken right now
- additions:
- TODO check for outer cluster. If found, check eccentricity larger e.g. 1.2. Then use long axis to create a track from center. Check: does it hit the center cluster? Maybe within 1.5 radius or something
- STARTED possibly use Hough trafo: compute lines between all pairs of points. Determine slope and intercept and put these onto a new plot: gives Hough space. If cluster there, correlated in euclidean space
DONE Non plot related things to do:
- get number of total events
- get number of events with any activity
- get number of events with any activity on outer chips
- get number of events with any activity on the outer chips iff there is activity in center
- get number of events with only activity on the center chip
26.30. Meeting Klaus about TPA analysis
Discussion about: ./../Mails/KlausUpdates/klaus_update_06_07_21/ (septem events PDFs as well as Org gen'd PDF)
About septem events:
- looks very reasonable now.
- algorithm seems to behave mostly as it should
- some events are "why did it veto this / not veto that?"
- definitely helps to have outer chips
- some low energy events have super small radii. Low energy photons almost certainly absorbed early. So should have very large diffusion!
DONE:
- colorize clusters as they are recognized as individual cluster in septem frame clustering! Why are some together and others aren't? Some bug in cluster finding?
TODO:
- What would DBSCAN give?
About general event information:
- Curious peak at 2.261 s and 2.4 s. The 2018 data only contains a peak at 2.261 s. Did the shutter length change?
- Events with 0 second shutter: what's going on here?
- Energy distribution of outer chips for 0 s events: difference is very strong.
TODO:
- check in raw data what shutter lengths are used
- check which runs have 2.261 s peaks and which 2.4 s event durations in 2017/18 dataset
- study events with 0 s duration more. What does this look like in calibration data? c/f events in which shutter didn't trigger
- compute:
- hits histogram for events with 0 s for all chips (same distribution as energy? So too low gain?)
- compute energy histogram for all chips for all events.
- compute hits histogram for all chips for all events.
Hough transformation:
- very hard to read plots. What we humans detect as patterns is not the only thing Hough connects. Multiple tracks are connected etc. And density plays much bigger role than for us humans
- problematic to deduce anything from it. Possibly not continue further.
TODO:
- read Simone Ziemmermann thesis about Hough trafo application. Maybe something can be applied here
- instead of Hough trafo: compute long axis & position of non center clusters. Draw a line of these. Create plots to see: do they pass through center?
General TODO:
- create a notion of "radius" for photons of low energy.
- compute histogram of that comparing for background clusters and CDL reference for everything ~< 2 keV or so.
- compute mean free path based on energy. To get an idea of how large each photon of an energy actually goes in the detector.
26.31. Meeting Klaus about TPA analysis
Meeting with Klaus was about the following files:
~/org/Mails/KlausUpdates/klaus_update_03_08_21/septemEvents_2017_logL_dbscan_eps_50.pdf~/org/Mails/KlausUpdates/klaus_update_03_08_21/septemEvents_2017_logL_dbscan_eps_65_w_lines.pdf
The first two files are a continuation of the septem events of all clusters that pass the logL cuts in the gold region.
Instead of using the old clustering algorithm we now use the DBSCAN clustering algorithm. The two files correspond to two different settings.
The minimum number of samples in a cluster is 5 in both cases. The ε parameter (something like the search radius) is 50 and 65 respectively. The latter gives the better results (but the final number needs to be determined, as that is still just a number from my ass).
In addition the ε = 65 case contains lines that go through the cluster centers along the slope corresponding to the rotation angle of the clusters. These lines however are currently not used for veto purposes, but will be in the future.
Looking at the clustering in the ε = 65 case we learn:
- some center clusters that are still separate are still not passing now. Why? Is this because DBSCAN drops some pixels changing the geometry or because of the energy computation?
- many clusters where something is found on the outside are now correctly identified as being connected to something.
- few clusters are not connected to the outside cluster. Might be caught with a slight ε modification?
- some clusters (of those still passing / not passing due to bug, see above?) can be removed if we use the lines drawn as an additional veto (e.g. line going through 1 σ of passing cluster).
With this the veto stuff is essentially done now.
- Scintillator vetoes are implemented and will be used as a straight cut if < 80 clock cycles or something like this
- septem veto has just been discussed
- FADC: FADC will be dropped as a veto, as it doesn't provide enough information, is badly calibrated, was often noisy and won't be able to provide a lot of things to help.
If one computes the background rate based on the DBSCAN clustering septem veto, we get the background rate shown in the beginning. The improvement in the low energy area is huge (but makes sense from looking at the clusters!).
DONE:
- write a mail to Igor asking about the limit computation method used in the Nature paper
26.32. Meeting Klaus about TPA analysis [/]
Discussion of: ./../Mails/KlausUpdates/klaus_update_07_09_21/klaus_update_07_09_21.html
The main takeaways are:
Regarding 1 (line through cluster septem veto): looks very convincing and probably like something one would want to remove manually, but be very careful what this implies for a dead time of the detector. TODO: compute this line veto for a large subset of all background data to see in how many cases this veto happens.
Regarding 2 (spark detection): Nothing to say, need to TODO work on it.
Regarding 3 (calculate limit using Nature paper method): KDE of background rate looks very nice. Bandwidth (currently 0.3 keV) should ideally be something like the detector resolution (first order approximation same over all energy).
Heatmap: the raytracing flux should ideally be encoded using a 2D smooth function as well. We can do this via two ways:
- extend the KDE function to 2D and use that based on the actual 'X-rays' that pass the ray tracer
- use a bicubic interpolation on the heatmap
The former is nicer, but needs to be implemented. The raytracing output is centered at exactly the middle of the chip. Use our geometer data & X-ray finger runs to determine better number. For systematic uncertainties we also need to think about applying a gaussian 'smearing' to this 2D distribution. Either vary in dx / dy or in dr / dφ. Can fit this directly in the logL fit as well (what Igor proposed).
Mainly: continue on with this work. First make sure the limit calculation works without systematics and using a 2D grid for the heatmap.
Regarding 4: Neural network. Looks extremely good and promising. Need to be very careful about validation, in particular in terms of energy dependence (learning the energy) of the input data, because the signal like data is strongly energy dependent. Need to look at the input distributions of:
- the CDL training dataset for all variables
- the background training dataset for all variables
- the same for the test validation dataset
Further: to make sure energy behavior is sane, train on a subset of data that has a "flat" distribution in energy (take much less events on the exact peaks and ~all next to it) essentially. See if the performance suffers. Also train without explicit energy and number of hits. Possibly even without length and width as they are slightly energy dependent. Personally I don't think this overblown. The likelihood method also has a direct energy dependence after all. More important is proper validation. Finally can try to feed for classification a modified background dataset that is perfectly flat. Do we see a shape after prediction in the events that pass? What does the logL method produce for the same data?
Finally: we have multiple sources of ~flat X-ray information:
- X-ray finger has a flat contribution!
- targets in CDL are not perfect
- make use of calibration runs as well
26.32.1. TODO compute line veto for many background events for dead time
26.32.2. TODO generalize heatmap to smooth 2D distribution
26.32.3. TODO make use of X-ray finger runs to determine center location of each run (2 and 3)
This needs to be used to:
- move the raytracing center there
- is a good reason why we should also treat Run 2 and 3 completely separate in the log likelihood method, same as the nature paper (different datasets \(d\))
26.32.4. TODO Look into geometer measurements for positioning
See what we can learn from that maybe.
26.32.5. TODO no systematics, 2D grid heatmap compute the limit based on nature logL
26.32.6. STARTED train NN without energy & hits (& without length / width)
Training without energy & hits actually seems to improve the network's performance!
26.32.7. TODO train NN on 'flat' signal like data, by taking out statistically events in peaks
26.32.8. TODO predict with NN on a 'flat' background distribution. What shape shows up? (also in logL?)
26.32.9. TODO make histograms of training / validation signal / background datasets for each variable
26.32.10. TODO predict the X-ray finger run, calibration run, study CDL data for cont. spectrum part
26.32.11. TODO can we naively generate lower energy data?
If the energy directly is causing issues, we can (same as in bachelor thesis) generate lower energy data from inputs by randomly removing N pixels and dropping the energy of those events. Than have our fake datasets at energies that we are possibly 'insensitive' to and predict those!
26.33. Meeting Klaus about TPA analysis
Discussion of .
The limit calculation is looking fine so far. The main issue in the discussed code is the normalization of the axion flux in form of the dΦ/dE, which is in 1/[keV m² yr]. Instead needs to be integrated over a single pixel (actually number of pixel in gold region / gold region size) and tracking time.
26.34. Meeting Klaus about TPA analysis
Discussion about the current state of the 2017 limit method calculation from section 29.
Essentially the problem at the moment is that the logL is a linear line for the case where all candidates are outside the signal region, i.e. have a ~0 weight in signal and a constant weight in background.
This is 'correct', but of course problematic, because then we cannot compute the global maximum (as it does not exist).
Things we can try:
- Compute a heatmap of S / B for each pixel in the gold region. This should be ~0 outside the axion image and > 1 inside the image. If this is not the case, something is amiss. Using a coupling constant near where we expect our limit to be.
- Take some simple cases where we can compute the logL analytically and see if this is reproducible in our code (e.g. constant background, etc).
- change the method away from our Gaussian sigma idea. Instead compute the limit directly from the logL distribution, i.e. go down from maximum by a certain amount corresponding to the 95% line (i.e. determine the numbers of sigma required for this. Compare with Cowan chapter 9.6). This we can do starting in the physical region. That should solve our problems for the case of no global maximum!
- Write a mail to Igor, asking whether using the 0 range is fine for the cases with ~ linear line or what other algorithm to determine the limit we can use. Given that we can compute the logL function, but simply cannot create large numbers of toy experiments without running into trouble sometimes.
26.35. Meeting Klaus about TPA analysis
Discussion of the current state of the Nature based limit calculation after Igor's mail from the .
Essentially saying that we simply integrate over and demand:
0.95 = ∫_-∞^∞ L(gae²) Π(gae²) / L0 d(gae²)
where L is the likelihood function (not the ln L!), Π is the prior that is used to exclude the unphysical region of the likelihood phase space, i.e. it is:
Π(gae²) = { 0 if gae² < 0, 1 if gae² >= 0 }
And L0 is simply a normalization constant to make sure the integral is normalized to 1.
Thus, the integral reduces to the physical range:
0.95 = ∫0^∞ L(gae²) / L0 d(gae²)
where the 0.95 is, due to normalization, simply the requirement of a 95% confidence limit.
With this out of the way I implemented this into the limit calculation
code as the lkBayesScan limit.
The resulting likelihood function in the physical region (for a single toy experiment) can be seen in fig. 455, whereas its CDF is shown in fig. 433 which is used to determine the 95% level.
After doing 1000 toy MC experiments, we get the distribution shown in fig. 434.
Thus the main TODOs after the meeting are as follows:
26.35.1. TODOs [/]
- TODO Do MCs w/ manually placed candidates in signal region, multi colored histogram
- TODO Verify usage of all efficiencies in the code
This is crucial, because it is still possible we are using the wrong efficiencies somewhere.
- TODO Use the newest background rate including the septem veto
This requires recalculating all of the numbers. But we can try with the septem veto only working on the existing data files (i.e. no DBSCAN for general clustering).
26.36. Meeting Klaus about TPA analysis [/]
First discussed some general stuff, including extending my contract to on a 50% position with the thought of me being done way earlier than that.
TPA wise, I told him about the following things done since last meeting:
- apply septem veto in likelihood and use resulting background rate without the ~1 keV peak as the background contribution to the likelihood function. Does indeed result in improved expected limits.
- changing the one free parameter we still have to improve the "no candidates in signal" background (RT limited), namely the software efficiency. Scaled this up to 100% and indeed this moves the limit down.
- created background clusters based on reduction over whole chip. Lead to interesting result, namely background with septem veto is now significantly lower than without. Chip scale background is almost as good as in gold region, maybe due to outer region being kind of dead.
Aside from these, we discussed the following (partially) TODOs:
26.36.1. TODO Document improvement to exp. limits using septem veto
Needs to be documented in the relevant section with plots comparing before and after & showing what the KDE background rate looks like.
26.36.2. TODO Document improvement to RT limited limit by changing software eff.
Modifying the software efficiency to 100% leads to a theoretical limit "limit" from gae² = ~4e-21 to something like gae² = ~2.8e-21 or maybe better. Not sure.
Needs to be documented.
26.36.3. TODO Study optimal software efficiencies
With the code "done" as it is now, we can try to optimize the software efficiency, by computing the expected limit for a fixed software efficiency and then determining what yields the best limit.
Need to retrace the steps done for that old computation in ./../../CastData/ExternCode/TimepixAnalysis/Tools/backgroundRateDifferentEffs/
Can focus in addition of changing global ε, also do a split of low energy ε and high energy ε.
26.36.4. TODO Document background clusters over full chip using septem veto
Recreate the plot for this and compute the background rate over the whole chip that way!
26.36.5. TODO Fix the inclusion of gold region focus in limit calc
Currently the code still assumes the gold region in parts. Ideally we don't want any kind of restriction to specific regions.
We can do this by:
compute an interpolation (k-d tree based?) of the background from
clusters over the whole chip. Then have a background model that now
does depend on x.
Then we don't have to look at specific regions anymore!
26.36.6. TODO Include systematics into limit calc
The systematics we care about for the beginning are the following 2:
- energy calibration & resolution (treated as one thing for now)
- the location of the axion signal. "Most likely" point should be X-ray finger run for each data taking campaign, then move from there.
Systematics just add an additional term we need to integrate over. But we do this by integrating out that systematic term first, before computing the 95% value along the axis of coupling constant.
26.37. Meeting Klaus about TPA analysis
We discussed the background interpolation for the limit calculation.
We saw before that the regions below 2 keV and above seem to have rather distinctly different backgrounds. The first idea was to simply cut the region in 2 pieces and interpolate on each separately. The problem with that however is that this leads to a rather difficult way to normalize the background (if one wants to keep using the known background rate w/o position dependence).
Instead we decided to try to use a full 3D interpolation. This means we need to treat the energy as a third dimension in the interpolation. It's important to keep the distance under which we interpolate in the energy rather small, as we know the width of the fluorescence lines. If we make it too big along that axis, we dilute the good detector properties of barely having any background in certain energy ranges.
The problem we face is two fold:
- interpolating in a "naive" way using a sphere / ellipsoid is problematic, because there is no reason why energy and position should be correlated in a "euclidean" sense. At the same time a euclidean interpolation also is very problematic for the corners of the chip. I.e. what is the volume cut out by two planes at the edges of the chip? Already rather complicated in the 2D case!
- we could interpolate using a cylinder. I.e. euclidean in the x/y plane, but purely linear in the Z (energy) direction. This means the distance in Z is strictly binned in a "linear" sense. It should be rather easy to achieve this by using a custom metric in the k-d tree of arraymancer! That's what custom metrics are for.
If we decide to use the second approach, it begs the question: what metric describes a cylinder using a single radius? It's like a weird generalization of a mix of Manhattan and Euclidean metric. i.e. Euclidean in 2 of 3 dimensions and Manhattan in the third dimension. It should be that trivial I think.
Another question is about visualization of this. Going this route means we throw away our typical notion of our "background rate". While we can of course do slices along one of these axes or simply integrate over all positions for a background rate, the real background that is used for our limit. Well, as long as all our data is there & things are explained well, it should be fine.
26.38. Meeting Klaus about TPA analysis
Last meeting before Christmas.
Two topics of interest:
- general limit calculation & background interpolation business
- send background rate plot to Esther
26.38.1. TODO Limit calculation
Few remaining open issues.
- Background interpolation. Interpolation is working, but missing normalization. Need to normalize by integral over the area that we take into account. Once normalization is done, compute the average background rate over e.g. the gold region. Do this for O(1000) slices and check the resulting background rate of the gold region. Compare to our known background rate.
- compute limit for the method including background interpolation. Maybe compare two metrics for the interpolation. Also gaussian in energy or a box.
- Once these things are done, start with systematic uncertainties as nuisance parameters. This is for January.
26.38.2. DONE prepare background rate for Esther
Simply a background rate plot. Ask Esther what she wants it for.
- histogram
- data points + error bars?
- raw data (as CSV?)
- Vega-Lite plot ?
- GridPix preliminary before
- add a few words about it. Using septem veto, CAST data taking time & area of gold region, background outside is also good, but of course worse, software efficiency.
Send to Klaus before.
26.39. Meeting Klaus about TPA analysis
I showed Klaus the background interpolation work I did over the holidays. That was mainly implementing the proper normalization of the gaussian weighted nearest neighbor code as well as computing a regular "gold region background rate" from the interpolation.
The main takeaway is that it looks good so far. The energy distance used was 0.3 keV ("radius") that seemed to be reasonable in terms of background spectrum features.
Of note was the peak below 2 keV that shouldn't be there (but shows up less extreme in the pure input data). I'm debugging this right now. The increase in the all chip logL file compared to the gold region one is the effect of the additional line veto it seems so far.
26.40. Meeting Klaus about TPA analysis
Discussion of the following plots:
~/org/Figs/statusAndProgress/backgroundRates/background_interpolation_plots.pdf
These are background interpolations at specific energies & search radii:
let Es = @[1.0, 2.0, 4.0, 5.0] #linspace(0.0, 12.0, 10) for (radius, sigma, eSigma) in [(100.0, 33.3333, 0.3), (75.0, 75.0 / 3.0, 0.3), (50.0, 50.0 / 3.0, 0.3), (33.333, 11.1111, 0.3), (25.0, 25.0 / 3.0, 0.3), (100.0, 33.3, 0.5), (50.0, 50.0 / 3.0, 0.5)]:
in particular here.
Our takeaway from these plots is:
There is certainly some variation visible at the 25 pixel search radius. But: the variation could be treated as a systematic uncertainty on the limit calculation, arising from statistical fluctuations in our background data that builds the background hypothesis.
The background rate looks great at 25 pixels. Still no fluctuations visible there & the background below 2 keV is small and doesn't pull in a bunch of background from the corners.
So our approach for now is rather: Use a small radius and try to work with the statistical fluctuations, than taking a large radius and ruining our background at low energies.
Note in particular: in the 25 pixel case at ~1 keV the max peak is at
about slightly >1.5e-5. In the corresponding background rate plot:
the peak is barely lower at maybe 1.4e-5.
This means that we do not actually pull in a lot of background from the sides there!
In particular the line veto which is not used in this input, will probably help quite a bit!
For the uncertainty: In theory the following holds: our background is based on a fixed number of counts. At each point, we pull in a number \(N\) of clusters. These vary based on Poisson statistics. From there we perform a transformation, essentially: \[ B(\vec{x}) = Σ_i \gauss(\dist(\vec{x}_i, \vec{x})) * C \] where \(\vec{x}_i\) is the position of each cluster.
So in principle the error on \(N\) is just \(\sqrt{N}\). From here we can perform error propagation to get the uncertainty on \(B\). Investigate the correct way to do this!
Klaus thinks it might be correct to just use the relative error \(1/sqrt{B}\) at each point.
26.40.1. TODO Investigate effect of line veto
Two cases:
- line veto only in gold region (should give same as gold only for things acting in gold region)
- attempt line veto over the whole chip!
26.40.2. TODO Study error on background rate
Investigate the error propagation that is at play (see eq. above).
Make use of Julia's measurements package maybe to see what it computes for an uncertainty on each \(x\) passed through a sum over gaussians of the \(x\).
26.40.3. STARTED Compute other radius etc. pairs
Look at values smaller than 25 pixels & larger energy radius for the small values!
26.41. Meeting Klaus about TPA analysis
We discussed the uncertainties associated with the background interpolation, in part my ideas in section 29.1.3.5.
The idea being that we have statistical fluctuations in the background, plainly from not having unlimited statistics.
If we restricted ourselves to a non-weighted nearest neighbor, we would have a number \(N\) of clusters within a search radius. These have \(\sqrt{N}\) errors associated with them.
This uncertainty can be propagated through the normalization process to yield the final uncertainty.
For the gaussian weighted nearest neighbor, things are slightly more
complicated, but not significantly so (I think). Instead of taking
1 ± 1 for each element and summing over it (what one implicitly does
for the unweighted case! That's where the \(√N\) comes from after all),
we should do the right thing, if we use 1 ± 1 * weight and error
propagate from there. This means points further away contribute less
to the total error than closer ones!
However, this is only the statistical uncertainty. There may be a systematic uncertainty, if the background itself varies over the full area. The statistical uncertainty is the full uncertainty, iff the rate at the center (i.e. the computed number) is the same as the mean value over the range. Otherwise, it is biased towards any side. If the background varies linearly, it should still be correct for this reason.
26.41.1. STARTED Fix the gaussian error propagation code above and explanation
Just add what we learned (and wrote here) to the code. Write to Klaus about it.
26.41.2. TODO Implement MC toy sampling for background uncertainty
We can cross check whether things work as we expect them to, by doing Monte Carlo experiments.
Do the following:
- draw clusters (like the background clusters we have) all over the chip (O(10000) for the whole chip)
- compute the background at a single point with our fixed radii & energy (note: energy can be left out and divide total clusters to reasonable number in that energy range)
- run many times, store resulting background level
- distribution of it should be according to our statistical uncertainty that we compute
- Further: apply a linear and nonlinear weighing to the background that is drawn and see what the result of it is on the uncertainty.
26.42. Meeting Klaus about TPA analysis
Discussion about singularities in uncertainty code. Relevant section 24.2.1.
The case for b * (1 + θ) leads to a singularity at \(θ = -1\) due to
appearing in the denominator.
Our takeaways were as follows:
- if no analytical solution found, we can still integrate numerically
- integration in the area \(θ = -1\) an smaller is rather unphysical anyway. It corresponds to having a modified background of 0 or even < 0. This implies a rather bad estimation of our background and in terms of σ away from the hypothesis of the "correct" background, this is extremely unlikely! Shouldn't contribute to integration.
- \(θ > -1\) can be treated as the lowest integration range for the
nuisance parameter integration? Attempt with
assume(θ > -1)or similar in sage and see if we get analytical solution.
There are of course methods to deal with integrals over many nuisance parameters (even some "analytical approximations according to Klaus), but much more complicated. If can be avoided, good!
So:
- attempt to integrate with sagemath
- if at least one of 2 parameters can be integrated analytically, lifts burden of second (that may be numerical still)
26.43. Meeting Klaus about TPA analysis [3/5]
First of all we discussed the idea of using a log-normal distribution instead of a gaussian for the nuisance parameters.
The issue with that approach however, is that it's non trivial to rewrite it such that the log-normal has a mean of 0 around our desired \(θ\). Doing a simple substitution doesn't result in anything sensible, see 1 about that. In that case maybe the issue is that we should use the mean of the distribution as the starting point, namely \(\exp{μ - σ²/2}\) instead of substituting to get a gaussian exponential.
In any case, the complexity required means we'll just use a normal distribution after all that we cut off at some specified value for the integration. This is understandable and well defined.
Maybe I'll play around with the log-normal by subtracting the mean, but for now it doesn't matter.
The following things are the next TODOs:
[X]create likelihoodb.pdf plot (scan of \(θ_b\)) for a larger \(σ\) to verify that a cutoff of~-0.8or so is justified[ ]implement uncertainty on the number of drawn candidates. How? XXX[X]add function to compute expected limit, defined by the median of all toy limits[X]create plots of behavior of expected limit under different uncertainties. Range of background uncert, range of signal uncert & both. Point / line plot of these might be nice.[ ]play around with log-normal after substitution based on mean of distribution
26.44. Meeting Klaus about TPA analysis [0/5]
Initially we discussed the study of the behavior of the expected limits on the size of the uncertainty, see sec. 24.2.1.10.
That behavior looks good in our eyes, meaning we'll go on from here keeping the \(θ_b = -0.8\) cutoff for the background case integration.
This implies the next step is to understand what values we should attribute to each uncertainty input. The aspects mentioned in sec. 24.1 all provide some input to an uncertainty. The idea is to create a table of these and assign a value to each. Then we can add the squares of these to get the weighted sum of uncertainties. This should give us a sane value to use as a single number for signal and background uncertainty.
There is however one uncertainty that is more problematic: namely the
position of the axion image on the chip (i.e. the result of the ray
tracing). The issue is that this does not directly affect the amount
of signal received on the chip, but only the distribution and thus the
local s / b at each candidates position.
We have the following data points to judge the position of the spot:
- laser alignment before installation. This is likely to be the most accurate data point. The spot was (iirc) in the center up to O(1 mm) accuracy. Installation of detector should be pretty much at the same place, as the mounting doesn't leave too much room for error.
- geometer measurements of the position. We slightly aligned the position after installation according to the geometers. We have access to these measurements and need to check a) what we measured against and b) how accurate our placement was compared to the target.
- X-ray finger runs: in theory the X-ray finger runs provide an exact measurement of the location of the telescope spot on the detector. However this comes with a big problem. Based on the studies done by Johanna for IAXO, many things impact the resulting image in the detector. It is unclear at this time what the effect of the emission characteristic (in angular size & direction) is and where the X-ray finger was even placed. As such it is very tricky to make a statement about the location based on the X-ray finger.
As such our idea is to implement the uncertainty of the raytracing signal location in x and y position. We will assume a center position as the "best guess" based on laser alignment. From there the position can be varied using a parameter \(θ\) with an added gaussian penalty term that penalizes any position away from the center.
TODOs:
[ ]create table of uncertainties and try to come up with reasonable values for each of the discussed uncertainties[ ]implement uncertainty for raytracing position varying in x/y. Needs first the different position & then a gaussian penalty term for each axis.
Secondary TODOs:
[ ]find pictures of the laser alignment[ ]analyze the X-ray finger runs again and compute the location of the center[ ]talk to Johanna about the X-ray finger. Maybe simulate the result for such a source? See how it varies?
26.45. Meeting Klaus about TPA analysis
Discussion of the table about the systematic uncertainties.
Going well so far, need to add mainly the things about background now.
Then add x/y movement of the raytracing signal as nuisance parameters now.
Not much to discuss, as I just told him what I'm working on right now. Good progress.
26.46. Meeting Klaus about TPA analysis [0/3]
Discussed the progress on the table of uncertainties.
The state as of now:
| Uncertainty | signal or background? | rel. σ [%] | bias? | note | reference |
|---|---|---|---|---|---|
| Earth <-> Sun distance | signal | 3.3456 | Likely to larger values, due to data taking time | 24.1.4.1 | |
| Window thickness (± 10nm) | signal | 0.5807 | none | 24.1.4.2 | |
| Solar models | signal | < 1 | none | unclear from plot, need to look at code |  |
| Magnet length (- 1cm) | signal | 0.2159 | likely 9.26m | 24.1.4.3 | |
| Magnet bore diameter (± 0.5mm) | signal | 2.32558 | have measurements indicating 42.x - 43 | 24.1.4.3 | |
| Window rotation (30° ± 0.5°) | signal | 0.18521 | none | rotation seems to be same in both data takings | 24.1.4.4 |
| Alignment (signal, related mounting) | signal (position) | 0.5 mm | none | From X-ray finger & laser alignment | 24.1.4.4 |
| Detector mounting precision (±0.25mm) | signal (position) | M6 screws in 6.5mm holes. Results in misalignment, above. | |||
| Gas gain time binning | background | 0.26918 | to 0 | Computed background clusters for different gas gain binnings | 24.1.7.1 |
| Reference dist interp (CDL morphing) | background | 0.0844 | none | 24.1.7.2 | |
| Gas gain variation | ? | Partially encoded / fixed w/ gas gain time binning. | |||
| Random coincidences in septem/line veto | |||||
| Background interpolation | background | ? | none | From error prop. But unclear interpretation. Statistical. | 24.1.6.1 |
The new numbers for alignment, window rotation, gas gain time binning and reference distribution interpolation all look good.
Variation in gas gain is already decoded in the gas gain time binning and not really important as a separate thing.
Random coincidences: Klaus and me agree that their impact will be an effective reduction in the real tracking time. Thus it affects both background and signal. Need to come up with the equation for random coincidences for such models & compute the random coincidence rate. That rate can be turned into an effective dead time of the detector.
Background interpolation: the background interpolation systematics are particularly difficult because they are a "convolution" of statistical and systematic effects. In the real data it is hard to estimate, because changing a parameter changes the statistical influence by chance. Hard to deconvolve. Instead: use the MC models with flat background hypothesis that we already did and compute many MC toys of different cases. Then compare the background rate over e.g. the gold region with different parameters and the variation visible will be a measure for the influence of systematics on the method. Many toy models than can be used to deduce syst. uncertainty (by making statistical influence negligible).
Further, the things we already did there for different models also plays a role as a systematic. Tell us if some variation due to wrong assumption about background distribution or not.
Finally, energy calibration as a topic itself. Energy calibration in particular as a total thing. What happens if energy miscalibrated? Compute energy of calibration runs after energy calibration. The remaining variation there is a good indication for the systematic influences.
[ ]compute energy of peaks of 55Fe spectra after energy calibration. The variation visible there should be a sign for the systematic effects beyond variation in gas gain that are not encoded in the gas gain / energy calibration factor fit.[ ]compute random coincidences from theory. Need shutter time, cosmic rate & effective length of a single event + size of chips.[ ]use the MC modeling of the background interpolation with multiple biases as a basis for a study of the systematics with different parameters.
26.47. Meeting Klaus about TPA analysis [0/8]
Good meeting, mainly discussion of the progress on the θx and θy nuisance parameters & having understood their behavior. Changing sigma & candidates leads to an understandable change in the likelihood values.
Main focus then was on how to proceed from here. The TODOs are as follows:
[ ]talk to Igor about state of analysis. To give him idea where we are etc. Ask him what he thinks we should do with our (expected) result. What kind of paper should this be?[ ]prepare explanation talk of expected limit methodology for 18 May CAST collaboration meeting. This would be the "can we unblind our data" presentation. Needs to explain everything we do to give people the last time to ask "should we do anything differently?"[ ]figure out if we can we combine our limit w/ old 2013 gae limit & 2014/15 GridPix data. "think 1h about how hard it would be" according to Klaus.[ ]to test upper limit on impact, run the current limit with double the solar tracking time. Need to adjust background time scaling, expected signal rates & number of drawn clusters
[ ]talk to Johanna about MPE telescope simulation[ ]check if there are more systematics we need to estimate.[ ]fix limit calculation for θx and θy to be more efficient. Instead of using a linear scan in \(g_{ae}\) do some kind of binary search (in log space) for example and/or use an extrapolation based on few points (a la newton method)- See also: https://arxiv.org/pdf/1007.1727.pdf
[ ]combine θx, θy nuisance parameters with θs and θb[ ]finish a draft of analysis paper & write as much as possible on thesis until mid May to have an idea of how much longer to extend my contract! :DEADLINE:
26.48. Meeting Klaus about TPA analysis
Discussion of the implementation of an adaptive limit calculation method (see sec. 24.2.1.14). Klaus agrees that it seems like a good solution.
Further, discussion of the different tracking time 'experiments' (i.e. just scaling from 180 to 360 h), see sec. 24.2.1.15. Seems reasonable, is a good upper limit on what might be achievable and motivation to try to combine the data.
Finally, next steps:
[ ]implement 4 fold integration over \(θ_x\), \(θ_y\), \(θ_s\) and \(θ_b\)[ ]if too slow, talk to Hugo & c-blake about ideas how to make it fast. Maybe Metropolis Markov Chain based integration?[ ]else, maybe we can sort of factorize the terms? Maybe not fully exact, but approximate?
[ ]software efficiency systematics. Investigate by taking calibration runs, applying simple cut on transverse RMS and eccentricity and then propagating through likelihood cut method. Compute ratio for an approximate number on systematic uncertainty on software efficiency, at least at those energies. (even 2 energies!)[ ]talk to Johanna again about implementing MPE telescope into raytracer and make everything work for the old code. This part mainly depends on one single thing: we need our "analyze everything" script that we can essentially just feed different input data & it "does the right thing™". Once that is done, just need to change minor things and be able to compute limit.
26.49. Meeting Klaus about TPA analysis
The meeting discussed the "fallout" of the "CDL mapping bug" (section 14.7).
The resulting software efficiencies finally look fine (though we still need to investigate why the first 3 calibration runs have lower effective efficiencies!).
Klaus convinced me that the software efficiency is only an uncertainty on the signal. He argued that the actual logL cut value remains unchanged, which is perfectly true. The only thing that is uncertain is the corresponding efficiency. I thought in the wrong direction, thinking of actually changing the ε, which is not what happens during an uncertainty after all!
With it this concludes our systematics study for now.
So the next TODOs are as follows:
[ ]compute combined uncertainties for signal & background[ ]compute the "final" expected limits based on MCMC using the correct σ values[ ]prepare outline & talk for the CAST CM
26.50. Meeting Klaus about TPA analysis [2/4]
The main topic was on the expected limit stuff.
There are 3 (~4) things to do next:
[X]determine the correct "desired" focal distance / distance at which to compute the raytracing image. This should be done by using Beer-Lambert's law and checking the average depth of X-rays in the relevant energy range (say 0.5 - 3 keV). Convolving the axion spectrum with the absorption coefficient at specific energies should yield us something like an effective "absorption distance" that should be the correct number.[X]The strongbacks of the windows must not be moved with the signal spot when working with the position uncertainty systematic. As such we need to separate the strongback "signal" and the raytracing signal w/o strongback and only combine them in the limit code.[ ]fix the limit code for multithreading. Currently causes all sorts of segfaults.[ ]write mail to CAST PC about talk at Patras.
26.51. Meeting Klaus about TPA analysis [2/3]
Showed the calculation for the effective absorption point in the detector, sec. 3.3.1 (a depth of 1.22cm in the detector).
Then the updated axion image for the new depth, see sec. 11.4 without the window strongback. The strongback then is handled separately via a simple computation based on what's done in the raytracer as well, see 2.8.1.
The things left to do now are:
[X]implement the strongback / signal separation into the limit code[X]validate the MCMC approach again properly[ ]maybe manage to fix the multithreading segfaults we get
26.52. Meeting Klaus about TPA analysis
- abstract for Patras. Send it in?
- implementation of strongback / signal separation in the code
- MCMC using multiple chains & longer burn in: results in ~7.55e-23 expected with very bad statistics
In the end this is precisely what we discussed.
The next step is simply to speed up the limit calc performance by
using procPool, i.e. a multi process approach.
Hopefully done by Tuesday.
26.53. Meeting Klaus about thesis [/]
[ ]write email to Igor asking about open data & if we are allowed to host the data on Zenodo for example[ ]after the above is done, we can also ask the new HISKP professor (?) Sebastian Neubert (was supposed to give colloquium today) about the same. He's apparently into open data @ CERN stuff and even has some positions at CERN about it. The idea being that maybe we can even get some publicity out of publishing all the data!
26.54. Meeting Klaus about thesis [/]
Questions for meeting with Klaus today:
- Did you hear something from Igor? -> Nope he hasn't either. Apparently Igor is very busy currently. But Klaus doesn't think there will be any showstoppers regarding making the data available.
- For reference distributions and logL morphing:
We morph bin wise on pre-binned data. This leads to jumps in the logL cut value.
Maybe a good idea after all not use a histogram, but a smooth KDE? Unbinned is not directly possible, because we don't have data to compute an unbinned distribution for everything outside main fluorescence lines! -> Klaus had a good idea here: We can estimate the systematic effect of our binning by moving the bin edges by half a bin width to the left / right and computing the expected limit based on these. If the effective limit changes, we know there is some systematic effect going on. More likely though, the expected limit remains unchanged (within variance) and therefore the systematic impact is smaller than the variance of the limit.
[ ]DO THIS
About septem veto and line veto: What to do with random coincidences? Is it honest to use those clusters? -> Klaus had an even better idea here: we can estimate the dead time by doing the following:
- read full septemboard data
- shuffle center + outer chip event data around such that we know the two are not correlated
- compute the efficiency of the septem veto.
In theory 0% of all events should trigger either the septem or the line veto. The percentage that does anyway is our random coincidence!
[X]DO THIS
26.55. Meeting Klaus about thesis [/]
We discussed the estimation of random coincidences in the septem and line veto. It's a bit unfortunate (see results of that in thesis & here), but seems realistic that given the long shutter times there is a significant random coincidence rate.
One important point we realized is that of course this does not only introduce a longer dead time relative to the tracking time, but also the background time! So the time we use to calculate the background rate from also changes.
Finally, it would be a good idea to see how big the impact is of the line veto alone. For this we need to refactor the code slightly to allow to apply the line veto without the septem veto. My argument for why this is non trivial was that the line veto needs all the septem board event reconstruction after all. And it brings up the important question: Which cluster does the line have to point towards? The original one or a potential one that is reconstructed using outer chip information?
There are good arguments for both kind of. Original one implies that one doesn't want to "trust" the septem veto. The issue is that the pixels that would otherwise become part of the new combined cluster could likely just point at the original one? Or rather it's just a bit weird on how to deal with those clusters.
There will be some trade off between efficiency and random coincidence rate here as well.
Anyhow, implement this first.
26.56. Meeting Klaus about thesis [0/0]
Points to talk about: Note in mail that properties on left are currently mismatched!
- FADC veto cut had significant bug in it, upper end was never used!
- fixed issues with FADC rise & fall times, improved stability, much cleaner rise / fall time data.
line veto:
- show example without correct layout, why important
- show example with correct septemboard layout
- show examples for each line veto kind
- explain line veto eccentricity cutoff
- show plot of line veto ecc cutoffs & ratio plot
- Q: What should we really look at in terms of real fraction passing vs. fake fraction passing?
-> Our conclusion for now: lvRegularNoHLC and εcut = 1.5
These plots together are in
Discussed these plots: Generally Klaus agreed with my thoughts, with two points to remark:
- The line veto kind "1" (lvRegular) might still be the correct choice of course (especially if no septem veto used).
The fraction of events passing is WEIRD: The number of events passing for the fake data is much lower than it was previously when using the line veto alone (after implementing that!). See the notes about that, but we had an acceptance of about 1570/2000 (or so) on average. MUCH HIGHER than the value we now get at any cluster eccentricity cut! The old setting should correspond to the following:
- line veto kind:
lvRegular - use real layout:
false - eccentricity cutoff: 1.0
Therefore: use these settings and run the code to see what kind of fraction passes here! Note: there is a chance on of our changes broke something! For example the changes to how a line veto is considered passing etc.
- [X]: I already checked the raw numbers found in the output
.txtfiles that contain the fraction numbers and e.g. for fake εcut=1.2 they are indeed only about 850/2000!
- line veto kind:
Generally: we don't want to spend time on optimizing any parameters nilly willy, instead look at the impact on the expected limit! So the idea is (aside from a short look if we can reproduce passing fractions) to just continue on and then build a setup that allows to combine:
- define a setting to use for parameters (i.e. a
config.tomlsetup or similar) - calculate the relevant likelihood output files
- calculate the expected limit based on these files
We need to investigate that once we're there, but the two tricky aspects:
- make sure the expected limit code does not contain any hard coded numbers that do not come from the H5 files
- the time it takes to evaluate an expected limit is rather long, i.e. we can't afford to run very large numbers of parameters! -> Ideally we'd do a non linear optimization of the perfect set of all parameters.
26.57. Meeting Klaus about thesis
The main talking point was the analysis of the line veto depending on the line veto kind (this time with correct results). The septem veto + line veto wasn't fully done yet! Only 3 points were ready.
Our take away from looking at the plot (see thesis / section in this file for the final plot) was that we care about one of these two cases:
- line veto kind
lvRegularwith eccentricity cut 1.0 - septem veto + line veto (either veto kind should be fine) with eccentricity cut 1.5
The septem veto does add quite some dead time (about 27% dead time compared to maybe 12-13% in the lvRegular only case).
So the question is a bit on what the impact on the expected limit is!
Further I told Klaus about my PyBoltz calculation & expectation of the rise time etc. He was happy about that (although said I shouldn't have wasted time with such things, haha).
Anyhow, the important bit is:
[ ]Apply thelikelihoodwith all different sets of vetoes etc, (including the two septem + line veto cases discussed above) and then calculate the expected limit for each of these files.
26.58. Meeting Klaus about thesis
I wasted some more time in the last week, thanks to FADC veto…
Plots:
- /tmp/playground/backgroundrate2017crGoldscintifadc160.pdf -> Background rate of Run-2 data using rise time cut of 160
- /tmp/playground/backgroundrate2018crGoldscintifadc120.pdf -> Background rate of Run-3 data using rise time cut of 120
- /tmp/playground/backgroundratecrGoldscintifadc120.pdf -> Background rate of all data using cut of 120
- /tmp/playground/backgroundratecrGoldscintifadc105.pdf -> Background rate of all data using cut of 105
What do rise times look like?
- ~/org/Figs/statusAndProgress/FADC/oldrisefallalgorithm/fadcriseTimekdesignalvsbackgroundrun3.pdf -> Signal vs background
- /tmp/Figs/statusAndProgress/FADC/oldrisefallalgorithm/fadcriseTimekdeenergydeprun3.pdf -> Photo vs escape peak
- /tmp/Figs/statusAndProgress/FADC/oldrisefallalgorithm/fadcriseTimeridgelinekdeenergydepless200riseCDL.pdf -> CDL data
Where do we expect differences to come from? Absorption length:
- /t/absorptionlengthargoncast.pdf -> Absorption length argon CAST conditions
Do we see this in data?
- /t/fadc95-thvsabsLengthbytfkind.pdf -> 95th percentile of rise times for CDL data against absorption length
- /t/fadcmeanvsabsLengthbytfkind.pdf -> mean of the same
- /t/fadcMPVvsabsLengthbytfkind.pdf -> most probable value (mode) of the data -> does NOT really show the expected behavior interestingly enough!
All plots combined here:
Big takeaways from meeting:
- Klaus agrees the FADC veto is something worthwhile to look into!
[ ]need to understand whether what we cut away for example in 3 keV region of background is real photons or not! -> Consider that MM detectors have Argon fluorescence in 1e-6 range! But they also have analogue readout of scintillator and thus higher veto efficiency![X]Change the definition of the FADC rise time to be not to the minimum, but rather to a moving average of minimum value - percentile (e.g. 5% like for baseline!) -> Klaus mentioned that a value too far away might introduce issues with "time walk" (here's that term again…!) -> Done.[X]Check the events which are removed in the background rate for riseTime cut of 105 that are below the FADC threshold! -> My current hypothesis for these events is that they are secondary clusters from events with main clusters above the FADC activation threshold! -> See sec. 8.4.1 for the reason (hypothesis correct). Having understood the origins of this, the big question remains:[ ]What do we do with this knowledge? Apply some additional filter on the noisy region in the top chip? For the events that look "reasonable" the FADC rise time there might precisely be the stuff that we want it for. The events are vetoed because they are too long. Given that there are tracks on the outside, this means they are definitely not X-rays, so fine to veto!
[X]It seems to us that the CDL data does indeed prove that we do not need to worry much about different energies / different absorption lengths (the ones where it is significantly different are below threshold anyways!)[X]We want to determine the correct cutoff for the different datasets (FADC settings) and different energies / absorption lengths from the calibration data. However, need to understand what is still going on there. Is the data as plotted above reliable? -> What are the events that have rise times towards the tail, e.g. >130? Maybe those disappear when changing the rise time definition? -> These were due to larger than anticipated noise levels, needed longer 'offset' from baseline. MaderiseTimemuch more narrow.[X]handle the different FADC settings in the Run-2 data. This will likely be done as a "side effect" of defining the cuts for each calibration run separately (maybe we use a mean later, but then the mean needs to be done by FADC setting) -> There is clearly a difference in the rise times of these and they will be handled separately (sec. 8.2.3).
26.59. Meeting Klaus about thesis [/]
(Note: this meeting was postponed to ).
[X]Check the events which are removed in the background rate for riseTime cut of 105 that are below the FADC threshold! -> My current hypothesis for these events is that they are secondary clusters from events with main clusters above the FADC activation threshold! -> See sec. 8.4.1 for the reason (hypothesis correct). Having understood the origins of this, the big question remains:[ ]What do we do with this knowledge? Apply some additional filter on the noisy region in the top chip? For the events that look "reasonable" the FADC rise time there might precisely be the stuff that we want it for. The events are vetoed because they are too long. Given that there are tracks on the outside, this means they are definitely not X-rays, so fine to veto!
All the plots mentioned below are here:
- Events, die in der background rate unterhalb der FADC Schwelle dennoch vetoed werden: Quasi alles Events, in denen es auf dem Chip oben rechts einen Spark gab, siehe Seite 1. /tmp/playground/figs/DataRuns2018Reco2023-02-2203-37-37/septemEvents/septemfadcrun274event21139regioncrAlltoaLength-0.020.0applyAllfalse.pdf
- Algorithmus für rise/fall time hat nun auch einen Offset vom Minimum. Seite 2 55Fe X-ray mit gestrichelten Linien, die zeigen bis wo / von wo die rise / fall time bestimmt wird. /t/exampleeventalgorithmupgrade.pdf Seite 3: Eins der Beispiele (wie Seite 1), die unterhalb der FADC Schwelle vetoed wurden. Hier sieht man jetzt gut anhand der ~/org/Figs/statusAndProgress/FADC/improveRiseFallCalc/septemfadcrun279event15997regioncrAll.pdf Linien, dass Start und Stopp sinnvoll sind.
- Als nächstes hab ich geschaut, was in den 55Fe Verteilungen der rise time noch im Tail (größer 140 in den Plots Anfang der Woche) noch vorhanden ist. Seite 4: Ursache ist, dass etwas stärkere Variation der baseline als erwartet manchmal dazu führt, dass wir zu spät wieder zu baseline - offset kommen und damit das Signal deutlich länger wird. /tmp/exampleeventriseTimetail55fe.pdf Lösung: größerer Offset von 10% der Amplitude (baseline ⇔ peak) Seite 5: gleiches Event mit neuem Offset. ~/org/Figs/statusAndProgress/FADC/improvedrisefallalgorithm/calibriseTimeabove140/eventrun239event106810percenttopoffset.pdf
- Wie sieht generell die rise time Verteilung nach beiden
Modifikationen aus?
Seite 6: Photo vs escape, generell natürlich bei noch kleineren
Werten (wir kürzen die Signale ja nun mal) und die "tail
contributions" sind quasi komplett weg!
Seite 7: Was bedeutet das aber für signal vs. background? In erster Linie sind die signal Daten deutlich besser definiert, aber die
Untergrund weiterhin sehr breit! Seite 8: Einfluss auf die CDL Daten. Vor allem die höheren Energies sind deutlich besser definiert. In den Fällen Ag-Ag-6kV, Mn-Cr-12kV und Cu-Ni-15kV sieht man einen deutlicheren "bump" bei kleineren Werten. Das müsste quasi der Einfluss der deutlich höheren Absorptionslänge in diesen Fällen sein! (Ti-Ti-9kV & Al-Al-4kV haben jeweils eine kurze Absorptionslänge)
- Wie sieht die Untergrundrate mit einem Veto auf Basis von recht
"loosen" Cuts auf die rise time aus? (40 - 70)
Seite 9: Run-3 Daten (wegen unterschiedlichen FADC Einstellungen in
Run-2, die ich noch einzeln betrachten muss).
Sehr ähnlich zu einem ähnlichen groben Cut vorher (background Run-3
in PDF der letzten Mail; in der Tat sind dort noch ein paar Cluster weniger).
Seite 10: ROC Kurve für die rise time auf Basis der Run-3 55Fe Daten gegen Untergrund. Damit wir eine Idee haben wie viel die rise time alleine etwa bringen sollte und bis wo man sehr viel gewinnt. (x: Signaleffizienz, y: Untergrundunterdrückung) /t/roc.pdf
26.60. Meeting Klaus about thesis [/]
The text describing the plots above will be the main discussion.
The explanations for what we do, why we do it and what the results show were satisfactory. Especially the improvement to the narrowing of the rise time distribution after the 10% offset from baseline change were seen very nicely. Klaus joked that maybe one day we do achieve our factor 30x improvement over the old detector, haha.
We agreed to set the cut values for the rise time based on a fixed percentile. So that we have multiple sets of FADC cut values that we might want to use to compute an expected limit for.
One important thing we discussed: Each veto has a different dead time associated to it (due to its signal efficiency or random coincidence). However, these can be correlated in theory. Therefore multiplying the efficiencies of each veto is a conservative estimate of the total time used! Ideally we would somehow compute how and whether they are correlated, but that seems difficult to do, because we don't have a known dataset that can be used for that.
Thought: generate fake events from known X-rays (via calibration data) and known outside events (via background data) and apply all vetoes to that?
[ ]THINK ABOUT THIS
From here the main way forward is:
[X]Finish the implementation of the FADC vetoes being dependent on a fixed percentile of the calibration data[X]Do this on a per FADC setting basis
[X]Implement allowing a fixed percentile from command line argument for the FADC rise time veto[X]Extend existing parameters set via environment variables to also be set-able via command line arguments[ ]Finish the limit calculation logic[X]Write some code to automate runninglikelihoodwith different parameters & then limit calculation to get different expected limits for each set of parameters. These parameters are all the different vetoes and their settings.
26.61. Meeting Klaus about thesis [/]
Discussion of the following table mail:
Hey,
ich hab am Wochenende testweise mal Limits für verschiedene Setups berechnet. In erster Linie, um zu testen, ob die Automatisierung prinzipiell funktioniert. Im eigentlichen Limit Code fehlen jetzt noch die Anpassungen bzgl. der Totzeit und FADC Effizienz, weshalb die Zahlen nicht wirklich aussagekräftig sind.
limit name 8.9583e-23 no vetoes 8.8613e-23 scinti 8.6169e-23 scinti + fadc 7.3735e-23 scinti + fadc + line 7.6071e-23 scinti + fadc + septem 7.3001e-23 scinti + fadc + septem + line Die Limit Spalte hier ist jetzt der Median aus 2000 expected limits jeweils und bereits als gae · gaγ.
Der Vergleich hierbei wäre jetzt immer noch die 8.1e-23.
Ob sich die Tatsache, dass das Line Veto alleine vergleichbar gut ist, wie Septem + Line veto bewahrheitet wenn man mehr als 2000 Limits berechnet wird sich zeigen. Aber durch die geringere Zufallskoinzidenzen wenn wir nur das Line Veto nutzen besteht dabei womöglich die Chance, dass es insgesamt darauf hinaus laufen könnte als bestes Limit.
Für den FADC muss ich dann auch noch 3 verschiedene Cut Werte auf Basis verschiedener Quantile berechnen (im oberen Fall ist das das 99. Quantil).
Viele Grüße und bis gleich, Sebastian
which are the different setups and results from sec. 29.1.11.5.
Klaus agreed that the line veto alone now does indeed look very promising.
In addition the 2000 toys seems reasonable. The main question to us is:
What is the variance / uncertainty on the median of the expected limits? That is what we care most about, in particular we want a certainty that sort of relates to how far we are away from the 8.1e-23.
[ ]Check how to compute uncertainty of the median of a distribution[ ]We could compute that ourselves by seeing how the expected limit changes, if we recompute 2000 toys with a different RNG seed of course!
[ ]Finish implementation of random coincidence dead time & FADC efficiency[ ]Compute 2 other FADC efficiencies, 95-th and 90-th percentiles[ ]Compute expected limits for all these cases
26.62. Meeting Klaus about thesis [/]
We discussed the following table, which I sent as a mail to him:
| \(ε_{\ln\mathcal{L}, \text{eff}}\) | Scinti | FADC | \(ε_{\text{FADC, eff}}\) | Septem | Line | Efficiency | Expected limit (nmc=1000) |
|---|---|---|---|---|---|---|---|
| 0.8 | x | x | - | x | x | 0.8 | 8.9258e-23 |
| 0.8 | o | x | - | x | x | 0.8 | 8.8516e-23 |
| 0.8 | o | o | 0.98 | x | x | 0.784 | 8.6007e-23 |
| 0.8 | o | o | 0.90 | x | x | 0.72 | 8.5385e-23 |
| 0.8 | o | o | 0.80 | x | x | 0.64 | 8.4108e-23 |
| 0.8 | o | o | 0.98 | o | x | 0.61 | 7.5671e-23 |
| 0.8 | o | o | 0.90 | o | x | 0.56 | 7.538e-23 |
| 0.8 | o | o | 0.80 | o | x | 0.50 | 7.4109e-23 |
| 0.8 | o | o | 0.98 | x | o | 0.67 | 7.3555e-23 |
| 0.8 | o | o | 0.90 | x | o | 0.62 | 7.2889e-23 |
| 0.8 | o | o | 0.80 | x | o | 0.55 | 7.2315e-23 |
| 0.8 | o | o | 0.98 | o | o | 0.57 | 7.3249e-23 |
| 0.8 | o | o | 0.90 | o | o | 0.52 | 7.2365e-23 |
| 0.8 | o | o | 0.80 | o | o | 0.47 | 7.1508e-23 |
| 0.8 | o | x | - | o | x | ||
| 0.8 | o | x | - | x | o | ||
| 0.8 | o | x | - | o | o |
--vetoSet {fkNoVetoes, fkScinti, fkFadc, fkSeptem, fkLineVeto, fkExclusiveLineVeto} # for the first chunk
--vetoSet {+fkScinti, fkSeptem, fkLineVeto, fkExclusiveLineVeto} # `+` indicates only always *added*, so does not recreate no vetoes or only scinti
We're both very happy with the results!
One thing that tickles us a bit is that the "all in" case is still better than any other. So we're still only seeing improvements. What Klaus mentioned is that one would expect that a lower efficiency should push the \(R_T = 0\) case further right, while the improvements done by the lower efficiency should push the average expected limit with candidates further left.
One thing that bothers us a tiny bit is that the FADC veto likely has the biggest systematic uncertainties, because it is the least studied, the FADC had the most problems and we have the least data to be certain of its energy dependence (+ absorption length dependence). This makes it questionable to run a "fixed" efficiency, because it will vary based on (effectively) energy.
[X]Calculate expected limits also for the following cases:[X]Septem, line combinations without the FADC[X]Best case (lowest row of below) with lnL efficiencies of:[X]0.7[X]0.9
-> See journal.org, but necessary stuff implemented and run, results
in:
./../resources/lhood_limits_automation_correct_duration/
[ ]Run the limit calculation on all combinations we care about now:
[ ]Verify that those elements with lower efficiency indeed have \(R_T = 0\) at higher values! -> Just compute \(R_T = 0\) for all input files and output result, easiest.
26.63. Meeting Klaus about expected limits and MLP [/]
We're generally in agreement about the state of things. Unfortunate that expected limit seems worse than 8.1e-23 now. But maybe can improve slightly by altering parameters a bit further.
[ ]day before unblinding: Zoom meeting with Klaus, Igor, our group, maybe Zaragoza group in which I present the full method that we use (maybe 30min) and ask for any input whether someone sees anything wrong with it.[ ]redo all expected limit calculations with the following new cases:- 0.9 lnL + scinti + FADC@0.98 + line
- 0.8 lnL + scinti + FADC@0.98 + line εcut:
- 1.0, 1.2, 1.4, 1.6
[ ]for above: implement eccentricity cut off into efficiency! Currently have hardcoded efficiency for line veto, but that is dependent on the eccentricity cutoff![ ]understand the MLP better… See journal of today.
26.64. Meeting Klaus about MLP veto & random coincidence rate
26.64.1. MLP veto
I explained the current status of the MLP veto etc to Klaus and he agreed it all sounds reasonable. Including the 3 keV != escape photon properties. He was happy to hear that the diffusion fake generation seems to yield authentic events.
I told him to send him results once I have them.
26.64.2. Random coincidence rate
Klaus brought up an interesting point about our current estimation of the random coincidence rate.
First up: Different events have different event durations (obviously).
The set of events from which we bootstrap fake events is a set that will (in maybe 80% of cases?) have event durations less than ~2.2 s due to having an FADC trigger. The set of 'outer ring' events from which we sample the outer part is a mix of all 'event duration' data (mostly full length, as most events are full duration events and some shorter).
Potentially the random coincidence rate is actually not the fraction of events vetoed by the line / septem veto when evaluating the bootstrapped efficiency, but rather:
\[ ε' = ε · t_{\text{bootstrap}} / t_{\text{average}} \]
that is the actual efficiency (or dead time) is actually scaled down by the average duration of those events that participate in the random coincidences compared to the average duration of all events. This is essentially saying "there is a shorter time scale involved in which a septem / line veto can even happen / provide a random coincidence rate than the total time during which the detector was live".
(There is a further 'complication' due to how some events have no FADC trigger and therefore are always full length. But this is taken into account due to the sampling including both types).
Secondly: Another modification needs to be made based on what the typical event duration will be for real axion events. The axion spectrum provides a different distribution of events in energy. This is further modified by the detection efficiencies of the spectrum, which we know. All events below the FADC activation threshold definitely have the full event duration. Those above the the activation threshold are expected to give us a uniform distribution in the [0, 2.2s] event durations (i.e. the mean of that being 1.1s).
This time is the only fraction that can be affected by the random coincidence rate. This will be further smaller than the ε' provided above.
We can easily compute this numerically by computing the average durations for the final X-ray spectrum of the axions in our detector. Below FADC threshold constant, above uniform distribution.
[ ]Implement code to compute this[ ]Write down analytical expression that describes this!!! The below is incomplete
\[ t_S = \frac{ ∫ t_D(E) · S(E) dE }{ ∫ S(E) dE } \] where \[ t_D(E) = Θ(0, ~1.5 keV) · 2.2 s + Θ(~1.5 keV, ∞) · U(0 s, 2.2 s) \] is the energy dependent duration with \(U\) being a uniform distribution within the range 0 and 2.2 s.
[ ]FINISH THIS
[ ]Signal[ ]Random estimation
26.65. Igor about limit calculation
I had a ~45 min meeting with Igor about the limit computation this morning.
See also the mail I wrote to Klaus about this meeting.
The question we had came down to: Should we use the same method as used in the Nature paper. A Bayesian based unbinned likelihood fit.
Uncertainty for 2 reasons:
- does the method also produce unphysical results in a range that are removed via a sort of "rescaling".
- are there better methods he knows about for this.
The main tl;dr of the discussion is:
- he would use the same approach
- it's simple enough so that one can implement it manually and understand what's going on
- incorporating systematics is really easy (just add as gaussian nuisance parameter and include in fit)
- the analysis is simple enough that there really isn't a point in using a complicated approach. Those should ideally give the same results anyway!
- complex methods also do unphysical things or make decisions for you. Here one simply sees that directly.
His biggest pain point looking back: Keep the likelihood "phase space" as a result! That's the main important result from the limit calculation. In the future to combine it with other results etc. that's what one needs (which they didn't do).
Talked about 2002 paper about something F - something method for limit calc? Was it Feldman & Cousins? Possible: https://iopscience.iop.org/article/10.1088/0954-3899/28/10/313 Maybe write a mail to ask.
Book Igor recommended: Statistical Methods for Data Analysis in Particle Physics - Luca Lista
- http://people.na.infn.it/~lista/Statistics/
- https://www.springer.com/it/book/9783319628394
- https://www.amazon.de/-/en/Luca-Lista/dp/3319628399/
Starts from Frequentist vs. Bayesian and goes from there over topics needed for actual particle physics. Including things like "look elsewhere effect", etc.
26.66. CCM
Main takeaway:
- gave a short ad hoc "talk" about what we did since last meeting
- Horst asked me until when we have results
- in particular: in June there's the next CAST presentation. Should have a preliminary axion electron as well as chameleon limit!
- tell Klaus that Horst would like GridPix data taking for the proposal of extension of CAST by 1 more year!
27. Thesis plots [0/2]
[X]need a plot comparing the different background rates for different logL signal efficiencies in a single plot[ ]histogram (or similar) showing the different possible absorption positions of X-rays in the Argon gas of the CAST detector. I.e. take the logic that we use to compute the effective conversion point in the detector from section 3.3.1 and expand it to make a plot showing the amount of X-rays converting at what depths. Can be done either per energy or averaged over all energies. If we do it for all energies it will just be an exponential with absorption length 1.22 cm, iiuc. Can also do a cross check by doing this for different energies, sampled by the axion flux distribution and then computing the mean of all samples (as a MC simulation).
28. 2013 axion electron limit CAST paper data
This is the data manually extracted from the plot in the paper in order to run our analysis on their data. Thas should give us an idea what to expect in terms of methodology.
Bin width: slightly less than 0.3 keV Number of bins: 20 Known bins:
- idx: 3 @ 1.8 keV
- idx: 17 @ 5.8 keV
import seqmath let start = 1.8 let stop = 5.8 let idxStart = 3 let idxStop = 17 let knownBins = linspace(start, stop, idxStop - idxStart + 1) let binWidth = knownBins[1] - knownBins[0] var idx = idxStart var tmp = start while idx > 0: tmp = tmp - binWidth dec idx let firstBin = tmp idx = idxStop tmp = stop while idx < 19: tmp = tmp + binWidth inc idx let lastBin = tmp echo "From: ", firstBin, " to: ", lastBin echo linspace(firstBin - binWidth / 2.0, lastBin - binWidth / 2.0, 20) echo "Binwidth = ", binWidth
Those numbers match perfectly to the visual points of the plot
| Energy | Energy, binCenter | Candidates | Background |
|---|---|---|---|
| 0.7999 | 0.94287 | 1 | 2.27 |
| 1.0857 | 1.22857 | 3 | 1.58 |
| 1.3714 | 1.51428 | 1 | 2.4 |
| 1.6571 | 1.8 | 1 | 1.58 |
| 1.9428 | 2.08571 | 1 | 2.6 |
| 2.2285 | 2.37142 | 2 | 1.05 |
| 2.5142 | 2.65714 | 1 | 0.75 |
| 2.7999 | 2.94285 | 2 | 1.58 |
| 3.0857 | 3.22857 | 0 | 1.3 |
| 3.3714 | 3.51428 | 2 | 1.5 |
| 3.6571 | 3.79999 | 0 | 1.9 |
| 3.9428 | 4.08571 | 1 | 1.85 |
| 4.2285 | 4.37142 | 0 | 1.67 |
| 4.5142 | 4.65714 | 2 | 1.3 |
| 4.7999 | 4.94285 | 2 | 1.15 |
| 5.0857 | 5.22857 | 0 | 1.67 |
| 5.3714 | 5.51428 | 2 | 1.3 |
| 5.6571 | 5.8 | 1 | 1.3 |
| 5.9428 | 6.08571 | 2 | 2.27 |
| 6.2285 | 6.37142 | 2 | 1.3 |
The results shown below use the same systematic errors as the general case in 17.2.2.1 does, which means. Of course this is by far the biggest assumption we make, given that we don't know their estimations for systematics. But since we have seen that the influence of systematics is not earth shattering this seems fine for such a simple cross check.
Another thing this comparison of course completely ignores and which is certainly more important is the signal hypothesis or rather the losses due to telescope + detector efficiencies.
The telescope is comparable and should not make a big difference. However, detector related losses could be significant. The thing to keep in mind is:
- we know the sensitivity curve of the 2013 PN-CCD detector from the paper. We can push the plot through our data extractior tool and then use the resulting data as input for the detector efficiencies in the ray tracer. That way we can simulate the below results even with a reasonably correct signal hypothesis!
NOTE: The limits computed in the following section are affected by the "bad" axion-photon conversion probability as discussed in sections 17.5.2 and 11.2. For newer limits, read further in section 17.5.1.5.
28.1. Optimize for CLs
This is the case directly comparable to our main result.
"Tel" : SystematicError(cand: 0.05, back: 0.05) "Window" : SystematicError(cand: 0.10, back: 0.10) "Software" : SystematicError(cand: 0.05, back: 0.05) "Stat" : SystematicError(cand: 0.3, back: 0.1)
file:///home/basti/org/Figs/statusAndProgress/limit_2013_cast_gae_opt_cls.pdf file:///home/basti/org/Figs/statusAndProgress/limit_2013_cast_gae_opt_cls_sb.pdf
CLb = 0.23143 CLs = 0.04584168785829175 CLsb = 0.01060914182104446 <CLb> = 0.50001 <CLsb> = 0.03172895323341283 <CLs> = 0.063456637334079
Result for the coupling constant g_ae: 8.129294390231371e-10
28.2. Optimize for CLsb
"Tel" : SystematicError(cand: 0.05, back: 0.05) "Window" : SystematicError(cand: 0.10, back: 0.10) "Software" : SystematicError(cand: 0.05, back: 0.05) "Stat" : SystematicError(cand: 0.3, back: 0.1)
file:///home/basti/org/Figs/statusAndProgress/limit_2013_cast_gae_opt_clsb.pdf file:///home/basti/org/Figs/statusAndProgress/limit_2013_cast_gae_opt_clsb_sb.pdf
CLb = 0.25037 CLs = 0.2025176997691754 CLsb = 0.05070435649120843 <CLb> = 0.50001 <CLsb> = 0.1366923495017895 <CLs> = 0.2733792314189508
Result for the coupling constant g_ae: 5.077156714042002e-10
29. 2017 Nature paper
Ref:
The approach they use seems to be very similar to what is done in the 2013 paper for the axion electron coupling.
Essentially:
\[ \log \mathcal{L} ∝ - R_T + Σ_i^m \log R(E_i, d_i, x_i) \]
where \(R_T\) is the total expected number of counts from axion photon conversion and the sum is over all candidates in the tracking.
\(R(E_i, d_i, x_i)\) is the expected rate:
\[ R(E, d, x) = B(E, d) + S(E, d, x) \]
where \(B(E, d)\) is the (assumed constant) background hypothesis and \(S\) the total expected rate for a specific cluster:
\[ S(E, d, x) = \frac{\mathrm{d}Φ_a}{\mathrm{d}E} P_{a↦γ} ε(d, E, x) \]
where we have the differential flux expected from the sun (depending on \(g_{aγ}\) and / or \(g_{ae}\), the axion-photon conversion \(P_{a↦γ}\) depending on \(g_{aγ}\) and finally the detection efficiency \(ε\), which includes:
- gas absorption
- window transmission
- X-ray telescope efficiency
- and most importantly here: the ray traced flux of axion induced X-rays at the cluster position \(x\) (as a weight)
Question: Why is this encoded in \(ε\) and not in the differential flux?
In order to compute these things, we need (in addition to the code written for the reproduction of the 2013 limit calculation):
29.1. Reproduce the limit calculation method
- a smooth description of the background rate so that we can query the rate at the exact energy that a given cluster has: -> compute a KDE of the background rate. Question: how to compute rate of a KDE result? Have no bin width. -> normalize to 1, then adjust by area, time & total counts in input.
- get the heatmap (effective flux) from the raytracer. Question: how to compute the actual flux from that? Convert from N input?
- rest can be adapted from 2013 limit, I think.
import nimhdf5, os, sequtils, math, unchained, cligen, random, strformat, seqmath import ingrid / tos_helpers import numericalnim except linspace, cumSum import arraymancer except read_csv, cumSum # the interpolation code import ./background_interpolation defUnit(keV⁻¹•cm⁻²) type ChipCoord = range[0.0 .. 14.0] Candidate = object energy: keV pos: tuple[x, y: ChipCoord] ## TODO: split the different fields based on the method we want to use? ## SamplingKind represents the different types of candidate from background sampling we can do SamplingKind = enum skConstBackground, ## uses the constant background over the gold region. Only allows sampling in the gold region. skInterpBackground ## uses the interpolated background over the whole chip. UncertaintyKind = enum ukCertain, # no uncertainties ukUncertainSig, # uncertainty only on signal (integrated analytically) ukUncertainBack, # uncertainty only on background (integrated numerically) ukUncertain # uncertainty on both. Analytical result of ukUncertainSig integrated numerically PositionUncertaintyKind = enum puCertain # no uncertainty puUncertain # use uncertainty on position ## Stores the relevant context variables for the interpolation method Interpolation = object kd: KDTree[float] ## we use a KDTree to store the data & compute interpolation on top of backCache: Table[Candidate, keV⁻¹•cm⁻²] ## cache for the background values of a set of candidates. Used to avoid ## having to recompute the values in a single MC iteration (within limit computation). ## Only the signal values change when changing the coupling constants after all. radius: float ## radius of background interpolation (σ is usually radius / 3.0) energyRange: keV ## energy range of the background interpolation nxy: int ## number of points at which to sample the background interpolation in x/y nE: int ## number of points at which to sample the background interpolation in E xyOffset: float ## Offset in x/y coordinates (to not sample edges). Is `coords[1] - coords[0] / 2` eOffset: float ## Offset in E coordinates (to not sample edges). Is `energies[1] - energies[0] / 2` coords: seq[float] ## the coordinates at which the background interpolation was evaluated to ## compute the the expected counts tensor energies: seq[float] ## the energy values at which the background interpolation was evaluated ## to compute the expected counts tensor expCounts: Tensor[float] ## the tensor containing the expected counts at different (x, y, E) pairs # these are always valid for a single `computeLimit` call! zeroSig: int ## counts the number of times the expected signal was 0 zeroBack: int ## counts the number of times the background was 0 zeroSigBack: int ## counts the number of times the signal & background was zero Context = ref object ## XXX: make ref object mcIdx: int # monte carlo index, just for reference axionModel: DataFrame integralBase: float # integral of axion flux using base coupling constants # interpolators axionSpl: InterpolatorType[float] efficiencySpl: InterpolatorType[float] raytraceSpl: Interpolator2DType[float] backgroundSpl: InterpolatorType[float] # background candidate sampling backgroundCDF: seq[float] # CDF of the background energyForBCDF: seq[float] # energies to draw from for background CDF totalBackgroundClusters: int # total number of background clusters in non-tracking time case samplingKind: SamplingKind # the type of candidate sampling we do of skInterpBackground: interp: Interpolation ## A helper object to store all interpolation fields else: discard # skConstant doesn't need # limit related couplings: Tensor[float] # the coupling range we scan couplingStep: float # a step we take in the couplings during a scan g_aγ²: float # the reference g_aγ (squared) g_ae²: float # the current g_ae value (squared) logLVals: Tensor[float] # the logL values corresponding to `couplings` maxIdx: int # index of the maximum of the logL curve case uncertainty: UncertaintyKind of ukUncertainSig: σs_sig: float # Uncertainty on signal in relative terms, percentage of ukUncertainBack: σb_back: float # Uncertainty on background in relative terms, percentage of ukUncertain: ## annoying.... σsb_sig: float σsb_back: float else: discard # uncertainty on the center position of the signal case uncertaintyPosition: PositionUncertaintyKind of puUncertain: σ_p: float # relative uncertainty away from the center of the chip, in units of # ??? θ_x: float θ_y: float of puCertain: discard # no uncertainty converter toChipCoords(pos: tuple[x, y: float]): tuple[x, y: ChipCoord] = result = (x: ChipCoord(pos.x), y: ChipCoord(pos.y)) converter toChipCoords(pos: Option[tuple[x, y: float]]): Option[tuple[x, y: ChipCoord]] = if pos.isSome: let p = pos.get result = some((x: ChipCoord(p.x), y: ChipCoord(p.y))) const DefaultRange = (-2e-44, 2e-44) #-2e-44, 2e-44) let TrackingTime = 180.h proc cdf(x: float, μ = 0.0, σ = 1.0): float = 0.5 * (1.0 + erf((x - μ) / (σ * sqrt(2.0)))) proc calcSigma95(): float = let res = block: var x = 0.0 while cdf(x) < 0.95: x += 0.0001 x result = res * res / 2.0 let sigma95 = calcSigma95() proc flatten(dfs: seq[DataFrame]): DataFrame = ## flatten a seq of DFs, which are identical by stacking them for df in dfs: result.add df.clone proc readFiles(path: string, s: seq[string]): DataFrame = var h5fs = newSeq[H5FileObj]() echo path echo s for fs in s: h5fs.add H5open(path / fs, "r") result = h5fs.mapIt( it.readDsets(likelihoodBase(), some((chip: 3, dsets: @["energyFromCharge", "centerX", "centerY"]))) .rename(f{"Energy" <- "energyFromCharge"})).flatten if result.isNil: quit("what the fuck") result = result.filter(f{`Energy` < 15.0}) for h in h5fs: discard h.close() defUnit(keV⁻¹•cm⁻²•s⁻¹) defUnit(keV⁻¹•m⁻²•yr⁻¹) defUnit(cm⁻²) defUnit(keV⁻¹•cm⁻²) proc readAxModel(): DataFrame = let upperBin = 10.0 proc convert(x: float): float = result = x.keV⁻¹•m⁻²•yr⁻¹.to(keV⁻¹•cm⁻²•s⁻¹).float result = readCsv("/home/basti/CastData/ExternCode/AxionElectronLimit/axion_diff_flux_gae_1e-13_gagamma_1e-12.csv") .mutate(f{"Energy / keV" ~ c"Energy / eV" / 1000.0}, f{"Flux / keV⁻¹•cm⁻²•s⁻¹" ~ convert(idx("Flux / keV⁻¹ m⁻² yr⁻¹"))}) .filter(f{float: c"Energy / keV" <= upperBin}) proc detectionEff(ctx: Context, energy: keV): UnitLess template toCDF(data: seq[float], isCumSum = false): untyped = var dataCdf = data if not isCumSum: seqmath.cumSum(dataCdf) let integral = dataCdf[^1] let baseline = dataCdf[0] dataCdf.mapIt((it - baseline) / (integral - baseline)) proc setupBackgroundInterpolation(kd: KDTree[float], radius, sigma: float, energyRange: keV, nxy, nE: int): Interpolation = ## Make sure to set the global variables (*ughhh!!!*) # set globals of interpolation Radius = radius # 33.3 Sigma = sigma # 11.1 EnergyRange = energyRange # 0.3.keV ## Need an offset to not start on edge, but rather within ## and stop half a step before let xyOffset = 14.0/(nxy).float / 2.0 ## XXX: fix this for real number ``within`` the chip let eOffset = 12.0/(nE).float / 2.0 let dist = (xyOffset * 2.0).mm let area = dist * dist # area of considered area echo area let ΔE = (eOffset * 2.0).keV echo ΔE let volume = area * ΔE let time = TrackingTime ## XXX: DON'T HARDCODE THIS HERE #echo "XXX: still using 3300 h of background!!! ", time defUnit(cm²•keV) echo volume var t = newTensor[float]([nxy, nxy, nE]) let coords = linspace(0.0 + xyOffset, 14.0 - xyOffset, nxy) let energies = linspace(0.0 + eOffset, 12.0 - eOffset, nE) for yIdx in 0 ..< nxy: for xIdx in 0 ..< nxy: for iE, E in energies: let y = coords[yIdx] let x = coords[xIdx] let tup = kd.queryBallPoint([x.toIdx.float, y.toIdx.float, E].toTensor, Radius, metric = CustomMetric) let val = compValue(tup) .correctEdgeCutoff(Radius, x.toIdx, y.toIdx) .normalizeValue(Radius, EnergyRange) let valCount = val * volume * time.to(Second) #echo val, " as counts: ", valCount, " at ", x, " / ", y, " E = ", E t[yIdx, xIdx, iE] = valCount echo t.sum() result = Interpolation(kd: kd, nxy: nxy, nE: nE, radius: radius, energyRange: energyRange, coords: coords, energies: energies, xyOffset: xyOffset, eOffset: eOffset, expCounts: t) proc initContext(path: string, files: seq[string], useConstantBackground: bool, # decides whether to use background interpolation or not radius, sigma: float, energyRange: keV, nxy, nE: int, σ_sig = 0.0, σ_back = 0.0, # depending on which `σ` is given as > 0, determines uncertainty σ_p = 0.0 ): Context = let samplingKind = if useConstantBackground: skConstBackground else: skInterpBackground let uncertain = if σ_sig == 0.0 and σ_back == 0.0: ukCertain elif σ_sig == 0.0: ukUncertainBack elif σ_back == 0.0: ukUncertainSig else: ukUncertain let uncertainPos = if σ_p == 0.0: puCertain else: puUncertain let axData = readAxModel() ## TODO: use linear interpolator to avoid going to negative? let axSpl = newCubicSpline(axData["Energy / keV", float].toRawSeq, axData["Flux / keV⁻¹•cm⁻²•s⁻¹", float].toRawSeq) let combEffDf = readCsv("/home/basti/org/resources/combined_detector_efficiencies.csv") let effSpl = newCubicSpline(combEffDf["Energy [keV]", float].toRawSeq, #combEffDf["Efficiency", float].toRawSeq) combEffDf["Eff • ε • LLNL", float].toRawSeq) # effective area included in raytracer let raySpl = block: let hmap = readCsv("/home/basti/org/resources/axion_image_heatmap_2017.csv") ggplot(hmap, aes("x", "y", fill = "z")) + geom_raster() + ggsave("/tmp/raster_what_old.pdf") var t = zeros[float]([256, 256]) let area = 1.4.cm * 1.4.cm let pixels = 256 * 256 let pixPerArea = pixels / area let zSum = hmap["z", float].sum let zMax = hmap["z", float].max for idx in 0 ..< hmap.len: let x = hmap["x", int][idx] let y = hmap["y", int][idx] #echo "X ", x, " and ", y let z = hmap["z", float][idx] t[y, x] = (z / zSum * pixPerArea).float #zMax / 784.597 # / zSum # TODO: add telescope efficiency abs. * 0.98 newBilinearSpline(t, (0.0, 255.0), (0.0, 255.0)) # bicubic produces negative values! var df = readFiles(path, files) let kdeSpl = block: var dfLoc = df.toKDE(true) newCubicSpline(dfLoc["Energy", float].toRawSeq, dfLoc["KDE", float].toRawSeq) let backgroundInterp = toNearestNeighborTree(df) let energies = linspace(0.071, 9.999, 10000).mapIt(it) # cut to range valid in interpolation let backgroundCdf = energies.mapIt(kdeSpl.eval(it)).toCdf() result = Context(samplingKind: samplingKind, axionModel: axData, axionSpl: axSpl, efficiencySpl: effSpl, raytraceSpl: raySpl, backgroundSpl: kdeSpl, backgroundCDF: backgroundCdf, energyForBCDF: energies, totalBackgroundClusters: df.len, g_aγ²: 1e-12 * 1e-12, uncertainty: uncertain, uncertaintyPosition: uncertainPos) let ctx = result # XXX: hack to workaround bug in formula macro due to `result` name!!! let axModel = axData .mutate(f{"Flux" ~ idx("Flux / keV⁻¹•cm⁻²•s⁻¹") * detectionEff(ctx, idx("Energy / keV").keV) }) echo axModel let integralBase = simpson(axModel["Flux", float].toRawSeq, axModel["Energy / keV", float].toRawSeq) result.integralBase = integralBase result.couplings = linspace(DefaultRange[0] / result.g_aγ², DefaultRange[1] / result.g_aγ², 1000).toTensor #result.couplings = linspace(-9e-45 / result.g_aγ², 9e-45 / result.g_aγ², 1000).toTensor result.couplingStep = result.couplings[1] - result.couplings[0] ## Set fields for interpolation if not useConstantBackground: ## initialize the variables needed for the interpolation let interp = setupBackgroundInterpolation( backgroundInterp, radius, sigma, energyRange, nxy, nE ) result.interp = interp ## Set fields for uncertainties case uncertain of ukUncertainSig: result.σs_sig = σ_sig of ukUncertainBack: result.σb_back = σ_back of ukUncertain: result.σsb_sig = σ_sig result.σsb_back = σ_back else: discard # nothing to do case uncertainPos of puUncertain: result.σ_p = σ_p else: discard # nothing to do proc rescale(x: float, new: float): float = ## `new` must already be squared! let old = 1e-13 # initial value is always 1e-13 result = x * new / (old * old) proc rescale(s: seq[float], g_ae²: float): seq[float] = ## rescaling version, which takes a `new` squared coupling constant ## to allow for negative squares result = newSeq[float](s.len) for i, el in s: result[i] = el.rescale(g_ae²) proc plotCandidates(cands: seq[Candidate]) = let dfC = toDf({ "x" : cands.mapIt(it.pos.x.float), "y" : cands.mapIt(it.pos.y.float), "E" : cands.mapIt(it.energy.float)}) ggplot(dfC, aes("x", "y", color = "E")) + geom_point() + ggsave("/tmp/candidates.pdf") import random / mersenne import alea / [core, rng, gauss, poisson] proc drawCandidates(#df: DataFrame, ctx: Context, rnd: var Random, posOverride = none(tuple[x, y: ChipCoord]), toPlot: static bool = false): seq[Candidate] = ## draws a number of random candidates from the background sample ## using the ratio of tracking to background ~19.5 # 1. clear the background cache of context, if we're using interpolation if ctx.samplingKind == skInterpBackground: ctx.interp.backCache.clear() when false: var df = df.filter(f{`Energy` <= 10.0}) # filter to < 10 keV for interpolation .mutate(f{float: "Random" ~ rand(1.0)}) .filter(f{`Random` <= 1.0 / TrackingBackgroundRatio}) # take the 1/19.5 subset case ctx.samplingKind of skConstBackground: let uni = uniform(0.0, 1.0) let goldUni = uniform(4.5, 9.5) # 0. create Poisson sampler based on expected number of clusters (λ = tracking cluster expectation) let pois = poisson(ctx.totalBackgroundClusters / TrackingBackgroundRatio) for i in 0 ..< rnd.sample(pois).int: # 1. draw energy based on background CDF let energy = ctx.energyForBCDF[ctx.backgroundCdf.lowerBound(rnd.sample(uni))].keV # 2. draw position within region of interest let pos = block: if posOverride.isSome: posOverride.get else: (x: ChipCoord(rnd.sample(goldUni)), y: ChipCoord(rnd.sample(goldUni))) result.add Candidate(energy: energy, pos: pos) of skInterpBackground: var pois = poisson(0.0) ## Will be adjusted for each grid point var uniXY = uniform(0.0, 0.0) ## Will be adjusted for each grid point var uniE = uniform(0.0, 0.0) result = newSeqOfCap[Candidate](10000) # 1. iterate over every position of the background tensor for iE in 0 ..< ctx.interp.energies.len: for ix in 0 ..< ctx.interp.coords.len: for iy in 0 ..< ctx.interp.coords.len: # 2. draw form a poisson with mean = the value at that tensor position (is normalized to expected counts) pois.l = ctx.interp.expCounts[iy, ix, iE] for _ in 0 ..< rnd.sample(pois).int: # 3. the resulting number of candidates will be created # 3a. for each candidate, smear the position & energy within the volume of the grid cell uniE.a = ctx.interp.energies[iE] - ctx.interp.eOffset uniE.b = ctx.interp.energies[iE] + ctx.interp.eOffset if posOverride.isSome: let pos = posOverride.get result.add Candidate(energy: rnd.sample(uniE).keV, pos: pos) else: uniXY.a = ctx.interp.coords[ix] - ctx.interp.xyOffset uniXY.b = ctx.interp.coords[ix] + ctx.interp.xyOffset let xpos = rnd.sample(uniXY) uniXY.a = ctx.interp.coords[iy] - ctx.interp.xyOffset uniXY.b = ctx.interp.coords[iy] + ctx.interp.xyOffset let ypos = rnd.sample(uniXY) result.add Candidate(energy: rnd.sample(uniE).keV, pos: (x: ChipCoord(xpos), y: ChipCoord(ypos))) when false: # sampling validation var Es = newSeq[float]() for i in 0 ..< 100_000: Es.add ctx.energyForBCDF[ctx.backgroundCdf.lowerBound(rnd.sample(uni))] ggplot(toDf(Es), aes("Es")) + geom_histogram(bins = 200) + ggsave("/tmp/sampled_background.pdf") if true: quit() when false: for row in df: if posOverride.isNone: result.add Candidate(energy: row["Energy"].toFloat, pos: (x: ChipCoord(row["centerX"].toFloat), y: ChipCoord(row["centerY"].toFloat))) else: let pos = posOverride.get result.add Candidate(energy: row["Energy"].toFloat, pos: (x: ChipCoord(pos.x), y: ChipCoord(pos.y))) when toPlot: plotCandidates(result) defUnit(cm²) defUnit(keV⁻¹) proc axionFlux(ctx: Context, energy: keV): keV⁻¹ = ## the absolute differential flux coming from the sun (depends on g_ae) let areaBore = π * (2.15 * 2.15).cm² # area of bore in cm² #echo "Spl ", ctx.axionSpl.eval(energy) #echo "Resc ", ctx.axionSpl.eval(energy).rescale(ctx.g_ae²).keV⁻¹•cm⁻²•s⁻¹ #echo "Area ", ctx.axionSpl.eval(energy).rescale(ctx.g_ae²).keV⁻¹•cm⁻²•s⁻¹ * areaBore #echo "Time ", ctx.axionSpl.eval(energy).rescale(ctx.g_ae²).keV⁻¹•cm⁻²•s⁻¹ * areaBore * 190.0.h.to(s) #echo "Rescaling flux ", ctx.axionSpl.eval(energy), " to ", ctx.g_ae², " is ", ctx.axionSpl.eval(energy).rescale(ctx.g_ae²) if energy < 0.001.keV or energy > 10.0.keV: return 0.0.keV⁻¹ result = ctx.axionSpl.eval(energy.float).rescale(ctx.g_ae²).keV⁻¹•cm⁻²•s⁻¹ * # missing keV⁻¹ areaBore * #1.0 / (8359.18367347) * # ratio of pixels in gold region #(5.mm * 5.mm).to(cm²).float * 1.0 / (8359.18367347) * # ratio of pixels in gold region TrackingTime.to(s) #* # tracking time #12.0.keV # 12 keV range used #echo "AXION FLUX ", result proc detectionEff(ctx: Context, energy: keV): UnitLess = # window + gas if energy < 0.001.keV or energy > 10.0.keV: return 0.0 result = ctx.efficiencySpl.eval(energy.float) proc raytracing(ctx: Context, pos: tuple[x, y: float]): cm⁻² = ## returns the 'flux likelihood' at the given point let x = pos.x * (1.0 + ctx.θ_x) let y = pos.y * (1.0 + ctx.θ_y) if x notin 0.0 .. 14.0 or y notin 0.0 .. 14.0: return 0.cm⁻² let px = x / 14.0 * 255.0 let py = y / 14.0 * 255.0 result = ctx.raytraceSpl.eval(px, py).cm⁻² proc detectionEfficiency(ctx: Context, energy: keV, pos: tuple[x, y: float]): cm⁻² = ## the total detection efficiency result = ctx.detectionEff(energy) * ctx.raytracing(pos) func conversionProbability(): UnitLess = ## the conversion probability in the CAST magnet (depends on g_aγ) ## simplified vacuum conversion prob. for small masses let B = 9.0.T let L = 9.26.m let g_aγ = 1e-12.GeV⁻¹ # ``must`` be same as reference in Context result = pow( (g_aγ * B.toNaturalUnit * L.toNaturalUnit / 2.0), 2.0 ) proc expectedSignal(ctx: Context, energy: keV, pos: tuple[x, y: float]): keV⁻¹•cm⁻² = ## TODO: conversion to detection area?? result = ctx.axionFlux(energy) * conversionProbability() * ctx.detectionEfficiency(energy, pos) # let m = MyMetric(radius: 10.0, sigma: 5.0, energyRange: 1.5.keV) proc toIntegrated(r: keV⁻¹•cm⁻²•s⁻¹): keV⁻¹•cm⁻² = ## Turns the background rate into an integrated rate over the tracking time #let area = 1.4.cm * 1.4.cm let t = TrackingTime.to(Second) result = r * t #x * area * t proc evalInterp(interp: var Interpolation, c: Candidate): keV⁻¹•cm⁻² = #echo "POSITION ", pos.x, " and ", pos.y #echo "INTERP: ", pos.x, " and ", pos.y ## NOTE: `pos.x/y` needs to be given as value [0, 255] to kd tree, but we get [0, 14]! template computeBackground(): untyped {.dirty.} = let px = c.pos.x.toIdx let py = c.pos.y.toIdx interp.kd.queryBallPoint([px.float, py.float, c.energy.float].toTensor, radius = interp.radius, metric = CustomMetric) .compValue() .correctEdgeCutoff(interp.radius, px, py) # this should be correct .normalizeValue(interp.radius, interp.energyRange).to(keV⁻¹•cm⁻²•s⁻¹) .toIntegrated() ## Either get the cached value or compute the value and place it into the table result = interp.backCache.getOrDefault(c, -Inf.keV⁻¹•cm⁻²) if classify(result.float) == fcNegInf: result = computeBackground() interp.backCache[c] = result proc background(ctx: Context, c: Candidate): keV⁻¹•cm⁻² = if ctx.samplingKind == skConstBackground: result = ctx.backgroundSpl.eval(c.energy.float).keV⁻¹•cm⁻² else: result = ctx.interp.evalInterp(c) proc background(ctx: Context, energy: keV, pos: tuple[x, y: ChipCoord]): keV⁻¹•cm⁻² = ## Convenience wrapper around background for the case of calling it with args instead ## of a candidate result = ctx.background(Candidate(energy: energy, pos: pos)) proc rate(ctx: Context, c: Candidate): float = let b = ctx.background(c) let s = ctx.expectedSignal(c.energy, c.pos) if s == 0.0.keV⁻¹•cm⁻² and b == 0.0.keV⁻¹•cm⁻²: if ctx.samplingKind == skInterpBackground: inc ctx.interp.zeroSigBack result = 1.0 elif b == 0.0.keV⁻¹•cm⁻²: if ctx.samplingKind == skInterpBackground: inc ctx.interp.zeroBack #echo "b == 0 : ", c # make a plot #ctx.interp.kd.plotSingleEnergySlice(c.energy.float, # &"/tmp/b_equal_0_energy_slice_E_{c.energy.float}_keV.pdf", # &"Candidate with b = 0 at (x/y) = ({c.pos.x:.2f} / {c.pos.y:.2f}), E = {c.energy}") #result = (1.0 + s / 0.095.keV⁻¹)#6.6e-8 result = 1.0 elif s == 0.0.keV⁻¹•cm⁻²: if ctx.samplingKind == skInterpBackground: inc ctx.interp.zeroSig result = 1.0 else: result = (1.0 + s / b) #if s > 0.0.keV⁻¹: # result = s + b # else leave 0 to not explode our `ln(s + b)`. All `b` without an `s` don't contribute anyway #echo "EXP SIGNAL ", ctx.expectedSignal(c.energy, c.pos), " and EXP BACK ", ctx.background(c.energy, c.pos), " RES ", result, " at position/energy ", c # 1. integrate flux # 2. rescale to cm⁻² keV⁻¹ s⁻¹ # 3. multiply bore area # 4. multiply tracking time # 5. apply conversion prob defUnit(cm⁻²•s⁻¹) defUnit(m⁻²•yr⁻¹) proc expRate(ctx: Context): UnitLess = ## TODO: only count the fraction of evnts expected in gold region! Extract inforamtion ## from heatmap by looking for ratio of sum inside gold / sum outside gold let trackingTime = TrackingTime let areaBore = π * (2.15 * 2.15).cm² ## TODO: only compute integral once and then rescale integral! ## UPDATE: done in the object now #let axModel = ctx.axionModel # .mutate(f{"Flux" ~ rescale(idx("Flux / keV⁻¹•cm⁻²•s⁻¹"), ctx.g_ae²) * # detectionEff(ctx, idx("Energy / keV")) * 0.78})# * areaBore * #let integralExp = simpson(axModel["Flux", float].toRawSeq, # axModel["Energy / keV", float].toRawSeq) let integral = ctx.integralBase.rescale(ctx.g_ae²) ## Rudimentary check that rescaling after integration == rescaling before # doAssert abs(integral - integralExp) < 10.0, " instead " & $integral & " vs " & $integralExp result = integral.cm⁻²•s⁻¹ * areaBore * trackingTime.to(s) * conversionProbability() #echo "Expected rate: ", result ## TODO: this also needs to include detector efficiency of course!! ## It is the number of expected signals after all, no? proc resetZeroCounters(ctx: Context) = ## sets the `zero*` fields of the interpolator to 0 ctx.interp.zeroSig = 0 ctx.interp.zeroBack = 0 ctx.interp.zeroSigBack = 0 proc printZeroCounters(ctx: Context, numCand: int) = echo "================================================================================" echo "g_aγ² = ", ctx.g_aγ² echo "g_ae² = ", ctx.g_ae² echo "Number of candidates: ", numCand echo "Number of zero signal candidates: ", ctx.interp.zeroSig echo "Number of zero background candidates: ", ctx.interp.zeroBack echo "Number of zero sig & back candidates: ", ctx.interp.zeroSigBack template L(s, s_c, b_c, θ_s, σ_s, θ_b, σ_b: untyped, θ_x = 0.0, σ_xp = 0.0, θ_y = 0.0, σ_yp = 0.0): untyped = ## `s`, `s_i` and `b_i` may be modified / unmodified depending on which uncertainty ## is selected ##: XXX: better to do exp( ln( ... ) ), or exp() * exp() * exp() ? when false: result = -s if σ_s > 0.0: ## NOTE: this isn't really "correct" for the case where we ## want to run it with σ = 0 (i.e. "no" nuisance parameter). In that ## case this should exactly be 1 only for θ_s = σ_s else, inf ## Problem is this breaks the case where we mean "ignore" by σ = 0. result -= pow(θ_s / σ_s, 2) if σ_b > 0.0: result -= pow(θ_b / σ_b, 2) if σ_xp > 0.0 and σ_yp > 0.0: result -= pow(θ_x / σ_xp, 2) - pow(θ_y / σ_yp, 2) echo "Initial res ", result, " at θ_s = ", θ_s, ", θ_b = ", θ_b, ", s = ", s, ", σ_s = ", σ_s, ", σ_b = ", σ_b, " θ_x = ", θ_x, ", θ_y = ", θ_y, ", σ_xyp = ", σ_xp for c {.inject.} in candidates: let s_c = s_i let b_c = b_i #echo "b_c = ", b_c, " s_c = ", s_c, " current result = ", result, " (1 + s_c / b_c) = ", 1 + s_c / b_c, " ln(1 + s_c / b_c) = ", ln(1 + s_c / b_c) ## XXX: how to deal with `s_c` or `b_c` negative? Results in negative arg to log if `s/b` is smaller ## than -1. In product this is not an issue. But well... if b_c.float != 0.0: result += ln(1 + s_c / b_c) echo "Result nonexp is ", result if result.isNan: quit("quitting from L") result = exp(result.float) else: result = exp(-s) #echo "-s ", s, " result ", result if σ_s > 0.0: result *= exp(-pow(θ_s / (sqrt(2.0) * σ_s), 2)) ## FIXME the normalization of denominator is wrong missing √2 elif σ_s == 0.0 and θ_s != 0.0: result = 0 if σ_b > 0.0: result *= exp(-pow(θ_b / (sqrt(2.0) * σ_b), 2)) elif σ_b == 0.0 and θ_b != 0.0: result = 0 if σ_xp > 0.0 and σ_yp > 0.0: result *= exp(-pow(θ_x / (sqrt(2.0) * σ_xp), 2)) * exp(-pow(θ_y / (sqrt(2.0) * σ_yp), 2)) #echo "current result ", result for (s_i {.inject.}, b_i {.inject.}) in cands: ## XXX: how to deal with `s_c` or `b_c` negative? Results in negative arg to log if `s/b` is smaller ## than -1. In product this is not an issue. But well... if b_c.float != 0.0: #echo "result at b_i ", b_i, " res = ", result result *= (1 + s_c / b_c) # log-normal (but wrong): / (b_c * σ_b * b_i) #if true: quit() #echo "Result exp is ", result, " for θ_s = ", θ_s, ", θ_b = ", θ_b if result.isNan: echo "WARNING WARNING NAN" #quit("quitting from L") proc logLUncertainSig(ctx: Context, candidates: seq[Candidate]): float = if ctx.samplingKind == skInterpBackground: resetZeroCounters(ctx) ## integration of L over `θ_s` using the current parameters for `s`, `b_i`, `s_i` ## is equivalent to integration & then evaluating integral at position of these params let s_tot = expRate(ctx) let σ_s = ctx.σs_sig var cands = newSeq[(float, float)](candidates.len) for i, c in candidates: cands[i] = (ctx.expectedSignal(c.energy, c.pos).float, ctx.background(c.energy, c.pos).float) proc likelihood(θ_s: float, nc: NumContext[float, float]): float = L(s_tot * (1 + θ_s), s_i * (1 + θ_s), b_i, θ_s, σ_s, 0.0, 0.0) if σ_s > 0.0: let res = adaptiveGauss(likelihood, -10, 10) #echo "Integration result: ", res, ", ln(res) = ", ln(res), " for ", ctx.g_ae², " compare ", logLCertain(ctx, candidates) if res.isNan: echo "Cands: ", cands var f = open("/tmp/bad_candidates.txt", fmWrite) f.write("E, x, y\n") for cnd in candidates: f.write(&"{cnd.energy.float},{cnd.pos.x},{cnd.pos.y}\n") f.close() #quit() return Inf result = ln( res ) else: L(s_tot, s_i, b_i, 0.0, 0.0, 0.0, 0.0) result = ln(result) proc logLUncertainBack(ctx: Context, candidates: seq[Candidate]): float = if ctx.samplingKind == skInterpBackground: resetZeroCounters(ctx) ## integration of L over `θ_b` using the current parameters for `s`, `b_i`, `s_i` ## is equivalent to integration & then evaluating integral at position of these params let s_tot = expRate(ctx) let σ_b = ctx.σb_back var cands = newSeq[(float, float)](candidates.len) for i, c in candidates: cands[i] = (ctx.expectedSignal(c.energy, c.pos).float, ctx.background(c.energy, c.pos).float) proc likelihood(θ_b: float, nc: NumContext[float, float]): float = L(s_tot, s_i, b_i * (1 + θ_b), # log-normal (but wrong): exp(b_i * (1 + θ_b)), 0.0, 0.0, θ_b, σ_b) ## Mark the point `-1` as a difficult point, so that it's not evaluated. We do not care ## about the singularity at that point for the integration let res = adaptiveGauss(likelihood, -0.80, 10.0) #, initialPoints = @[-1.0]) #echo "Integration result: ", res, ", ln(res) = ", ln(res), " for ", ctx.g_ae² #, " compare ", logLCertain(ctx, candidates) if res.isNan: quit() result = ln( res ) proc logLUncertain(ctx: Context, candidates: seq[Candidate]): float = if ctx.samplingKind == skInterpBackground: resetZeroCounters(ctx) ## integration of L over `θ_b` using the current parameters for `s`, `b_i`, `s_i` ## is equivalent to integration & then evaluating integral at position of these params let s_tot = expRate(ctx) let σ_b = ctx.σsb_back let σ_s = ctx.σsb_sig var cands = newSeq[(float, float)](candidates.len) for i, c in candidates: cands[i] = (ctx.expectedSignal(c.energy, c.pos).float, ctx.background(c.energy, c.pos).float) var count = 0 proc likeBack(θ_b: float, nc: NumContext[float, float]): float = proc likeSig(θ_s: float, nc: NumContext[float, float]): float = L(s_tot * (1 + θ_s), s_i * (1 + θ_s), b_i * (1 + θ_b), θ_s, σ_s, θ_b, σ_b) result = adaptiveGauss(likeSig, -2.0, 2.0) #echo "Result of inner integral: ", result, " for θ_b = ", θ_b, " at call ", count inc count ## There is a singularity at `-1`. Everything smaller is irrelevant and the singularity is ## unphysical for us. Start above that. let res = adaptiveGauss(likeBack, -0.80, 2.0, maxintervals = 9999) #, initialPoints = @[-1.0]) #echo "Integration result: ", res, ", ln(res) = ", ln(res), " for ", ctx.g_ae², " compare ", logLCertain(ctx, candidates) if res.isNan: quit() result = ln( res ) proc logLPosUncertain(ctx: Context, candidates: seq[Candidate]): float = if ctx.samplingKind == skInterpBackground: resetZeroCounters(ctx) ## integration of L over `θ_b` using the current parameters for `s`, `b_i`, `s_i` ## is equivalent to integration & then evaluating integral at position of these params var cands = newSeq[(float, float)](candidates.len) let SQRT2 = sqrt(2.0) let σ_p = ctx.σ_p let s_tot = expRate(ctx) for i, c in candidates: let sig = ctx.detectionEff(c.energy) * ctx.axionFlux(c.energy) * conversionProbability() cands[i] = (sig.float, ctx.background(c.energy, c.pos).float) proc likeX(θ_x: float, nc: NumContext[float, float]): float = ctx.θ_x = θ_x proc likeY(θ_y: float, nc: NumContext[float, float]): float = ctx.θ_y = θ_y let P1 = exp(-s_tot) let P2 = exp(-pow(θ_x / (SQRT2 * σ_p), 2)) * exp(-pow(θ_y / (SQRT2 * σ_p), 2)) var P3 = 1.0 for i in 0 ..< cands.len: let (s_init, b_c) = cands[i] if b_c.float != 0.0: let s_c = (s_init * ctx.raytracing(candidates[i].pos)).float P3 *= (1 + s_c / b_c) result = 1.0 when true: result *= P1 when true: result *= P2 when true: result *= P3 result = romberg(likeY, -1.0, 1.0, depth = 6) result = romberg(likeX, -1.0, 1.0, depth = 6) # result = simpson(likeY, -1.0, 1.0, N = 100)#, N = 500) #result = ln(simpson(likeX, -1.0, 1.0, N = 100))#, N = 500)) proc logLFullUncertain(ctx: Context, candidates: seq[Candidate]): float = if ctx.samplingKind == skInterpBackground: resetZeroCounters(ctx) var cands = newSeq[(float, float)](candidates.len) let SQRT2 = sqrt(2.0) let σ_p = ctx.σ_p let s_tot = expRate(ctx) let σ_b = ctx.σsb_back let σ_s = ctx.σsb_sig for i, c in candidates: let sig = ctx.detectionEff(c.energy) * ctx.axionFlux(c.energy) * conversionProbability() cands[i] = (sig.float, ctx.background(c.energy, c.pos).float) proc likeX(θ_x: float, nc: NumContext[float, float]): float = ctx.θ_x = θ_x proc likeY(θ_y: float, nc: NumContext[float, float]): float = ctx.θ_y = θ_y proc likeSig(θ_s: float, nc: NumContext[float, float]): float = proc likeBack(θ_b: float, nc: NumContext[float, float]): float = let s_tot_p = s_tot * (1 + θ_s) let P1 = exp(-s_tot_p) let P2 = exp(-pow(θ_x / (SQRT2 * σ_p), 2)) * exp(-pow(θ_y / (SQRT2 * σ_p), 2)) * exp(-pow(θ_s / (SQRT2 * σ_s), 2)) * exp(-pow(θ_b / (SQRT2 * σ_b), 2)) var P3 = 1.0 for i in 0 ..< cands.len: let (s_init, b_i) = cands[i] let s_i = s_init * (1 + θ_s) let b_c = b_i * (1 + θ_b) if b_c.float != 0.0: let s_c = (s_i * ctx.raytracing(candidates[i].pos)).float P3 *= (1 + s_c / b_c) #echo P1, " ", P2, " ", P3 result = 1.0 when true: result *= P1 when true: result *= P2 when true: result *= P3 result = adaptiveGauss(likeBack, -0.8, 2.0) #, depth = 6) result = romberg(likeSig, -2.0, 2.0, depth = 5) result = romberg(likeY, -1.0, 1.0, depth = 5) result = romberg(likeX, -1.0, 1.0, depth = 5) #result = ln( res ) if result.isNan: echo "!!!" if true: quit() # # result = trapz(likeBack, -0.8, 2.0, N = 30)#, depth = 2) # result = trapz(likeSig, -2.0, 2.0, N = 30)# , depth = 2) # result = romberg(likeY, -1.0, 1.0, depth = 3) #result = ln(romberg(likeX, -1.0, 1.0, depth = 3)) proc logLCertain(ctx: Context, candidates: seq[Candidate]): float = if ctx.samplingKind == skInterpBackground: resetZeroCounters(ctx) when false: let s_tot = expRate(ctx) let σ_b = ctx.σsb_back let σ_s = ctx.σsb_sig var cands = newSeq[(float, float)](candidates.len) for i, c in candidates: cands[i] = (ctx.expectedSignal(c.energy, c.pos).float, ctx.background(c.energy, c.pos).float) L(s_tot, s_i, b_i, 0.0, 0.0, 0.0, 0.0) result = ln(result) when true: result = -expRate(ctx)# * 0.002 #echo "ExpRate ", result for c in candidates: let rt = ctx.rate(c) #echo "PURE RATE ", rt, " and ln ", ln(rt), " at position ", c.pos, " at g_ae ", ctx.g_ae², " result: ", result result += ln(rt) #if rt > 0.0: #echo "CURRENT RESULT ", result #echo "after ln ", result, " for g_ae² = ", ctx.g_ae² #printZeroCounters(ctx, candidates.len) proc logL(ctx: Context, candidates: seq[Candidate]): float = if ctx.uncertaintyPosition == puCertain: case ctx.uncertainty of ukCertain: result = logLCertain(ctx, candidates) of ukUncertainSig: result = logLUncertainSig(ctx, candidates) of ukUncertainBack: result = logLUncertainBack(ctx, candidates) of ukUncertain: result = logLUncertain(ctx, candidates) else: case ctx.uncertainty of ukCertain: result = logLPosUncertain(ctx, candidates) of ukUncertain: result = logLFullUncertain(ctx, candidates) else: doAssert false, "Not implemented mixed uncertainties w/o all" proc linearScan(ctx: Context, cands: seq[Candidate], range = DefaultRange): DataFrame = let couplings = linspace(range[0] / ctx.g_aγ², range[1] / ctx.g_aγ², 1000) #let couplings = linspace(range[0], range[1], 1000) var ctx = ctx let vals = block: var res = newSeq[float](couplings.len) for i, el in couplings: ctx.g_ae² = el res[i] = ctx.logL(cands) res echo "LINEAR SCAN DONE" result = toDf({"CouplingsRaw" : couplings, "logL" : vals}) .mutate(f{"Couplings" ~ `CouplingsRaw` * ctx.g_aγ²}) ggplot(result, aes("CouplingsRaw", "logL")) + geom_line() + ggsave("/tmp/linear_scan.pdf") proc coarseScan(ctx: Context, cands: seq[Candidate], couplings: seq[float]): Option[int] = ## returns an int if we find a suitable index corresponding to a maximum ## in `couplings`. If `couplings` is monotonically decreasing (increasing?) ## we return none, or if all values are NaN var curLog = NaN var lastLog = curLog var idx = 0 var ctx = ctx while classify(lastLog) != fcNormal and idx < couplings.len: lastLog = curLog ctx.g_ae² = couplings[idx] curLog = ctx.logL(cands) #echo "Current: ", curLog, " at ", idx, " is ", ctx.g_ae², " and last ", lastLog # check if this or last is larger if classify(lastLog) == fcNormal and curLog < lastLog: dec idx break inc idx if idx != couplings.len: result = some(idx) proc scan(ctx: Context, cands: seq[Candidate]): tuple[σ, μ, logMin, logMax: float] = # binary search for the NaN cutoff on the left side var logMin = -1e-10 var logMax = 1e-10 var ctx = ctx template evalAt(val: float): untyped = ctx.g_ae² = val ctx.logL(cands) # find cut value from NaN -> fcNormal while (logMax + logMin) / 2.0 != logMax and (logMax - logMin) > 1e-32: let logMid = (logMax + logMin) / 2.0 let curMid = evalAt(logMid) if classify(curMid) != fcNormal: logMin = logMid else: logMax = logMid doAssert classify(evalAt(logMin)) != fcNormal #echo "current ", logMin, " nad ", logMax, " and diff ", logMax - logMin echo "log Min ", logMin, " and max ", logMax, " vals ", evalAt(logMin), " an d ", evalAt(logMax) # using logMin we can now scan to find the maximum to the right of this var step = abs(logMax / 1000.0) var curLog = logMax var curVal = evalAt(curLog) echo "CIUR VAL ", curVal var lastVal = -Inf while curVal > lastVal: lastVal = curVal curLog += step curVal = evalAt(curLog) #if abs(curVal - lastVal) > 1.0: # curLog -= step # curVal = evalAt(curLog) # lastVal = -Inf # step /= 10.0 #echo "Current value ", curVal, ", compared last ", lastVal, ", at ", curLog echo "Found value of max at ", curLog let maxLog = curLog - step echo "Max coup value ", maxLog, " and ", evalAt(maxLog) # now for 1 sigma step = step / 10.0 curVal = evalAt(maxLog) curLog = maxLog let maxVal = curVal const cutoff = 1.0 # left while abs(curVal - maxVal) < cutoff: curLog -= step curVal = evalAt(curLog) echo "found left" var sigmaLeft = curLog var coupSigmaLeft = curVal if classify(coupSigmaLeft) != fcNormal: coupSigmaLeft = evalAt(logMax) sigmaLeft = logMax # right curLog = maxLog curVal = maxVal while abs(curVal - maxVal) < cutoff: curLog += step let newVal = evalAt(curLog) if abs(newVal - curVal) < 1e-3: step *= 2.0 curVal = newVal let sigmaRight = curLog let coupSigmaRight = curVal echo "Sigma region is between ", sigmaLeft, " and ", sigmaRight, " are ", coupSigmaLeft, " and ", coupSigmaRight result = (σ: abs(sigmaRight - sigmaLeft), μ: maxLog, logMin: sigmaLeft, logMax: sigmaRight) #var coarseCutVal = 5.0 ## first determine maximum in negative or positive ## 1. scan from large negative # # #var logCoup = logspace(-25.0, -10.0, 50).reversed.mapIt(-it) #var idxOpt = ctx.coarseScan(cands, logCoup) #if idxOpt.isSome: # # in negative #elif idxOpt.isNone: # # try positive side # logCoup = logCoup.mapIt(-it).reversed # idxOpt = ctx.coarseScan(cands, logCoup) # if idxOpt.isNone: # echo "Candidates: ", cands, " at ", ctx # quit("Could not find anything, what") #doAssert idxOpt.isSome ## logCoup now contains range to recurse in #var idxStart = log(abs(logCoup[idx-1]), 10.0) #var idxStop = log(abs(logCoup[idx]), 10.0) #while abs(evalAt(idxStart) - evalAt(idxStop)) > coarseCutVal: proc findMaximum(ctx: Context, cands: seq[Candidate]): Context = # first do a log scan over a very large range negative & positive numbers # get first NaN -> valid number range & check if next number is larger # 1. scan from large negative var logCoup = logspace(-25.0, -10.0, 50).reversed.mapIt(-it) var dfScan: DataFrame var idxOpt = ctx.coarseScan(cands, logCoup) if idxOpt.isSome: let idx = idxOpt.get doAssert idx > 0 echo logCoup[idx-1], " ", logCoup[idx] echo log(abs(logCoup[idx-1]), 10.0), " ", log(abs(logCoup[idx]), 10.0) if true: quit() #logCoup = logspace(log(logCoup[idx-1]), log(logCoup[idx]), 50) #idxOpt = ctx.coarseScan(cands, logCoup) dfScan = ctx.linearScan(cands, range = (logCoup[idx-1] * ctx.g_aγ², logCoup[idx] * ctx.g_aγ²)) elif idxOpt.isNone: # try positive side logCoup = logCoup.mapIt(-it).reversed idxOpt = ctx.coarseScan(cands, logCoup) if idxOpt.isNone: echo "Candidates: ", cands, " at ", ctx quit("Could not find anything, what") let idx = idxOpt.get doAssert idx > 0 dfScan = ctx.linearScan(cands, range = (logCoup[idx-1] * ctx.g_aγ², logCoup[idx] * ctx.g_aγ²)) # else found it on the negative side, nothing to do doAssert not dfScan.isNil echo dfScan.pretty(-1) #echo "First non NaN value: ", curLog, " at ", idx, " is ", ctx.g_ae² if true: quit() let dfMax = ctx.linearScan(cands)#, range = (-1e-21, 1e-21)) .filter(f{classify(`logL`) == fcNormal}) #echo dfMax #echo dfMax.pretty(-1) ggplot(dfMax, aes("CouplingsRaw", "logL")) + geom_line() + #scale_y_log10() + ggsave("/tmp/linear_scan.pdf") result = ctx result.logLVals = dfMax["logL", float] result.couplings = dfMax["CouplingsRaw", float] result.maxIdx = result.logLVals.toRawSeq.argmax proc findConstantCutLimit(ctx: Context, cands: seq[Candidate], cutValue = 1.0, searchLeft = false): float = ## Finds the the coupling constant such that `logL = start + cutValue`. Our scan starts ## at the coupling corresponding to `start`. var ctx = ctx ctx.g_ae² = ctx.couplings[ctx.maxIdx] var couplingStep = ctx.couplings[1] - ctx.couplings[0] let startVal = ctx.logL(cands) var curLogL = startVal while curLogL > startVal - cutValue: # and classify(curLogL) == fcNormal: # compute next coupling step and logL value if searchLeft: ctx.g_ae² -= couplingStep else: ctx.g_ae² += couplingStep let newLogL = ctx.logL(cands) # possibly readjust coupling step echo "New log ", newLogL, " vs old ", curLogL, " diff ", newLogL - curLogL, " target ", cutValue, " and ", startVal, " to search left ?? ", searchLeft, " coupling ", ctx.g_ae² #if abs(newLogL - curLogL) < 0.001: # couplingStep *= 2.0 #elif abs(newLogL - curLogL) > 0.1: # couplingStep /= 2.0 if curLogL > 200.0 and classify(newLogL) != fcNormal: ctx.g_ae² -= couplingStep couplingStep /= 10.0 curLogL = newLogL result = ctx.g_ae² proc computeSigma(ctx: Context, cands: seq[Candidate]): float = ## Computes the 1σ value of the current `logL` "distribution" let limitLeft = ctx.findConstantCutLimit(cands, searchLeft = true) let limitRight = ctx.findConstantCutLimit(cands, searchLeft = false) let gaussSigma = limitRight - limitLeft result = gaussSigma proc calcSigmaLimit(μ, σ: float, ignoreUnphysical = false): tuple[limit, cdf: float] = ## Computes the limit based on a 1 σ gaussian distrubition around the computed logL ## results. The 1 σ range is determined based on the coupling range covered by ## logL_min + 1. ## The limit is then the gaussian CDF at a value of 0.95. Either in the full ## data range (`ignoreUnphysical = false`) or the CDF@0.95 only in the physical ## range (at x = 0). var x = μ var offset = 0.0 if ignoreUnphysical: x = 0.0 offset = x.cdf(μ, σ) while x.cdf(μ, σ) < (1.0 - (1.0 - offset) * 0.05): x += (σ / 1000.0) #echo "Current cdf = ", x.cdf(μ, σ) result = (limit: x, cdf: x.cdf(μ, σ)) proc simpleLimit(ctx: Context, cands: seq[Candidate]): float = ## starting at coupling of 0, walk by `sigma95` to the right downwards ## (as long as it only goes down). Use that point as our limit var coupling = 0.0 var couplingStep = 1e-24 var ctx = ctx template evalAt(val: float): untyped = ctx.g_ae² = val ctx.logL(cands) let logL0 = evalAt(0.0) var curLogL = logL0 var logLMax = logL0 while curLogL >= logLMax - sigma95: coupling += couplingStep ## ADJUST let lastLogL = curLogL curLogL = evalAt(coupling) if abs(lastLogL - curLogL) < 1e-4: couplingStep *= 2.0 elif abs(lastLogL - curLogL) > 1e-1: coupling -= couplingStep curLogL = evalAt(coupling) couplingStep /= 2.0 if curLogL > logLMax: echo "We're going up!!! ", curLogL, " vs ", logLMax logLMax = curLogL #echo "Current coupling ", coupling, " at logL ", curLogL, " from max ", logLMax, " using ", sigma95, " x - y ", logLMax - sigma95 echo "This limit ", coupling result = coupling template evalAt(ctx: Context, cands: seq[Candidate], val: untyped): untyped = ctx.g_ae² = val ctx.logL(cands) import /home/basti/org/Misc/sorted_seq type Likelihood = object coupling: float computed: bool # whether L has been computed L: float LimitHelper = object ctx: Context # storing a ref obj of the context cands: seq[Candidate] Ls: SortedSeq[Likelihood] cdf: seq[float] # CDF based on couplings & L in `Ls` deriv: seq[float] # 2nd 'derivative' (sort of) of CDF to check where to insert more points dy: float # relative value need larger to compute more points in CDF tail dDeriv: float # relative value needed to compute more points in derivative proc `<`(l1, l2: Likelihood): bool = result = l1.coupling < l2.coupling proc `==`(l1, l2: Likelihood): bool = l1.coupling == l2.coupling proc cumSumUnequal(y, x: seq[float]): seq[float] = result = newSeq[float](y.len) doAssert x.len > 1 var dx = x[1] - x[0] var cum = y[0] * dx # 0.0 #y[0] * dx for i in 0 ..< x.len: if i > 0: dx = x[i] - x[i-1] cum += y[i] * dx result[i] = cum # (cum - result[i]) * dx + result[i] proc cdfUnequal(y, x: seq[float]): seq[float] = let cumS = cumSumUnequal(y, x) let integral = cumS[^1] let baseline = cumS[0] doAssert integral != baseline, "what? " & $cumS result = cumS.mapIt((it - baseline) / (integral - baseline)) proc couplings(lh: LimitHelper): seq[float] = result = newSeq[float](lh.Ls.len) for i in 0 ..< lh.Ls.len: result[i] = lh.Ls[i].coupling proc likelihoods(lh: LimitHelper): seq[float] = result = newSeq[float](lh.Ls.len) for i in 0 ..< lh.Ls.len: assert lh.Ls[i].computed result[i] = lh.Ls[i].L proc computeCdf(lh: LimitHelper): seq[float] = # get xs and ys let xs = lh.couplings() let ys = lh.likelihoods() result = cdfUnequal(ys, xs) proc gradientSecond(xs, cdf: seq[float]): seq[float] = result = newSeq[float](xs.len) let xMax = xs[^1] let xMin = xs[0] for i in 1 ..< xs.high: let s1 = (cdf[i-1] - cdf[i]) / (xs[i-1] - xs[i]) * (xMax - xMin) let s2 = (cdf[i+1] - cdf[i]) / (xs[i+1] - xs[i]) * (xMax - xMin) ## NOTE: we do *not* want to normalize to the distance between points! That defeats the ## purpose. We care about making the slopes similar in absolute terms. Normalizing we ## get the real second derivative, but we want the slopes to become "similar enough" instead, ## i.e. to define a smoothness result[i] = (abs(s2) - abs(s1)) # / ((xs[i+1] - xs[i-1]) / 2.0) proc computeDeriv(lh: LimitHelper): seq[float] = let xs = lh.couplings() let cdf = lh.cdf doAssert cdf.len == xs.len, "CDF must be up to date!" result = gradientSecond(xs, cdf) proc insert(lh: var LimitHelper, c: float) = ## Inserts the given coupling into the heapqueue and computes the likelihood value ## associated to the coupling constant for the given `Context` let L = lh.ctx.evalAt(lh.cands, c) #echo "L: ", L, " at ", c let cL = Likelihood(coupling: c, computed: true, L: L) lh.Ls.push cL proc initLimitHelper(ctx: Context, cands: seq[Candidate], couplings: seq[float]): LimitHelper = var h = initSortedSeq[Likelihood]() result = LimitHelper(ctx: ctx, cands: cands, Ls: h, dy: 0.005, dDeriv: 0.05) # insert into the heapqueue for c in couplings: result.insert(c) result.cdf = result.computeCdf() result.deriv = result.computeDeriv() proc derivativesLarger(lh: LimitHelper, than: float): bool = ## Checks if any derivatives are larger `than`. result = lh.deriv.anyIt(abs(it) > than) proc computeCouplings(lh: var LimitHelper) = let xs = lh.couplings() let cdf = lh.cdf var x = xs[0] var y = cdf[0] let der = lh.deriv var i = 0 var j = 0 #var done: set[uint16] while i < xs.high: let derv = if der[min(der.high, j)] > 0: der[min(der.high, j)] else: 1.0 #if i > 0 and abs(cdf[j] - y) > lh.dy: # echo "CASE 1 \n" # lh.insert((xs[i] + x) / 2.0) # inc i #TODO: add back above to avoid points after 10 if i > 0 and abs(derv) > lh.dDeriv and abs(cdf[j] - y) > lh.dy: #echo "DIFFERENCE : ", abs(cdf[j] - y), " for ", x, " at j ", j, " of ", cdf.len, " and i ", i, " of ", xs.len let xi = xs[i] let xi1 = xs[i+1] lh.insert((xi + x) / 2.0) lh.insert((xi1 + xi) / 2.0) #done.incl j.uint16 if i > 0: inc i inc j x = xs[i] y = cdf[j] inc i inc j proc genplot(lh: LimitHelper, title = "", outname = "/tmp/ragged_cdf.pdf") = let xs = lh.couplings() let Ls = lh.likelihoods() let cdf = lh.cdf let lSum = Ls.max let df = toDf({ "x" : xs, "L [norm]" : Ls.mapIt(it / lSum), "cdf" : cdf }) .gather(["cdf", "L [norm]"], key = "Type", value = "val") let xm = xs.max #df.showbrowser() ggplot(df, aes("x", "val", color = "Type")) + geom_line() + geom_point(size = 1.0) + #ylim(0.9, 1.0) + geom_linerange(aes = aes(y = 0.95, xMin = 0.0, xMax = xm), lineType = ltDashed, color = "purple") + ggtitle(title) + ggsave(outname) proc plotSecond(lh: LimitHelper) = let der = lh.deriv let xx = lh.couplings() let df = toDf(xx, der) ggplot(df, aes("xx", "der")) + geom_line() + ggsave("/tmp/cdf_second_der.pdf") import os, flatty proc bayesLimit(ctx: Context, cands: seq[Candidate], toPlot: static bool = false): float = # {.gcsafe.} = ## compute the limit based on integrating the posterior probability according to ## Bayes theorem using a prior that is zero in the unphysical range and constant in ## the physical # 1. init needed variables var ctx = ctx const nPoints = 10000 var Ls = newSeqOfCap[float](nPoints) var cdfs = newSeqOfCap[float](nPoints) var couplings = newSeqOfCap[float](nPoints) var coupling = 0.0 let couplingStep = 5e-22 var idx = 0 # 2. compute starting values and add them when false: let L0 = ctx.evalAt(cands, 0.0) cdfs.add L0 Ls.add L0 couplings.add coupling var curL = L0 echo "Cur L ", curL #echo "L0 = ", L0, " and curL = ", curL, " abs = ", abs(ln(L0) / ln(curL)), " is nan ?? ", abs(ln(L0) / ln(curL)).isNan #if true: quit() # 3. walk from g_ae² = 0 until the ratio of the `ln` values is 0.9. Gives us good margin for CDF # calculation (i.e. make sure the CDF will have plateaued var lastL = curL var cdfVal = lastL var decreasing = false let stopVal = curL / 500.0 # if curL < 5e-3: curL / 200.0 else: 5e-3 while curL > stopVal: # and idx < 1000: #ln(curL) >= 0.0: echo "Limit step ", idx, " at curL ", curL, " at g_ae²: ", ctx.g_ae², " decreasing ? ", decreasing, " curL < lastL? ", curL < lastL coupling += couplingStep curL = ctx.evalAt(cands, coupling) cdfVal += curL cdfs.add cdfVal Ls.add curL couplings.add coupling if decreasing and # already decreasing curL > lastL: # rising again! Need to stop! echo "Breaking early!" #break if lastL != curL and curL < lastL: # decreasing now! decreasing = true lastL = curL inc idx else: couplings = linspace(0.0, 2e-20, 10) var lh = initLimitHelper(ctx, cands, couplings) let ε = 0.005 #1e-3 # with in place, compute derivatives & insert until diff small enough var diff = Inf var at = 0 #echo lh.deriv genplot(lh, title = "MC Index: " & $ctx.mcIdx) plotSecond(lh) #echo lh.derivativesLarger(0.5) while diff > ε and lh.derivativesLarger(0.5): computeCouplings(lh) lh.cdf = lh.computeCdf() lh.deriv = lh.computeDeriv() at = lh.cdf.lowerBound(0.95) diff = lh.cdf[at] - 0.95 #echo "XS : ", xs #echo "Diff ", diff, " at ", lh.cdf[at], " x ", lh.Ls[at] genplot(lh, title = "MC Index: " & $ctx.mcIdx) plotSecond(lh) #sleep(300) #echo "Final x: ", xs, " of length: ", xs.len, " and dervs ", dervs echo "Diff: ", diff if classify(diff) == fcInf: writeFile("/tmp/bad_candidates.bin", cands.toFlatty()) Ls = lh.likelihoods() couplings = lh.couplings() # 4. renormalize the CDF values from 0 to 1 let cdfsNorm = lh.cdf #toCdf(cdfs, isCumSum = true) # 5. now find cdf @ 0.95 #let idxLimit = cdfsNorm.lowerBound(0.95) # 6. coupling at this value is limit result = couplings[at] #couplings[idxLimit] when true:# false: # toPlot: let df = toDf({"Ls" : Ls, "cdfs" : cdfs, "cdfsNorm" : cdfsNorm, "couplings" : couplings}) #df.showBrowser() ggplot(df.mutate(f{"logL" ~ ln(`Ls`)}), aes("couplings", "logL")) + geom_line() + ggsave("/tmp/couplings_vs_ln_likelihood.pdf") ggplot(df, aes("couplings", "cdfsNorm")) + geom_line() + ggsave("/tmp/couplings_vs_cdfsNorm_ln_likelihood.pdf") ggplot(df, aes("couplings", "Ls")) + geom_line() + ggsave("/tmp/couplings_vs_likelihood.pdf") #ggplot(df, aes("couplings", "cdfs")) + # geom_line() + ggsave("/tmp/couplings_vs_cdf_of_likelihood.pdf") #if couplings[idxLimit] == 0.0: # echo "CANDS " #, cands # #if true: quit() type LimitKind = enum lkSimple, ## purely physical region going down to 95% equivalent lkScan, ## proper scan for maximum using a binary approach lkLinearScan, ## limit based on linear scan in pre defined range lkBayesScan ## limit based on integrating bayes theorem (posterior prob.) proc computeLimit(ctx: Context, cands: seq[Candidate], limitKind: LimitKind, toPlot: static bool = false): float =# {.gcsafe.} = when false: case limitKind of lkSimple: result = ctx.simpleLimit(cands) of lkScan: let (σ, μ, logMin, logMax) = ctx.scan(cands) let (limit, cdf) = calcSigmaLimit(μ, σ, ignoreUnphysical = false) let (limitPhys, cdfPhys) = calcSigmaLimit(μ, σ, ignoreUnphysical = true) echo "Limit at = ", limit, " , cdf = ", cdf echo "Physical limit at = ", limitPhys, " , cdf = ", cdfPhys result = limitPhys of lkLinearScan: var ctx = ctx.findMaximum(cands) let σ = ctx.computeSigma(cands) let (limit, cdf) = calcSigmaLimit(ctx.couplings[ctx.maxIdx], σ, ignoreUnphysical = false) let (limitPhys, cdfPhys) = calcSigmaLimit(ctx.couplings[ctx.maxIdx], σ, ignoreUnphysical = true) echo "Limit at = ", limit, " , cdf = ", cdf echo "Physical limit at = ", limitPhys, " , cdf = ", cdfPhys result = limitPhys of lkBayesScan: discard ## compute the limit based on integrating the posterior probability according to ## Bayes theorem using a prior that is zero in the unphysical range and constant in ## the physical result = ctx.bayesLimit(cands, toPlot = toPlot) echo "Limit at = ", result when false: #if toPlot: const g²_aγ = 1e-12 * 1e-12 let μ = ctx.couplings[ctx.maxIdx] let xs = linspace(μ - 3 * σ, μ + 3 * σ, 2000) let df = toDf(xs) .mutate(f{float: "gauss" ~ smath.gauss(`xs`, μ, σ)}, f{"xs" ~ `xs` * g²_aγ}) let range = DefaultRange # (-9e-45, 9e-45) let couplings = linspace(range[0] / g²_aγ, range[1] / g²_aγ, 5000) let dfLogL = ctx.linearScan(cands, range) .filter(f{ `logL` <= 200.0 and classify(`logL`) == fcNormal }) let lim = limit let limPhys = limitPhys let xLimLow = (μ - 3 * σ) * g²_aγ let xLimHigh = (μ + 3 * σ) * g²_aγ let logLMaxVal = ctx.logLVals[ctx.maxIdx] let physProduct = (sqrt(limitPhys) * 1e-12) ggmulti([ggplot(df, aes("xs", "gauss")) + geom_line() + geom_linerange(aes(x = lim * g²_aγ, yMin = 0.0, yMax = 1.0)) + geom_linerange(aes(x = limPhys * g²_aγ, yMin = 0.0, yMax = 1.0)) + xlim(xlimLow, max(xlimHigh, dflogL["Couplings", float].max)) + xlab("g²_ae g²_aγ") + annotate(&"Limit at: {limit:.2e} (g_ae²)\nCorresponds to CDF cut @{cdf:.2f}", x = lim * g²_aγ, bottom = 0.5) + annotate(&"Physical limit at: {limitPhys:.2e} (g_ae²)\nCorresponds to CDF cut @{cdfPhys:.2f}", x = limPhys * g²_aγ, bottom = 0.3) + ggtitle(&"Physical g_ae * g_aγ = {physProduct:.2e} @ g_aγ = 1e-12 GeV-¹"), ggplot(dflogL, aes("Couplings", "logL")) + geom_line() + geom_linerange(aes(y = logLMaxVal - 1.0, xMin = xlimLow, xMax = xLimHigh)) + geom_linerange(aes(y = logLMaxVal - 4.0, xMin = xlimLow, xMax = xLimHigh)) + geom_linerange(aes(x = lim * g²_aγ, yMin = logLMaxVal, yMax = logLMaxVal - 4.0)) + geom_linerange(aes(x = limPhys * g²_aγ, yMin = logLMaxVal, yMax = logLMaxVal - 4.0)) + annotate("logL_min - 1", y = logLMaxVal - 1.1, left = 0.2) + annotate("logL_min - 4", y = logLMaxVal - 4.1, left = 0.2) + xlim(xlimLow, max(xlimHigh, dflogL["Couplings", float].max)) + xlab("g²_ae g²_aγ") + ggtitle("Scan of g²_ae g²_aγ for g_aγ = 1e-12 GeV⁻¹")], &"/tmp/test_multi.pdf", 1200, 500) proc plotContextLines(ctx: Context, cands: seq[Candidate]) = var ctx = ctx ctx.g_ae² = 8.1e-11 * 8.1e-11 let energies = linspace(0.1, 10.0, 1000) let dfT = toDf({ "expSignal" : energies.mapIt(ctx.expectedSignal(it.keV, (x: 7.0, y: 7.0)).float) }) echo simpson(dfT["expSignal", float].toRawSeq, energies) echo "vs ", expRate(ctx) block Facet: var df = toDf({ "axion" : energies.mapIt(ctx.axionFlux(it.keV).float), "convProb" : conversionProbability(), "detEff@center" : energies.mapIt(ctx.detectionEfficiency(it.keV, (x: 7.0, y: 7.0)).float), "eff" : energies.mapIt(ctx.efficiencySpl.eval(it).float), #"expSignal" : energies.mapIt(ctx.expectedSignal(it.keV, (x: 7.0, y: 7.0)).float), #"raytrace" : energies.mapIt(ctx.raytraceSpl.eval(127.0, 127.0)), "background" : energies.mapIt(ctx.backgroundSpl.eval(it)), "energy" : energies }) .mutate(f{"axSignal" ~ `axion` * `convProb` * idx("detEff@center")}) if ctx.samplingKind == skInterpBackground: df["backgroundInterp"] = energies.mapIt(ctx.background(it.keV, (x: 7.0, y: 7.0)).float) df = df.gather(["axSignal", "background", "detEff@center", "eff", "axion", "backgroundInterp"], "Type", "Value") else: df = df.gather(["axSignal", "background", "detEff@center", "eff", "axion"], "Type", "Value") #echo df.pretty(-1) let conv = conversionProbability() ggplot(df, aes("energy", "Value")) + facet_wrap("Type", scales = "free") + geom_line() + ggtitle(&"Conversion probability = {conv}, raytrace @ center = {ctx.raytraceSpl.eval(127.0, 127.0)}") + ggsave("/tmp/plot_facet_context_lines.pdf", width = 1200, height = 800) block Facet: let df = toDf({ "axion" : energies.mapIt(ctx.axionFlux(it.keV).float), "convProb" : conversionProbability(), "detEff" : energies.mapIt(ctx.detectionEfficiency(it.keV, (x: 7.0, y: 7.0)).float), #"eff" : energies.mapIt(ctx.efficiencySpl.eval(it)), #"raytrace" : energies.mapIt(ctx.raytraceSpl.eval(it)), "background" : energies.mapIt(ctx.backgroundSpl.eval(it)), "energy" : energies }) .mutate(f{"axSignal" ~ `axion` * `convProb` * `detEff`}) .gather(["axSignal", "background"], "Type", "Value") #echo df.pretty(-1) ggplot(df, aes("energy", "Value", color = "Type")) + geom_line() + ggsave("/tmp/plot_context_lines.pdf") proc plotSignalOverBackground(ctx: Context, cands: seq[Candidate]) = ## creates a plot of the signal over background for each pixel on the chip. ## Uses a limit of 8.1e-23 var ctx = ctx ctx.g_ae² = 8.1e-11 * 8.1e-11 var xs = newSeq[float](256 * 256) var ys = newSeq[float](256 * 256) var sb = newSeq[float](256 * 256) let energy = 1.5 for y in 0 ..< 256: for x in 0 ..< 256: xs[y * 256 + x] = x.float ys[y * 256 + x] = y.float let xp = x.float / 256.0 * 14.0 let yp = y.float / 256.0 * 14.0 ## TODO: Turn it around, why? #sb[y * 256 + x] = ctx.expectedSignal(energy, (x: yp, y: xp)) / ctx.background(energy) let pos = (x: yp, y: xp) let back = ctx.background(energy.keV, pos) let sig = ctx.expectedSignal(energy.keV, pos) #echo "Sig: ", sig, " vs ", back sb[y * 256 + x] = ln(1 + sig / back) let df = toDf(xs, ys, sb) template low: untyped = 4.5 / 14.0 * 256.0 template hih: untyped = 9.5 / 14.0 * 256.0 #showBrowser(df) ggplot(df, aes("xs", "ys", fill = "sb")) + geom_raster() + xlim(0, 256) + ylim(0, 256) + scale_x_continuous() + scale_y_continuous() + geom_linerange(aes = aes(x = low(), yMin = low(), yMax = hih()), color = some(parseHex("FF0000"))) + geom_linerange(aes = aes(x = hih(), yMin = low(), yMax = hih()), color = some(parseHex("FF0000"))) + geom_linerange(aes = aes(y = low(), xMin = low(), xMax = hih()), color = some(parseHex("FF0000"))) + geom_linerange(aes = aes(y = hih(), xMin = low(), xMax = hih()), color = some(parseHex("FF0000"))) + #ggtitle("Signal / Background for E = 1.5 keV & g_ae = 8.1e-11") + ggtitle("ln(1 + S / B) for E = 1.5 keV & g_ae = 8.1e-11") + ggsave("/tmp/raster_signal_over_background.pdf") proc integrateSignalOverImage(ctx: Context) = ## integrate the signal contribution over the whole image to see if we recover ## the ~O(10) axion induced signals var ctx = ctx ctx.g_ae² = pow(8.1e-11, 2.0) echo "Expected number of signals in total = ", expRate(ctx) # now integrate over full area var integral = 0.0 var intBack = 0.0 var integralGold = 0.0 var intBackGold = 0.0 let energies = linspace(0.071, 9.999, 100).mapIt(it.keV) # cut to range valid in interpolation let eWidth = energies[1].keV - energies[0].keV let pix = 256 * 256 let area = 1.4.cm * 1.4.cm let pixArea = area / pix for idx, energy in energies: var sumOfRT = 0.0 var sumOfGold = 0.0 for y in 0 ..< 256: for x in 0 ..< 256: let xp = x.float / 256.0 * 14.0 let yp = y.float / 256.0 * 14.0 let pos = (x: xp, y: yp) let sig = ctx.expectedSignal(energy, pos) * eWidth * pixArea let back = ctx.background(energy, pos) * eWidth * pixArea integral += sig intBack += back sumOfRT += ctx.raytraceSpl.eval(x.float, y.float) if xp in 4.5 .. 9.5 and yp in 4.5 .. 9.5: sumOfGold += ctx.raytraceSpl.eval(x.float, y.float) integralGold += sig intBackGold += back echo "Total sum of RT contribution = ", sumOfRT echo "Total sum of RT gold contribution = ", sumOfGold echo "Ratio ", sumOfGold / sumOfRT #if true: quit() echo "Total integral of signal: ", integral, " (integrated over the whole chip!)" echo "Total integral of background: ", intBack, " (integrated over the whole chip!)" echo "Total integral of signal: ", integralGold, " (integrated over gold region!)" echo "Total integral of background: ", intBackGold, " (integrated over gold region!)" echo "Normalization factor: ", integral / expRate(ctx) #if true: quit() proc candsInSens(ctx: Context, cands: seq[Candidate], cutoff = 0.5): int = var ctx = ctx # use a fixed g_ae² for the computation here ctx.g_ae² = pow(8.1e-11, 2.0) for c in cands: let sig = ctx.expectedSignal(c.energy, c.pos) if ln(1 + sig / ctx.background(c.energy, c.pos)) >= cutoff: inc result proc plotLikelihoodCurves(ctx: Context, candidates: seq[Candidate]) = ## Plots the likelihood curves at a specific coupling constant in θ let s_tot = expRate(ctx) var cands = newSeq[(float, float)](candidates.len) for i, c in candidates: cands[i] = (ctx.expectedSignal(c.energy, c.pos).float, ctx.background(c.energy, c.pos).float) case ctx.uncertainty of ukUncertain: let σ_b = ctx.σsb_back let σ_s = ctx.σsb_sig block θ_signal: proc likeBack(θ_b: float): float = proc likeSig(θ_s: float, nc: NumContext[float, float]): float = L(s_tot * (1 + θ_s), s_i * (1 + θ_s), b_i * (1 + θ_b), θ_s, σ_s, θ_b, σ_b) result = adaptiveGauss(likeSig, -10.0, 10.0) let θs = linspace(-0.99, 10.0, 1000) let df = toDf({"θs" : θs, "L" : θs.mapIt(likeBack(it))}) .filter(f{`L` > 1e-6}) #df.showBrowser() ggplot(df, aes("θs", "L")) + geom_line() + scale_y_log10() + ggtitle("L(θ_s) = ∫_{-∞}^∞ L(θ_s, θ_b) dθ_b, at σ_s = " & &"{ctx.σsb_sig}") + ggsave("/tmp/likelihood_θs_integrated_θb.pdf") block θ_background: proc likeSig(θ_s: float): float = proc likeBack(θ_b: float, nc: NumContext[float, float]): float = L(s_tot * (1 + θ_s), s_i * (1 + θ_s), b_i * (1 + θ_b), θ_s, σ_s, θ_b, σ_b) result = adaptiveGauss(likeBack, -0.9, 10.0) let θs = linspace(-1.5, 1.5, 1000) let df = toDf({"θb" : θs, "L" : θs.mapIt(likeSig(it))}) .filter(f{`L` > 1e-6}) #df.showBrowser() ggplot(df, aes("θb", "L")) + geom_line() + scale_y_log10() + ggtitle("L(θ_b) = ∫_{-∞}^∞ L(θ_s, θ_b) dθ_s, at σ_b = " & &"{ctx.σsb_back}") + ggsave("/tmp/likelihood_θb_integrated_θs.pdf") of ukUncertainSig: let σ_s = ctx.σs_sig proc likeSig(θ_s: float): float = L(s_tot * (1 + θ_s), s_i * (1 + θ_s), b_i, θ_s, σ_s, 0.0, 0.0) let θs = linspace(-0.99, 10.0, 1000) let df = toDf({"θ" : θs, "L" : θs.mapIt(likeSig(it))}) .filter(f{`L` > 1e-6}) #df.showBrowser() ggplot(df, aes("θ", "L")) + geom_line() + scale_y_log10() + ggtitle(&"L(θ_s), at σ_s = {ctx.σs_sig}") + ggsave("/tmp/likelihood_θs.pdf") of ukUncertainBack: let σ_b = ctx.σb_back proc likeBack(θ_b: float): float = L(s_tot, s_i, b_i * (1 + θ_b), # log-normal (but wrong): exp(b_i * (1 + θ_b)), 0.0, 0.0, θ_b, σ_b) let θs = linspace(-0.98, 1.0, 1000) var df = toDf({"θ" : θs, "L" : θs.mapIt(likeBack(it))}) echo df #df = df # .filter(f{`L` > 1e-6}) #echo df #df.showBrowser() ggplot(df, aes("θ", "L")) + geom_line() + scale_y_log10() + ggtitle(&"L(θ_b), at σ_b = {ctx.σb_back}") + ggsave("/tmp/likelihood_θb.pdf") else: if ctx.uncertaintyPosition == puUncertain: when false: #block TX: let s_tot = expRate(ctx) proc likeX(θ_x: float): float = ctx.θ_x = θ_x proc likeY(θ_y: float, nc: NumContext[float, float]): float = ctx.θ_y = θ_y for i, c in candidates: cands[i] = (ctx.expectedSignal(c.energy, c.pos).float, ctx.background(c.energy, c.pos).float) L(s_tot, s_i, b_i, 0.0, 0.0, # signal 0.0, 0.0, # background θ_x, ctx.σ_p, θ_y, ctx.σ_p) result = adaptiveGauss(likeY, -1.0, 1.0, maxIntervals = 100) let θx = linspace(-1.0, 1.0, 1000) var df = toDf({"θ" : θx, "L" : θx.mapIt(likeX(it))}) echo df df = df .filter(f{`L` > 1e-24}) #echo df #df.showBrowser() ggplot(df, aes("θ", "L")) + geom_line() + scale_y_log10() + ggtitle(&"L(θ_x), at σ_p = {ctx.σ_p} integrated over θ_y") + ggsave("/tmp/likelihood_θx.pdf") when false: #block TY: let s_tot = expRate(ctx) proc likeY(θ_y: float): float = ctx.θ_y = θ_y proc likeX(θ_x: float, nc: NumContext[float, float]): float = ctx.θ_x = θ_x for i, c in candidates: cands[i] = (ctx.expectedSignal(c.energy, c.pos).float, ctx.background(c.energy, c.pos).float) L(s_tot, s_i, b_i, 0.0, 0.0, # signal 0.0, 0.0, # background θ_x, ctx.σ_p, θ_y, ctx.σ_p) result = adaptiveGauss(likeX, -1.0, 1.0, maxIntervals = 100) let θy = linspace(-1.0, 1.0, 1000) var df = toDf({"θ" : θy, "L" : θy.mapIt(likeY(it))}) echo df df = df .filter(f{`L` > 1e-24}) #echo df #df.showBrowser() ggplot(df, aes("θ", "L")) + geom_line() + scale_y_log10() + ggtitle(&"L(θ_y), at σ_p = {ctx.σ_p} integrated over θ_x") + ggsave("/tmp/likelihood_θy.pdf") block Test: let s_tot = expRate(ctx) var cands = newSeq[(float, float)](candidates.len) let SQRT2 = sqrt(2.0) for i, c in candidates: let sig = ctx.detectionEff(c.energy) * ctx.axionFlux(c.energy) * conversionProbability() cands[i] = (sig.float, ctx.background(c.energy, c.pos).float) let σ_p = ctx.σ_p proc likeX(θ_x: float): float = ctx.θ_x = θ_x proc likeY(θ_y: float, nc: NumContext[float, float]): float = ctx.θ_y = θ_y result = exp(-s_tot) result *= exp(-pow(θ_x / (SQRT2 * σ_p), 2)) * exp(-pow(θ_y / (SQRT2 * σ_p), 2)) for i in 0 ..< cands.len: let (s_init, b_c) = cands[i] if b_c.float != 0.0: let s_c = (s_init * ctx.raytracing(candidates[i].pos)).float result *= (1 + s_c / b_c) result = simpson(likeY, -1.0, 1.0) let θx = linspace(-1.0, 1.0, 1000) var df = toDf({"θ" : θx, "L" : θx.mapIt(likeX(it))}) echo df df = df .filter(f{`L` > 1e-24}) ggplot(df, aes("θ", "L")) + geom_line() + scale_y_log10() + ggtitle(&"L(θ_x), at σ_p = {ctx.σ_p} integrated over θ_y") + ggsave("/tmp/likelihood_θx_alternative.pdf") block TestXY: let s_tot = expRate(ctx) var cands = newSeq[(float, float)](candidates.len) let SQRT2 = sqrt(2.0) for i, c in candidates: let sig = ctx.detectionEff(c.energy) * ctx.axionFlux(c.energy) * conversionProbability() cands[i] = (sig.float, ctx.background(c.energy, c.pos).float) let σ_p = ctx.σ_p proc like(θ_x, θ_y: float): float = ctx.θ_x = θ_x ctx.θ_y = θ_y result = exp(-s_tot) result *= exp(-pow(θ_x / (SQRT2 * σ_p), 2)) * exp(-pow(θ_y / (SQRT2 * σ_p), 2)) for i in 0 ..< cands.len: let (s_init, b_c) = cands[i] if b_c.float != 0.0: let s_c = (s_init * ctx.raytracing(candidates[i].pos)).float result *= (1 + s_c / b_c) let θs = linspace(-1.0, 1.0, 1000) var θx = newSeq[float]() var θy = newSeq[float]() var val = newSeq[float]() for x in θs: for y in θs: θx.add -x θy.add -y val.add like(x, y) var df = toDf({"θx" : θx, "θy" : θy, "L" : val}) echo df #df = df # .filter(f{`L` > 1e-24}) ggplot(df, aes("θx", "θy", fill = "L")) + geom_raster() + ggtitle(&"L(θ_x, θ_y), at σ_p = {ctx.σ_p}") + ggsave("/tmp/likelihood_θx_θy.pdf") else: quit("not va") proc plotLikelihoodParts(ctx: Context, candidates: seq[Candidate]) = ## Plots the behavior of the different parts of the position uncertainty likelihood ## for increasing `g_ae²` due to the weird exponential rise for some candidates var cands = newSeq[(float, float)](candidates.len) let SQRT2 = sqrt(2.0) let σ_p = ctx.σ_p template genIt(g_ae²: float, UseP1, UseP2, UseP3: static bool): untyped = block: ctx.g_ae² = g_ae² let s_tot = expRate(ctx) for i, c in candidates: let sig = ctx.detectionEff(c.energy) * ctx.axionFlux(c.energy) * conversionProbability() cands[i] = (sig.float, ctx.background(c.energy, c.pos).float) proc likeX(θ_x: float, nc: NumContext[float, float]): float = ctx.θ_x = θ_x proc likeY(θ_y: float, nc: NumContext[float, float]): float = ctx.θ_y = θ_y let P1 = exp(-s_tot) let P2 = exp(-pow(θ_x / (SQRT2 * σ_p), 2)) * exp(-pow(θ_y / (SQRT2 * σ_p), 2)) var P3 = 1.0 for i in 0 ..< cands.len: let (s_init, b_c) = cands[i] if b_c.float != 0.0: let s_c = (s_init * ctx.raytracing(candidates[i].pos)).float P3 *= (1 + s_c / b_c) result = 1.0 when UseP1: result *= P1 when UseP2: result *= P2 when UseP3: result *= P3 result = romberg(likeY, -1.0, 1.0) let res = romberg(likeX, -1.0, 1.0) res var logL = 0.0 #ctx.g_ae² = 5e- let step = 1e-24 var p1s = newSeq[float]() var p2s = newSeq[float]() var p3s = newSeq[float]() var pAs = newSeq[float]() var gaes = newSeq[float]() var g_ae² = 5e-23 while logL < 0.02: echo "At step: ", ctx.g_ae², " logL = ", logL let p1 = genIt(g_ae², true, false, false) #let p2 = genIt(g_ae², false, true, false) let p3 = genIt(g_ae², false, false, true) logL = genIt(g_ae², true, true, true) p1s.add p1 #p2s.add p2 p3s.add p3 pAs.add logL gaes.add g_ae² g_ae² += step let penalty = genIt(g_ae², false, true, false) let df = toDf({"expRate" : p1s, "penalty" : penalty, "1+s/b" : p3s, "L" : pAs, "g_ae" : gaes}) .gather(["expRate", "1+s/b", "L"], key = "Type", value = "Val") df.showBrowser() ggplot(df, aes("g_ae", "Val")) + geom_line() + facet_wrap("Type", scales = "free") + facet_margin(0.5) + margin(top = 1.75) + ggtitle(&"Behavior of _seperately integrated_ (!!!) terms. Penalty term = {penalty}") + ggsave("/tmp/behavior_theta_xy_gae_for_terms.pdf") proc expectedLimit(limits: seq[float]): float = ## Returns the expected limit of a set of MC toy experiment limits. ## Currently it's just defined as the median of the determined limits. result = limits.median(q = 50) proc monteCarloLimits(ctx: Context, rnd: var Random, limitKind: LimitKind): float = # 1. determine limit of no signal let candsNoSignal = newSeq[Candidate]() #ctx.drawCandidates(rnd, posOverride = some((x: 14.0, y: 14.0))) let limitNoSignal = ctx.computeLimit(candsNoSignal, limitKind) # 2. perform regular limit calc using simple limit const nmc = 1_000 var limits = newSeq[float](nmc) var candsInSens = newSeq[int](nmc) for i in 0 ..< nmc: #if i mod 10 == 0: echo "MC index ", i, "\n\n" ctx.mcIdx = i let cands = ctx.drawCandidates(rnd) limits[i] = ctx.computeLimit(cands, limitKind) candsInSens[i] = candsInSens(ctx, cands) let expLimit = limits.expectedLimit() when true: echo "Expected limit: ", expLimit let dfL = toDf(limits, candsInSens) .filter(f{`limits` < 2e-19}) let uncertainSuffix = case ctx.uncertainty of ukCertain: &"uncertainty_{ctx.uncertainty}" of ukUncertainSig: &"uncertainty_{ctx.uncertainty}_σs_{ctx.σs_sig}" of ukUncertainBack: &"uncertainty_{ctx.uncertainty}_σb_{ctx.σb_back}" of ukUncertain: &"uncertainty_{ctx.uncertainty}_σs_{ctx.σsb_sig}_σb_{ctx.σsb_back}" ggplot(dfL, aes("limits", fill = "candsInSens")) + geom_histogram(bins = 35, hdKind = hdOutline, position = "identity", alpha = some(0.5)) + geom_linerange(aes = aes(x = limitNoSignal, y = 0.0, yMin = 0.0, yMax = 30.0), color = some(parseHex("FF0000"))) + annotate(text = "Limit w/o signal, only R_T", x = limitNoSignal - 0.01e-21, y = 10, rotate = -90.0, font = font(color = parseHex("FF0000")), backgroundColor = color(0.0, 0.0, 0.0, 0.0)) + scale_x_continuous() + scale_y_continuous() + ggsave(&"/tmp/mc_limit_bayes_sampling_{ctx.samplingKind}_{uncertainSuffix}_position_{ctx.uncertaintyPosition}.pdf") result = expLimit when false: #import weave import std / threadpool #import taskpools var chan: Channel[tuple[σ_s, σ_b: float]] var chanRes: Channel[tuple[σ_s, σ_b, limit: float]] proc singleLimit(tup: tuple[ctx: Context, limitKind: LimitKind, id: int]) {.thread.} = let (ctx, limitKind, id) = tup var rnd = wrap(initMersenneTwister(id.uint32)) var nMsg = 0 while nMsg >= 0: # break if channel closed (peek returns -1) if nMsg == 0: sleep(100) else: # get a message & process let (σ_s, σ_b) = chan.recv() ctx.σsb_sig = σ_s ctx.σsb_back = σ_b echo "Thread ", id, " computing limit ", σ_s, ", ", σ_b let res = ctx.monteCarloLimits(rnd, limitKind) chanRes.send((σs, σb, res)) nMsg = chan.peek() echo "Thread ", id, " shutting down!" proc computeSigmaLimits(ctx: Context, limitKind: LimitKind): seq[tuple[σ_s, σ_b, limit: float]] = var expLimits = newSeq[float]() var σVals = @[0.05, 0.1, 0.15, 0.2, 0.25, 0.3] var σ_pairs = newSeq[(float, float)]() for σ_s in σVals: for σ_b in σVals: σ_pairs.add (σ_s, σ_b) chan.open() chanRes.open() # create threadpool const nThreads = 32 var thr = newSeq[Thread[tuple[ctx: Context, limitKind: LimitKind, id: int]]](nThreads) for i in 0 ..< nThreads: let ctxL = ctx.deepCopy() createThread(thr[i], singleLimit, (ctxL, limitKind, i)) for p in σ_pairs: chan.send(p) while result.len != σ_pairs.len: let res = chanRes.recv() echo "Received ", res result.add res chan.close() chanRes.close() when isMainModule: #let path = "/home/basti/CastData/ExternCode/TimepixAnalysis/Tools/backgroundRateDifferentEffs/out/" #let backFiles = @["lhood_2017_eff_0.8.h5", # "lhood_2018_eff_0.8.h5"] #let path = "/tmp/" #let backFiles = @["lhood_2017_septemveto_all_chip_dbscan.h5", # "lhood_2018_septemveto_all_chip_dbscan.h5"] let path = "/home/basti/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/" let backFiles = @["lhood_2017_all_chip_septem_dbscan.h5", "lhood_2018_all_chip_septem_dbscan.h5"] var dfBacks = readFiles(path, backFiles) echo dfBacks let radius = 40.0 #33.3#33.3 40.0 let σ = radius / 3.0 let energyRange = 0.6.keV#0.3.keV #0.6.keV let nxy = 10 let nE = 20 let useConstantBackground = false var ctx = initContext( path, backFiles, useConstantBackground = useConstantBackground, radius = radius, sigma = σ, energyRange = energyRange, nxy = nxy, nE = nE, σ_sig = 0.15, σ_back = 0.15, σ_p = 0.05) # large values of σ_sig cause NaN and grind some integrations to a halt! ## XXX: σ_sig = 0.3) #echo ctx.interp.expCounts #if true: quit() echo "drawing" #ctx.interp.kd.plotSingleEnergySlice(1.0) var rnd = wrap(initMersenneTwister(299792458 + 2)) let cands = drawCandidates(ctx, rnd, toPlot = true) ctx.g_ae² = 1e-10 * 1e-10 #limit when false: import flatty let candsF = fromFlatty(readFile("/tmp/bad_candidates.bin"), seq[Candidate]) plotCandidates(candsF) #ctx.σs_sig = 0.15 ## set manually to 0 ctx.plotLikelihoodCurves(candsF) #if true: quit() echo "calling limit" echo ctx.computeLimit(candsF, lkBayesScan) echo "calling certain limit" #ctx.uncertainty = ukCertain #echo ctx.computeLimit(candsF, lkBayesScan) if true: quit() plotCandidates(cands) ctx.plotContextLines(cands) if true: quit() echo ctx.computeLimit(cands, lkbayesScan) #ctx.plotLikelihoodCurves(cands) #if true: quit() #let candsNoSignal = newSeq[Candidate]() #echo ctx.computeLimit(candsNoSignal, lkBayesScan) #if true: quit() #let candsNoSignal = ctx.drawCandidates(rnd, posOverride = some((x: 14.0, y: 14.0))) #ctx.plotLikelihoodParts(candsNoSignal) #if true: quit() #echo ctx.logL(cands) #echo ctx.scan(cands) # #ctx.plotSignalOverBackground(cands) #echo ctx.g_ae² #plotContextLines(ctx, cands) #echo "AGAIN LIMIT ", limit #ctx.integrateSignalOverImage() if true: quit() #echo ctx.linearScan(cands) #if true: quit() # MC limit const limitKind = lkBayesScan #let limit = computeLimit(ctx, cands, limitKind, toPlot = true) #const nmc = 1_000 #var limits = newSeq[float](nmc) #for i in 0 ..< nmc: # if i mod 100 == 0: # echo "MC index ", i, "\n\n" # let cands = drawCandidates(dfBacks) # limits[i] = ctx.computeLimit(cands, lkScan) #var dfL = toDf(limits) # .filter(f{`limits` < 2e-20}) #ggplot(dfL, aes("limits")) + # geom_histogram(bins = 100) + # ggsave("/tmp/mc_limit_binaryscan.pdf") # now create the plot for the simple scan in the physical range & adding a line for # the case of no signal #block SimpleLimit: # discard ctx.monteCarloLimits(rnd, limitKind) when false: ## debug NaN in uncertain signal let df = readCsv("/tmp/bad_candidates.txt") var cnds = newSeq[Candidate]() for row in df: cnds.add Candidate(energy: row["E"].toFloat.keV, pos: (x: row["x"].toFloat, y: row["y"].toFloat)) plotCandidates(cnds) ctx.σs_sig = 0.225 #ctx.g_ae² = 5e-20 ctx.plotLikelihoodCurves(cnds) # looks fine. Larger g_ae² shift maximum to negative values discard ctx.computeLimit(cnds, limitKind, toPlot = true) if true: quit() when false: # `ctx` must be `ukUncertainBack` block ScanSigmaBack: var expLimits = newSeq[float]() var σbs = @[0.05, 0.075, 0.1, 0.125, 0.15, 0.175, 0.2, 0.225, 0.25, 0.275, 0.3] for σ in σbs: ctx.σb_back = σ expLimits.add ctx.monteCarloLimits(rnd, limitKind) let df = toDf({"σ_b" : σbs, "expLimits" : expLimits}) ggplot(df, aes("σ_b", "expLimits")) + geom_point() + ggtitle("Expected limit after 1000 MC toys for different σ_b") + ggsave("expected_limits_σ_b.pdf") when true: # `ctx` must be `ukCertain` and `puUncertain` block ScanSigmaXY: echo ctx.monteCarloLimits(rnd, limitKind) when false: var expLimits = newSeq[float]() var σbs = @[0.05, 0.075, 0.1, 0.125, 0.15, 0.175, 0.2, 0.225, 0.25, 0.275, 0.3] for σ in σbs: ctx.σb_back = σ expLimits.add ctx.monteCarloLimits(rnd, limitKind) let df = toDf({"σ_b" : σbs, "expLimits" : expLimits}) ggplot(df, aes("σ_b", "expLimits")) + geom_point() + ggtitle("Expected limit after 1000 MC toys for different σ_b") + ggsave("expected_limits_σ_b.pdf") when false: # `ctx` must be `ukUncertainSig` block ScanSigmaSig: var expLimits = newSeq[float]() var σss = @[0.05, 0.075, 0.1, 0.125, 0.15, 0.175, 0.2, 0.225, 0.25, 0.275, 0.3] for σ in σss: ctx.σs_sig = σ expLimits.add ctx.monteCarloLimits(rnd, limitKind) let df = toDf({"σ_s" : σss, "expLimits" : expLimits}) ggplot(df, aes("σ_s", "expLimits")) + geom_point() + ggtitle("Expected limit after 1000 MC toys for different σ_s") + ggsave("expected_limits_σ_s.pdf") when false: # `ctx` must be `ukUncertain` block ScanSigmaSigBack: let expLimits = ctx.computeSigmaLimits(limitKind) let df = toDf({ "σ_s" : expLimits.mapIt(it.σ_s), "σ_b" : expLimits.mapIt(it.σ_b), "expLimits" : expLimits.mapIt(it.limit)}) ggplot(df, aes("σ_s", "σ_b", color = "expLimits")) + geom_point() + geom_text(text = expLimits) + ## XXX: finish this! text annotation below each point xMargin(0.05) + yMargin(0.05) + ggtitle("Expected limit after 1000 MC toys for different σ_s, σ_b") + ggsave("expected_limits_σ_s_σ_b.pdf") when false: # `ctx` must be `ukUncertain` block ScanSigmaSigBack: var expLimits = newSeq[float]() var σVals = @[0.05, 0.1, 0.15, 0.2, 0.25, 0.3] var σss = newSeq[float]() var σsb = newSeq[float]() for σ_s in σVals: for σ_b in σVals: ctx.σsb_sig = σ_s ctx.σsb_back = σ_b σss.add σ_s σsb.add σ_b expLimits.add ctx.monteCarloLimits(rnd, limitKind) let df = toDf({"σ_s" : σss, "σ_b" : σsb, "expLimits" : expLimits}) ggplot(df, aes("σ_s", "σ_b", color = "expLimits")) + geom_point(size = 3.0) + xMargin(0.05) + yMargin(0.05) + ggtitle("Expected limit after 1000 MC toys for different σ_s, σ_b") + ggsave("expected_limits_σ_s_σ_b.pdf") when false: #block SimpleLimitParallel: # 1. determine limit of no signal let candsNoSignal = ctx.drawCandidates(rnd, posOverride = some((x: 14.0, y: 14.0))) let limitNoSignal = ctx.computeLimit(candsNoSignal, limitKind) # 2. perform regular limit calc using simple limit let nmc = if ctx.samplingKind == skInterpBackground: 100 else: 1_000 var limits = newSeq[float](nmc) var candsSens = newSeq[int](nmc) init Weave var limBuf = cast[ptr UncheckedArray[float]](limits[0].addr) var cInSBuf = cast[ptr UncheckedArray[int]](candsSens[0].addr) parallelFor i in 0 ..< nmc: captures: {limBuf, cInSBuf, limitKind, ctx} var rnd2 = wrap(initMersenneTwister(1234)) let cands = ctx.drawCandidates(rnd2) limBuf[i] = ctx.computeLimit(cands, lkBayesScan) cInSBuf[i] = candsInSens(ctx, cands) exit Weave if true: quit() block NumInSensitiveRegionLimits: # 1. determine limit of no signal let candsNoSignal = ctx.drawCandidates(rnd, posOverride = some((x: 14.0, y: 14.0))) let limitNoSignal = ctx.computeLimit(candsNoSignal, limitKind) # 2. perform regular limit calc using simple limit const numSensTotal = 10 const nmc = 100 var limits = newSeq[float](numSensTotal * nmc) var numInSens = newSeq[int](numSensTotal * nmc) let uni = uniform(0.5, 4.0) for num in 0 ..< numSensTotal: for i in 0 ..< nmc: if i mod 10 == 0: echo "MC index ", i, " for number ", num, "\n\n" let cands = block: var res = newSeq[Candidate]() for j in 0 ..< 30: if j < num: res.add Candidate(energy: rnd.sample(uni).keV, pos: (x: 7.0, y: 7.0)) else: res.add Candidate(energy: rnd.sample(uni).keV, pos: (x: 14.0, y: 14.0)) res let idx = nmc * num + i limits[idx] = ctx.computeLimit(cands, limitKind, toPlot = true) #if limits[idx] < 3.8e-21: # echo "INVALID LIMIT AT num ", num # echo cands # echo ctx.linearScan(cands) # echo ctx.bayesLimit(cands, toPlot = true) # if true: quit() numInSens[idx] = num let dfL = toDf(limits, numInSens) .filter(f{`limits` < 2e-19}) ggplot(dfL, aes("limits", fill = "numInSens")) + geom_histogram(bins = 100) + geom_linerange(aes = aes(x = limitNoSignal, y = 0.0, yMin = 0.0, yMax = 20.0), color = some(parseHex("FF0000"))) + annotate(text = "Limit w/o signal, only R_T", x = limitNoSignal - 0.01e-21, y = 20, rotate = -90.0, font = font(color = parseHex("FF0000")), backgroundColor = color(0.0, 0.0, 0.0, 0.0)) + scale_x_continuous() + scale_y_continuous() + ggsave("/tmp/mc_limit_bayes_num_in_sens.pdf")
The code gives us now a multitude of things.
Starting with a facet plot of the different aspects that affect the logL function, fig. 423. WARNING: this plot and the following still have bugs about axion flux units I think!
/tmp/plotfacetcontextlines.pdf
Next is a similar plot, showing the comparison of the background hypothesis to the axion flux at the limit in fig. 424.
/tmp/plotcontextlines.pdf
Further, a plot showing the logL space as well as the corresponding gaussian that is used to compute the 95% (physical / unphysical) CDF in fig. 425.
/tmp/testmulti.pdf
And a histogram of 2000 monte carlo toy experiments showing only the limits obtained in each, fig. 426.
/tmp/mclimit.pdf
29.1.1. TODO
- add energy slice at 1 keV plot cut to limit of 2e-5
- add section describing all inputs & how they need to be normalized
- add discussion of numbers below (sanity checks)
- describe that we now use keV⁻¹•cm⁻² units & how this is done for ray tracer etc
- add facet plot showing center stuff
- add a candidates sampling plot
- add plots of ln(1 + s/b) comparing constant & interpolation
- add ~/org/Figs/statusAndProgress/limitCalculation/mclimitbayesbackgroundinterpolation.pdf and equivalent for no interpolation now
29.1.2. Sanity checks
Total sum of RT contribution = 33436.73469387749 Total sum of RT gold contribution = 28111.71122738584 Ratio 0.8407433167370049 Total integral of signal: 6.594339003685158 (integrated over the whole chip!) Total integral of background: 472.1077604287829 (integrated over the whole chip!) Total integral of signal: 5.571734416737382 (integrated over gold region!) Total integral of background: 25.58172308274701 (integrated over gold region!) Normalization factor: 1.004019241348128
- Checks to write / add
[X]background data[X]plot / distributions of clusters[X]plot background clusters without noisy clusters[ ]plot background clusters usingplotBackgroundClustersviashell[X]number of background clusters[X]total background rate as cm⁻² s⁻¹ over whole chip[X]background rate in gold of those clusters, as number and plot[X]plot background rate usingplotBackgroundRateviashell[X]total time of background data- ?
[X]raytracing[X]plot of read raytracing image (w/o window)[X]plot of raytracing image with added window[X]plot of raytracing image with applied θx,θy
[X]background interpolation[X]create plot at 3 energies (very low, intermediate, high)[X]with multiple color scale ranges
[X]compare raw interpolation with normalized & edge corrected[X]integral of background interp over the full chip (as part ofintegrateSignalOverImage)[ ] maybe reuse existing "studyBackgroundInterpolation" procedure[X]compute background rate in gold region based on background interpolation[X]using same binning as regular background rate plot[ ]using a smooth representation[ ]using same binning as regular background rate plot, but using an energy range of only 0.1 keV
[X]candidate sampling[X]visualization of the x/y grid in which we sample, maybe also x/E, y/E[X]integral of all boxes * volume over whole chip, should match with background interp integral (barring statistical variation)[X]plot of sample candidates (energy as color)
[X]signal[X]plot of pure signal over whole chip At multiple energies? Is a smooth scaling, so shouldn't be needed. At different coupling constants?[X]"integral" of signal over the full chip Current result of normalization constant:
- INFO: Normalization factor: 0.8592522483146823
implies what we already knew: the normalization currently normalizes without the window strongback. Which means that once we include the strongback we see too little signal!
[X]plot signal over background
[X]energy dependent inputs[X]efficiency (split by component taken from external tool & combined from interpolation)[X]conversion probability in title[X]axion flux
[ ]likelihood[X]plot of likelihood for a set of candidates w/o nuisance parameters[ ]same plot as above, but including nuisance parameters?[ ]same plot as above, but smaller & larger nuisance parameters?[ ]think about plots / numbers to validate behavior of likelihood against coupling constant & number of clusters & # sensitive clusters[ ]compute likelihood without candidates w/ realistic parameters. How to represent? Plot of likelihood phase space? If so, add MCMC & integration for this case.
[ ]1. likelihood scan w/o candidates w/o uncertainties (MCMC & analytical?)[X]2a. draw a set of candidates, such that few in signal region[X]2b. plot of cands in sensitive region (i.e. plot of candidates with color being s/b)[X]2c. likelihood scan w/ set of candidates (few in signal region)[X]3. same as 2, but with multiple candidates in signal region[ ]4. effect of nuisance parameter on signal / background. Increasing them shifts[ ]likelihood curve to right. Same candidates, change syst. and compare likelihood?[ ]5. effect of position uncertainty. Changing value causes different points to be[ ]in sensitive region?[ ]6. example likelihood of realistic systematics and 2 different sets of candidates (few & many in signal)[ ]??
[ ]Limit without candidates -> Show multiple limit calls usinglkMCMCundergo MCMC related variance as well! Not a perfectly fixed number.[ ]systematic uncertainties[ ]list of used values[ ]study of behavior. how impacting things?
Functional aspects:
[ ]write simple unit tests for all procs being used for signal / background calc. Check for unit conversions, correct units & correct normalizations[ ]combine all PDFs and output into a single file for easy review
- Raytracing sanity checks with window strongback
One of the last things we had on our list was to separate the position uncertainty nuisance parameter of the axion flux from the window strongback.
This has since been implemented. As a sanity check we created two plots of the axion image for 2 different cases.
- no nuisance parameter for θ
- a nuisance parameter of θ = 0.6 for both axes
Before that though, first the axion image as it is used now (using 1470 + 12.2 mm distance, see 3.3.1) without the strongback as we use it as a base now:
From here now the two θ cases:
We can see that the strongback does not move with the movement of the signal position, which is precisely what we expect.
As a reference, the next plot shows what happens if the position is moved in addition to the strongback:
- Sanity checks documentation
This document contains notes about the different sanity checks we perform, includes all generated plots, and logging snippets as well as explanations of what one should take away from them to reason about a sane limit calculation.
We'll now go through each of the different sanity checks we perform one by one.
The sanity checks log to a logging file called
sanity.log.- Input data overview
The input data and background as well as tracking time can be taken straight from the log:
[2022-07-27 - 18:15:47] - INFO: =============== Input =============== [2022-07-27 - 18:15:47] - INFO: Input path: /home/basti/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/ [2022-07-27 - 18:15:47] - INFO: Input files: @[(2017, "lhood_2017_all_chip_septem_dbscan.h5"), (2018, "lhood_2018_all_chip_septem_dbscan.h5")] [2022-07-27 - 18:15:47] - INFO: =============== Time =============== [2022-07-27 - 18:15:47] - INFO: Total background time: 3318 Hour [2022-07-27 - 18:15:47] - INFO: Total tracking time: 169 Hour [2022-07-27 - 18:15:47] - INFO: Ratio of tracking to background time: 0.0509343 UnitLess
so a total of 3318 h of background time and 169 h of tracking time were assumed in the context of the sanity checks. This is currently still based on the numbers read from the files "by hand". In the near future this will be replaced by values read straight from the files. The reason it's not done yet is that the data files containing the tracking data are not read yet.
- Detection efficiency
Next up is the combined detection efficiency of the detector and the solar model data resulting in a certain amount of solar flux. Additionally, the conversion probability is covered.
From the log file:
[2022-07-27 - 18:15:48] - INFO: =============== Detection efficiency =============== [2022-07-27 - 18:15:48] - INFO: Maximum detection efficiency = 0.4157505595505261 at energy = 1.52152 KiloElectronVolt [2022-07-27 - 18:15:48] - INFO: Average detection efficiency (0-10 keV) = 0.1233761000155432 [2022-07-27 - 18:15:48] - INFO: Average detection efficiency (0.5-4 keV) = 0.261024542003623 [2022-07-27 - 18:15:48] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/sanity_detection_eff.pdf
which shows us the average detection efficiency in two energy ranges as well as the maximum efficiency and its specific energy. Especially the average numbers are low, but this is expected based on the telescope efficiency towards higher energies and the window transmission at lower energies.
The generated figure is fig. 405
Figure 405: Axion flux at a specific coupling constant and the combined detection efficiency of the detector without the window strongback affecting it. The conversion probability of an axion into a photon is shown in the title at a fixed coupling constant \(g_{aγ}\). Note that the axion flux is integrated over the tracking time and magnet bore given as per keV. - Background
The background checks include the number of clusters found in the background data (with and without filtering of noisy pixels) as well as a list of pixels that were filtered and for what input file.
[2022-07-27 - 18:15:48] - INFO: =============== Background =============== [2022-07-27 - 18:15:48] - INFO: Number of background clusters = 8106 [2022-07-27 - 18:15:48] - INFO: Total background time = 3318 Hour [2022-07-27 - 18:15:48] - INFO: Ratio of background to tracking time = 19.6331 UnitLess [2022-07-27 - 18:15:48] - INFO: Expected number of clusters in tracking time = 412.873417721519 [2022-07-27 - 18:15:48] - INFO: Background rate over full chip = 0.000346236 CentiMeter⁻²•Second⁻¹ [2022-07-27 - 18:15:48] - INFO: Background rate over full chip per keV = 2.8853e-05 CentiMeter⁻²•Second⁻¹•KiloElectronVolt⁻¹ [2022-07-27 - 18:15:48] - INFO: Pixels removed as noisy pixels: @[(64, 109), (64, 110), (67, 112), (65, 108), (66, 108), (67, 108), (65, 109), (66, 109), (67, 109), (68, 109), (65, 110), (66, 110), (67, 110), (65, 111), (66, 111), (67, 111), (68, 110), (68, 109), (68, 111), (68, 108), (67, 107), (66, 111), (69, 110)] [2022-07-27 - 18:15:48] - INFO: Number of pixels removed as noisy pixels: 23 [2022-07-27 - 18:15:48] - INFO: Percentage of total pixels: 0.03509521484375 % [2022-07-27 - 18:15:48] - INFO: in input files: @["lhood_2017_all_chip_septem_dbscan.h5"] [2022-07-27 - 18:15:48] - INFO: Background rate in gold region = 0.000166097 CentiMeter⁻²•Second⁻¹ [2022-07-27 - 18:15:48] - INFO: Background rate in gold region per keV = 1.38414e-05 CentiMeter⁻²•Second⁻¹•KiloElectronVolt⁻¹ [2022-07-27 - 18:15:48] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/background_clusters.pdf [2022-07-27 - 18:15:48] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/background_rate_gold.pdf
The first plot fig. 406 shows the background clusters, colored by their energy, including the noisy pixels filtered out. The select pixels are seen in the logging output and are only filtered for the 2017 / beginning 2018 data.
Figure 406: Background cluster centers as read from the input files including filtering of noisy pixels in the 2017 data file, each cluster colored by its energy in keV. The comparison without filtered pixels is seen in fig. 407, so the noise pixel filtering reduces the number of clusters by about 1300.
Figure 407: Background cluster centers as read from the input files as raw clusters without any removed noisy pixels, colored by energy in keV. We can see about 1300 clusters are removed due to the noisy activity in one dataset. From the background clusters it is interesting to compute the constant background rate in the gold region (center 5x5mm² of the chip) to see if we reproduce the known background rate, that we would normally plot using our background plotting tools.
This is shown in fig. 408, which is simply computed by assigning a weight to each cluster of:
let weight = 1.0 / (ctx.totalBackgroundTime.to(Second) * 0.5.cm * 0.5.cm * 0.2.keV)
i.e. dividing out the normalization factor from counts to keV⁻¹•cm⁻²•s⁻¹ and then generating a histogram based on the clusters and their energy using a bin width of 0.2 keV.
The background rate reaches similar levels to our expectation, i.e. 1e-6 range outside of the argon fluorescence peak and about 4e-5 on the argon peak using this approach.
The integrated background rates can also be easily compared and they are 1.384e-5 keV⁻¹•cm⁻²•s⁻¹ over the whole range from 0 to 12 keV.
Figure 408: Background rate as computed by taking the number of clusters in the gold region and normalizing each cluster's weight to the aforementioned factor. The background rate reaches similar levels to our expectation, i.e. 1e-6 range outside of the argon fluorescence peak and about 4e-5 on the argon peak. To compare it with the "real" background rate, i.e. the logic we normally use to compute our background rate (as it's a separate script it is sensible to compare to lower the chance of a bug in either) we see it in fig. 409.
Looking at both figures in comparison, they agree rather well.
Figure 409: Background rate as computed from the same input files as above, but using the regular background rate plotting script, simply called from the limit calculation code directly. - Raytracing
The raytracing checks are rather straight forward. We evaluate the raytracing interpolator at every pixel on the chip and simply plot the values as a heatmap.
We look at three different cases.
- the raytracing interpolator without any systematics and without the window strongbac
- the raytracing interpolator without any systematics and the window strongback
- the raytracing interpolator with large positional systematic values of \(θ_x = θ_y = 0.6\)
For each case we also compute the sum of the raytracing contribution, as seen in the log.
A note on the meaning of the raytracing interpolator numbers: The interpolator is normalized such that each point corresponds to one pixel size of the detector and the sum of all pixels simply reflects the number of pixels per square centimeter. As such each pixels value is given in "relative flux per square centimeter". If a value is e.g. 20 cm⁻² it means the flux at that point corresponds to a factor 20 of the total raytracing flux if it were integrated over the whole chip. In that sense it is purely a weighting of each pixel to the whole flux. This is valid, as the rest of the signal computation already computes the absolute flux and thus only needs to be scaled accordingly.
TODO: really think about this explanation again after writing about the signal!
[2022-07-28 - 12:48:12] - INFO: =============== Raytracing checks =============== [2022-07-28 - 12:48:12] - INFO: Raytracing sanity check for: ignoreWindow = true, θ_x = θ_y = 0.0 [2022-07-28 - 12:48:12] - INFO: Sum of raytracing contributions over the whole chip: 33436.7 CentiMeter⁻² [2022-07-28 - 12:48:12] - INFO: corresponds to number of pixels per cm⁻² [2022-07-28 - 12:48:12] - INFO: Raytracing contributions over the whole chip normalized to chip area: 1 UnitLess [2022-07-28 - 12:48:12] - INFO: where the normalization is 33436.7 CentiMeter⁻², the number of pixel per cm² [2022-07-28 - 12:48:12] - INFO: meaning the raytracing contribution is normalized. [2022-07-28 - 12:48:12] - INFO: At a single pixel position the value thus corresponds to the amount of flux over unity one [2022-07-28 - 12:48:12] - INFO: would receive if taken over whole chip. [2022-07-28 - 12:48:12] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/axion_image_limit_calc_no_window_no_theta.pdf [2022-07-28 - 12:48:12] - INFO: Raytracing sanity check for: ignoreWindow = false, θ_x = θ_y = 0.0 [2022-07-28 - 12:48:12] - INFO: Sum of raytracing contributions over the whole chip: 27978.1 CentiMeter⁻² [2022-07-28 - 12:48:12] - INFO: corresponds to number of pixels per cm⁻² [2022-07-28 - 12:48:12] - INFO: Raytracing contributions over the whole chip normalized to chip area: 0.836748 UnitLess [2022-07-28 - 12:48:12] - INFO: where the normalization is 33436.7 CentiMeter⁻², the number of pixel per cm² [2022-07-28 - 12:48:12] - INFO: meaning the raytracing contribution is normalized. [2022-07-28 - 12:48:12] - INFO: At a single pixel position the value thus corresponds to the amount of flux over unity one [2022-07-28 - 12:48:12] - INFO: would receive if taken over whole chip. [2022-07-28 - 12:48:12] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/axion_image_limit_calc_no_theta.pdf [2022-07-28 - 12:48:12] - INFO: Raytracing image at θ_x = θ_y = 0.6 [2022-07-28 - 12:48:12] - INFO: Raytracing sanity check for: ignoreWindow = false, θ_x = θ_y = 0.6 [2022-07-28 - 12:48:12] - INFO: Sum of raytracing contributions over the whole chip: 29760.1 CentiMeter⁻² [2022-07-28 - 12:48:12] - INFO: corresponds to number of pixels per cm⁻² [2022-07-28 - 12:48:12] - INFO: Raytracing contributions over the whole chip normalized to chip area: 0.890043 UnitLess [2022-07-28 - 12:48:12] - INFO: where the normalization is 33436.7 CentiMeter⁻², the number of pixel per cm² [2022-07-28 - 12:48:12] - INFO: meaning the raytracing contribution is normalized. [2022-07-28 - 12:48:12] - INFO: At a single pixel position the value thus corresponds to the amount of flux over unity one [2022-07-28 - 12:48:12] - INFO: would receive if taken over whole chip. [2022-07-28 - 12:48:12] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/axion_image_limit_calc_theta_0_6.pdf
The raytracing signal in fig. 410 shows the case without any window strongbacks or systematics. It's the pure result of the raytracer normalized.
Figure 410: Raytracing interpolator evaluated over the chip without any systematics and no window strongback. The values are simply the values of the interpolator at that point. Figure 411: Raytracing interpolator evaluated over the chip without any systematics including the window strongback. The values are simply the values of the interpolator at that point. Figure 412: Raytracing interpolator evaluated over the chip with a position systematic value of \(θ_x = θ_y = 0.6\), showing that theta does indeed move the position of the signal around, but keeps the window strongback in its place. As one can see in fig. 412 the position systematic does indeed move the center of the signal spot by 0.6 times the chip size, as one would expect.
- Background interpolation
The next part is about the background interpolation. How it is computed from the background clusters and via a k-d tree and the corrections and normalizations applied. We'll look at different slices of the x/y detector plane at different energies for the setups from raw interpolation, edge correction and finally the fully normalized interpolation.
The log information:
[2022-07-28 - 15:05:30] - INFO: =============== Background interpolation =============== [2022-07-28 - 15:05:30] - INFO: Radius for background interpolation in x/y: 40.0 [2022-07-28 - 15:05:30] - INFO: Clusters are weighted with normal distribution dependent on distance using σ: 13.33333333333333 [2022-07-28 - 15:05:30] - INFO: Energy range for background interpolation in x/y: 0.6 KiloElectronVolt [2022-07-28 - 15:05:30] - INFO: Energy range is a fixed interval ± given value without weighting [2022-07-28 - 15:05:30] - INFO: --------------- Background interpolation slice @ 0.5 KiloElectronVolt --------------- [2022-07-28 - 15:05:30] - INFO: Generating background interpolation slices at energy: [2022-07-28 - 15:05:30] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/raw_interpolation_at_0.5keV_ymax_15.0.pdf [2022-07-28 - 15:05:40] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/raw_interpolation_at_0.5keV_ymax_0.0.pdf [2022-07-28 - 15:05:49] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/interpolation_edge_correct_at_0.5keV_ymax_15.0.pdf [2022-07-28 - 15:06:00] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/interpolation_edge_correct_at_0.5keV_ymax_0.0.pdf [2022-07-28 - 15:06:10] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/normalized_interpolation_at_0.5keV_ymax_5e-05.pdf [2022-07-28 - 15:06:20] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/normalized_interpolation_at_0.5keV_ymax_0.0.pdf [2022-07-28 - 15:06:30] - INFO: --------------- Background interpolation slice @ 1 KiloElectronVolt --------------- [2022-07-28 - 15:06:30] - INFO: Generating background interpolation slices at energy: [2022-07-28 - 15:06:30] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/raw_interpolation_at_1.0keV_ymax_15.0.pdf [2022-07-28 - 15:06:40] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/raw_interpolation_at_1.0keV_ymax_0.0.pdf [2022-07-28 - 15:06:50] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/interpolation_edge_correct_at_1.0keV_ymax_15.0.pdf [2022-07-28 - 15:07:01] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/interpolation_edge_correct_at_1.0keV_ymax_0.0.pdf [2022-07-28 - 15:07:11] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/normalized_interpolation_at_1.0keV_ymax_5e-05.pdf [2022-07-28 - 15:07:21] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/normalized_interpolation_at_1.0keV_ymax_0.0.pdf [2022-07-28 - 15:07:31] - INFO: --------------- Background interpolation slice @ 3 KiloElectronVolt --------------- [2022-07-28 - 15:07:31] - INFO: Generating background interpolation slices at energy: [2022-07-28 - 15:07:31] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/raw_interpolation_at_3.0keV_ymax_15.0.pdf [2022-07-28 - 15:07:39] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/raw_interpolation_at_3.0keV_ymax_0.0.pdf [2022-07-28 - 15:07:46] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/interpolation_edge_correct_at_3.0keV_ymax_15.0.pdf [2022-07-28 - 15:07:54] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/interpolation_edge_correct_at_3.0keV_ymax_0.0.pdf [2022-07-28 - 15:08:02] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/normalized_interpolation_at_3.0keV_ymax_5e-05.pdf [2022-07-28 - 15:08:09] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/normalized_interpolation_at_3.0keV_ymax_0.0.pdf [2022-07-28 - 15:08:17] - INFO: --------------- Background interpolation slice @ 8 KiloElectronVolt --------------- [2022-07-28 - 15:08:17] - INFO: Generating background interpolation slices at energy: [2022-07-28 - 15:08:17] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/raw_interpolation_at_8.0keV_ymax_15.0.pdf [2022-07-28 - 15:08:25] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/raw_interpolation_at_8.0keV_ymax_0.0.pdf [2022-07-28 - 15:08:33] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/interpolation_edge_correct_at_8.0keV_ymax_15.0.pdf [2022-07-28 - 15:08:40] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/interpolation_edge_correct_at_8.0keV_ymax_0.0.pdf [2022-07-28 - 15:08:48] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/normalized_interpolation_at_8.0keV_ymax_5e-05.pdf [2022-07-28 - 15:08:56] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/normalized_interpolation_at_8.0keV_ymax_0.0.pdf
The interpolation works by placing all clusters in a k-d tree for performant lookup of nearest neighbors and their distances from the given point within a certain radius under a custom metric. The interpolation is then simply the number of points found in that search radius under the metric, normalized such that
The important parameters for the base of the interpolation are:
- search radius around interpolation point: 40.0 pixel
- sigma of gaussian weighting associated with distance: 13.33333333333333
- energy range: 0.6 KiloElectronVolt
In the following we'll look at the energy slices [0.5, 1.0, 3.0, 8.0] keV. For each we check these plots:
raw interpolation output (essentially number of clusters found), with weighting applied in two different cases:
- maximum color value at 15
- maximum color value based on data
the former is meant to show the rate in the center of the chip, as there the background is lowest. We should be able to see a flat distribution in this range. The latter is for the distribution over the whole chip.
interpolation with edge correction (at the edges there will of course be less neighbors found. Edge correction corrects the number of found neighbors by the fraction of area lost in the search radius due to being "off" chip)
- same maximum colors as above, 15 and data
This is mainly to see that in the corners the apparent "drop off" becomes less pronounced than otherwise
- final interpolation taking into account the correct normalization necessary to convert the value to a background rate in keV⁻¹•cm⁻²•s⁻¹. This is based on an integral of the area under the gaussian weighting metric. Again two different ranges. One to 5e-5 and the other to max data. It is essentially expected to find backgrounds of the order of 1e-6 to 5e-5 in the background interpolation.
- Energy slice @ 0.5 keV
- Energy slice @ 1.0 keV
- Energy slice @ 3.0 keV
- Energy slice @ 8.0 keV
- Background rate from interpolation
The background rate is also computed from the interpolation in the gold region, by averaging points in the gold region.
Due to the energy range being larger than the binning used in the regular background rate plot, the rate is smeared out a bit. Lows become a bit higher and highs become a bit lower.
Figure 413: The background rate in the gold region computed from the interpolation by averaging points in the gold region. The background does reproduce the background rate reasonably well (compare fig. 409), but what is evident from a closer inspection is that parts with low background are a bit overestimated and parts with higher background underestimated. This however is expected, as we use an energy range for the interpolation of 0.6 keV (which effectively means looking at 1.2 keV around each point) for enough statistics. This causes a smearing of the rate.
- Candidate sampling from background
Next up is sampling of candidates from the background interpolation. This is done by generating a grid of cubes in which the background rate is assumed constant
[2022-07-28 - 15:09:03] - INFO: =============== Candidate sampling =============== [2022-07-28 - 15:09:03] - INFO: Sum of background events from candidate sampling grid (`expCounts`) = 426.214328925721 [2022-07-28 - 15:09:03] - INFO: Expected number from background data (normalized to tracking time) = 412.873417721519 [2022-07-28 - 15:09:03] - INFO: Number of grid cells for x/y: 10 [2022-07-28 - 15:09:03] - INFO: Number of grid cells for E: 20 [2022-07-28 - 15:09:03] - INFO: Offset in x/y to center points at: 0.7 [2022-07-28 - 15:09:03] - INFO: Offset in E to center points at: 0.25 [2022-07-28 - 15:09:03] - INFO: Coordinates in x/y: @[0.7, 2.1, 3.5, 4.9, 6.300000000000001, 7.700000000000001, 9.1, 10.5, 11.9, 13.3] [2022-07-28 - 15:09:03] - INFO: Coordinates in E: @[0.25, 0.75, 1.25, 1.75, 2.25, 2.75, 3.25, 3.75, 4.25, 4.75, 5.25, 5.75, 6.25, 6.75, 7.25, 7.75, 8.25, 8.75, 9.25, 9.75] [2022-07-28 - 15:09:03] - INFO: Sampling is smeared within grid volumes [2022-07-28 - 15:09:03] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/candidate_sampling_grid_index_2.pdf [2022-07-28 - 15:09:03] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/candidate_sampling_grid_index_5.pdf [2022-07-28 - 15:09:03] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/candidate_sampling_grid_index_16.pdf [2022-07-28 - 15:09:03] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/limitSanityChecks/candidate_sampling_grid_vs_energy.pdf
- Signal
- Likelihood without systematics
- Likelihood with systematics
- Scan of different systematic values and effect on limits
- Input data overview
- Notes on sanity checks
The cone tracing was updated in https://github.com/jovoy/AxionElectronLimit/pull/19 which resulted in a slightly modified axion image with a slightly higher focus in the center, see 11.4 for an update.
We can compare the impact on the expected limit, maybe best by looking at the expected limit scan for different σ values.
Note: both of these use an arbitrary (but the same!) input file for the background rate. Not in any way the "final" one.
We can see that the lowest systematic value (0.05) reduces from 5.428e-21 to 5.29e-21 and the highest one from (0.3) from 6.88e-21 to 6.316e-21. So a nice improvement!
29.1.3. Background interpolation
This section describes the computation of a background interpolation based on the cluster center information
import std / [math, strformat, sequtils, os] import ggplotnim, unchained import arraymancer except linspace import numericalnim except linspace from seqmath import gauss, linspace #from ingrid / tos_helpers import geometry const TrackingBackgroundRatio* = 19.6 ## TODO: replace by actual time!! const EnergyCutoff* = 12.0 proc toKDE*(df: DataFrame, toPlot = false, outname = ""): DataFrame = echo "[KDE] Number of clusters in DF: ", df echo "[KDE] Number of clusters in DF < ", EnergyCutoff, " keV: ", df.filter(f{`Energy` <= EnergyCutoff}) let dfFiltered = df.filter(f{`Energy` <= 12.0}, f{float -> bool: `centerX` in 4.5 .. 9.5 and `centerY` in 4.5 .. 9.5} ) let energy = dfFiltered["Energy", float] #dfFiltered.showBrowser() #if true: quit() echo "[KDE] Number of clusters ", energy.size, " normalized to tracking ", energy.size.float / TrackingBackgroundRatio let xs = linspace(energy.min, energy.max, 1000) echo "MAX ENERGY ", energy.max var kde = kde(energy, bw = 0.3, normalize = true) defUnit(cm²) let scale = energy.size.float / ( TrackingBackgroundRatio ) / #3318.h.to(Second) / 190.0.h.to(Second) ) / ( (5.0.mm * 5.0.mm).to(cm²) ) #.to(cm²) # normalize to cm⁻² #/ # * (175.0 * 3600.0) / #/ # the total background time (the detector was live in) #( (5.0.mm * 5.0.mm) / (14.0.mm * 14.0.mm) ) # scale counts up from gold region to equivalent in full chip #1.0 / (8359.18367347) # ratio of pixels in gold region #pow(0.95 - 0.45, 2.0) / # area of gold region! #pow(1.4, 2.0) / #12.0 # * # to get to / keV #(pow(1.4, 2.0) / 65535.0) echo "[KDE] SCALE: ", scale kde = kde.map_inline: x * scale.float #echo kde[0 .. 500] result = toDf({"Energy" : xs, "KDE" : kde}) let integral = simpson(result["KDE", float].toSeq1D, result["Energy", float].toSeq1D) echo "XXX: think about this INTEGRAL OF KDE!!!" echo "[KDE] ", integral, " and ratio to naive ", (integral / (energy.size.float / TrackingBackgroundRatio)) if toPlot: let outname = if outname.len == 0: "/home/basti/org/Figs/statusAndProgress/backgroundRates/background_2017_2018_kde_rate.pdf" else: outname ggplot(result, aes("Energy", "KDE")) + geom_line() + ggtitle("KDE of the background clusters, normalized to keV⁻¹•cm⁻² in 190 h of tracking") + ggsave(outname) #if true: quit() ## XXX: make them compile time variables and only allow modification from static vars? var Radius* = 40.0 # 100.0 #33.0 var Sigma* = 40.0 / 3.0 #33.3 #11.111 var EnergyRange* = 0.6.keV # Inf.keV #type # MyMetric* = object # radius*: float # sigma*: float # energyRange*: keV # ### A dummy proc that identifies our `MyMetric` as a custom metric #proc toCustomMetric(m: MyMetric): CustomMetric = CustomMetric() #let mym = MyMetric() proc distance*(metric: typedesc[CustomMetric], v, w: Tensor[float]): float = #echo "Metric ", metric #doAssert v.squeeze.rank == 1 #doAssert w.squeeze.rank == 1 doAssert EnergyRange.float != Inf, "WARNING: You did not set the `EnergyRange` (and likely `Sigma` and `Radius`) "& "variables! It is required to set them, as we cannot pass these variables to the `distance` procedure. They " & "are defined as globals above the procedure body!" #result = Euclidean.distance(v, w) #let diff = abs(v -. w) #let arg1 = diff[0, 0] #abs(v[0] - w[0]) #let arg2 = diff[0, 1] #abs(v[1] - w[1]) #let arg3 = diff[0, 2] #abs(v[2] - w[2]) # NOTE: this is the fastest way to compute the distance # - no squeeze # - no temp tensor allocation let arg1 = abs(v[0] - w[0]) let arg2 = abs(v[1] - w[1]) let arg3 = abs(v[2] - w[2]) let xyDist = arg1*arg1 + arg2*arg2 ##echo "xy dist ", xyDist, " vs ", Euclidean.distance(v, w) let zDist = arg3*arg3 if zDist <= (EnergyRange * EnergyRange).float: result = xyDist else: result = (2 * Radius * Radius).float # just some value larger than Radius² #pow(sqrt(zDist) * 6.0, 2.0) #if xyDist > zDist: # result = xyDist #elif xyDist < zDist and zDist <= Radius * Radius: # result = xyDist #else: # result = zDist ## XXX: In order to treat the energy as pure distance without gaussian behavior, we can do: ## - compute distance in both xy and z as currently ## - use `zDist` *only* (!!!) as an early "return" so to say. I.e. if larger than a cutoff we ## define, return it. Needs to be a global, as we don't know that cutoff from `v`, `w`, or rather ## hardcode into distance proc ## - else *always* return `xyDist`. This guarantees to give us the distance information of the points ## *always* along the xy, which is important for the weighing, but *not* along energy #proc distance(metric: typedesc[CustomMetric], v, w: Tensor[float]): float = # MyMetric.distance(v, w) import helpers/circle_segments proc correctEdgeCutoff*(val, radius: float, x, y: int): float {.inline.} = ## Corrects the effects of area being cut off for the given `val` if it is ## positioned at `(x, y)` and the considered radius is `radius`. ## ## TODO: for our normal weighted values, this edge cutoff is not correct. We need to ## renormalize by the *weighted* area and not the unweighted one... let refArea = PI * radius * radius let areaLeft = areaCircleTwoLinesCut(radius, min(x, 256 - x).float, min(y, 256 - y).float) result = val * refArea / areaLeft proc correctEdgeCutoff(t: var Tensor[float], radius: float) = ## Applies the edge correction for every point in the given tensor for y in 0 ..< 256: for x in 0 ..< 256: t[y, x] = correctEdgeCutoff(t[y, x], radius, x, y) proc correctEdgeCutoff3D(t: var Tensor[float], radius: float) = ## Applies the edge correction for every point in the given tensor for y in 0 ..< t.shape[0]: for x in 0 ..< t.shape[1]: for E in 0 ..< t.shape[2]: t[y, x, E] = correctEdgeCutoff(t[y, x, E], radius, x, y) proc plot2d[T](bl: T) = let pix = 256 var xs = newSeq[int](pix * pix) var ys = newSeq[int](pix * pix) var cs = newSeq[float](pix * pix) var idx = 0 for y in 0 ..< pix: for x in 0 ..< pix: xs[idx] = x ys[idx] = y cs[idx] = bl.eval(y.float, x.float)#t[y, x] inc idx ggplot(toDf(xs, ys, cs), aes("xs", "ys", fill = "cs")) + geom_raster() + #scale_fill_continuous(scale = (low: 0.0, high: 10.0)) + ggsave("/tmp/test.pdf") proc plot2dTensor*(t: Tensor[float], outname = "/tmp/test_tensor.pdf", title = "", yMax = 0.0) = var xs = newSeq[int](t.size) var ys = newSeq[int](t.size) var cs = newSeq[float](t.size) var idx = 0 for y in 0 ..< t.shape[0]: for x in 0 ..< t.shape[1]: xs[idx] = x ys[idx] = y #if t[y, x] > 5.0: # echo "Noisy pixel: ", x, " and ", y, " have count ", t[y, x] # inc sumNoise, t[y, x].int cs[idx] = t[y, x] inc idx #echo "Total noisy things: ", sumNoise template low: untyped = 4.5 / 14.0 * 256.0 template hih: untyped = 9.5 / 14.0 * 256.0 let df = toDf(xs, ys, cs) ggplot(df, aes("xs", "ys", fill = "cs")) + geom_raster() + geom_linerange(aes = aes(x = low(), yMin = low(), yMax = hih()), color = some(parseHex("FF0000"))) + geom_linerange(aes = aes(x = hih(), yMin = low(), yMax = hih()), color = some(parseHex("FF0000"))) + geom_linerange(aes = aes(y = low(), xMin = low(), xMax = hih()), color = some(parseHex("FF0000"))) + geom_linerange(aes = aes(y = hih(), xMin = low(), xMax = hih()), color = some(parseHex("FF0000"))) + scale_fill_continuous(scale = (low: 0.0, high: yMax)) + xlim(0, 256) + ylim(0, 256) + margin(top = 1.5) + ggtitle(title) + ggsave(outname) proc plot3DTensor(t: Tensor[float], outname = "/tmp/test_tensor_3d.pdf", title = "") = var xs = newSeq[int](t.size) var ys = newSeq[int](t.size) var Es = newSeq[int](t.size) var cs = newSeq[float](t.size) var idx = 0 var sumNoise = 0 for y in 0 ..< t.shape[0]: for x in 0 ..< t.shape[1]: for E in 0 ..< t.shape[2]: xs[idx] = x ys[idx] = y Es[idx] = E #if t[y, x] > 5.0: # echo "Noisy pixel: ", x, " and ", y, " have count ", t[y, x] # inc sumNoise, t[y, x].int cs[idx] = t[y, x, E] inc idx echo "Total noisy things: ", sumNoise when false: ggplot(toDf(xs, ys, Es, cs), aes("xs", "ys", fill = "cs")) + facet_wrap("Es", scales = "free") + geom_raster() + #scale_fill_continuous(scale = (low: 0.0, high: 10.0)) + ggtitle(title) + ggsave(outname, width = 1900, height = 1500) else: for tup, subDf in groups(toDf(xs, ys, Es, cs).group_by("Es")): ggplot(subDf, aes("xs", "ys", fill = "cs")) + geom_raster() + #scale_fill_continuous(scale = (low: 0.0, high: 10.0)) + ggtitle(title & " Energy: " & $tup[0][1].toFloat) + ggsave(&"/tmp/back_plot_energy_{tup[0][1].toFloat}.pdf") proc plotDf(df: DataFrame, title, outname: string) = ggplot(df, aes("centerX", "centerY")) + geom_point() + ggtitle(title) + ggsave(outname) template compValue*(tup: untyped, byCount = false, energyConst = false): untyped = ## Computes the weighted (`byCount`) / unweighted (`not byCount`) value associated ## with a position from the given neighbors (`tup` is a return of `query_ball_point` ## on a k-d tree) if byCount: tup.idx.size.float else: # weigh by distance using gaussian of radius being 3 sigma let dists = tup[0] var val = 0.0 for d in items(dists): val += seqmath.gauss(d, mean = 0.0, sigma = Sigma) val proc compDistance(t: var Tensor[float], kd: KDTree[float], radius: float, byCount = false) = for y in 0 ..< 256: for x in 0 ..< 256: let tup = kd.query_ball_point([x.float, y.float].toTensor, radius) let val = compValue(tup) t[y, x] = val proc compValueTree(kd: KDTree[float], x, y, E: float, radius: float, metric: typedesc[AnyMetric], byCount = false): float {.inline.} = ## Queries the tree at the given coordinate and energy and returns the correctly ## weighted value at the point. let tup = kd.query_ball_point([x, y, E].toTensor, radius, metric = metric) if x == 127 and y == 127: toDf({"dists" : tup[0]}).writeCsv("/tmp/distances_127_127.csv") #let df = seqsDoDf(dists) result = compValue( tup, byCount = byCount ) proc compDistance3D(t: var Tensor[float], Es: seq[float], kd: KDTree[float], radius: float, byCount = false, metric = Euclidean) = for y in 0 ..< 256: echo "Starting y ", y for x in 0 ..< 256: for E in 0 ..< Es.len: t[y, x, E] = kd.compValueTree(x.float, y.float, Es[E], radius, metric, byCount) defUnit(keV⁻¹•cm⁻²•s⁻¹, toExport = true) proc normalizeValue*(x, radius: float, energyRange: keV, backgroundTime: Hour): keV⁻¹•cm⁻²•s⁻¹ = let pixelSizeRatio = 65536 / (1.4 * 1.4).cm² when false: # case for regular circle with weights 1 let area = π * radius * radius # area in pixel else: let σ = Sigma ## This comes for integration with `sagemath` over the gaussian weighting. See the notes. let area = -2*π*(σ*σ * exp(-1/2 * radius*radius / (σ*σ)) - (σ*σ)) let energyRange = energyRange * 2.0 # we look at (factor 2 for radius) ## NOTE: for an *expected limit* this time must be the full background time, as it ## is the time that describes the number of clusters we have in the input! Thus, ## if we change it to `t_back - t_track`, we artificially increase our background! #let backgroundTime = 3318.h.to(Second) #(3318.h - 169.h).to(Second) let factor = area / pixelSizeRatio * # area in cm² energyRange * backgroundTime.to(Second) result = x / factor proc normalizeTensor(t: var Tensor[float], energies: int, radius: float) = ## Normalizes the tensor to units of /keV /cm^2 /s echo "Normalizing tensor by time: \n\n\n" for y in 0 ..< 256: for x in 0 ..< 256: for E in 0 ..< energies: t[y, x, E] = normalizeValue(t[y, x, E], radius, EnergyRange, 3318.Hour).float proc compNormalized(kd: KDTree[float], x, y: int, E: keV, radius: float, energyRange: keV, backgroundTime: Hour, metric: typedesc[AnyMetric] ): float = ## Computes a correctly normalized value for the given position and energy, ## using the `radius` from the given tree `kd`. result = compValueTree(kd, x.float, y.float, E.float, radius, metric) .correctEdgeCutoff(radius, x, y) .normalizeValue(radius, energyRange, backgroundTime).float template fillChip(body: untyped): untyped = var t {.inject.} = zeros[float]([256, 256]) for y {.inject.} in 0 ..< 256: for x {.inject.} in 0 ..< 256: body t proc compInterEnergy(t: var Tensor[float], kd: KDTree[float], energy: keV, radius: float, energyRange: keV, backgroundTime: Hour, metric: typedesc[AnyMetric], byCount = false) = t = fillChip: t[y, x] = kd.compNormalized(x, y, energy, radius, energyRange, backgroundTime, metric) if x == 128 and y == 128: echo "Rate at center: ", t[y, x] func toIdx*(arg: float): int = (arg / 14.0 * 256.0).round.int.clamp(0, 255) func toInch*(arg: float|int): float = (arg.float / 256.0 * 14.0).clamp(0.0, 14.0) proc plotGoldRegionBackgroundRate(kd: KDTree[float], outfile: string, title: string, backgroundTime = 3318.Hour) = var num = 25 let coords = linspace(4.5, 9.5, num) # the gold region var energies = linspace(0.0, 12.0, 75) var rates = newSeq[float](energies.len) for i, E in energies: var val = 0.0 for y in coords: for x in coords: val += compNormalized(kd, x.toIdx, y.toIdx, E.keV, Radius.float, EnergyRange, backgroundTime, metric = CustomMetric) rates[i] = val / (num * num).float echo "At energy ", E, " of index ", i, " rate: ", rates[i] let dfL = toDf(energies, rates) ggplot(dfL, aes("energies", "rates")) + geom_point() + ggtitle(title) + ggsave(outfile) #"/tmp/background_gold_region_from_interp.pdf") template plotEnergySlice*(outfile, title: string, yMax: float, body: untyped): untyped = let tr = fillChip: body tr.plot2dTensor(outfile, title, yMax) proc plotSingleEnergySlice*(kd: KDTree[float], energy: keV, backgroundTime = 3318.Hour, outfile = "", title = "") = let title = if title.len > 0: title else: &"Background interpolation at {energy} keV" let outfile = if outfile.len > 0: outfile else: &"/tmp/back_interp_energy_{energy}.pdf" var tr = zeros[float]([256, 256]) tr.compInterEnergy(kd, energy, Radius.float, EnergyRange, backgroundTime, byCount = false, metric = CustomMetric) tr.plot2dTensor(outfile, title) proc toNearestNeighborTree*(df: DataFrame): KDTree[float] = ## calls the correct interpolation function and returns the interpolated data echo "[INFO]: Building tree based on ", df.len, " background clusters in input" let tTree = stack([df["centerX", float].map_inline(toIdx(x).float), df["centerY", float].map_inline(toIdx(x).float), df["Energy", float].map_inline(x)], axis = 1) #df["Energy", float].map_inline(x * 25.6)], axis = 1) result = kdTree(tTree, leafSize = 16, balancedTree = true) proc studyBackgroundInterpolation*(df: DataFrame, toPlot = false): DataFrame = ## generates a kd tree based on the data and generates multiple plots ## we use to study the interpolation and determine good parameters var t = zeros[float]([256, 256]) for idx in 0 ..< df.len: let x = toIdx df["centerX", float][idx] let y = toIdx df["centerY", float][idx] t[y, x] += 1 t.plot2dTensor() #if true: quit() block Bilinear: var bl = newBilinearSpline(t, (0.0, 255.0), (0.0, 255.0)) # bicubic produces negative values! bl.plot2d() #block kdTree: # let tTree = stack([df["centerX", float].map_inline(toIdx(x).float), # df["centerY", float].map_inline(toIdx(x).float)], # axis = 1) # let kd = kdTree(tTree, leafSize = 16, balancedTree = true) # var treeDist = zeros[float]([256, 256]) # # for radius in [30]: #arange(10, 100, 10): # treeDist.compDistance(kd, radius.float, byCount = true) # treeDist.plot2dTensor("/tmp/background_radius_byenergy_" & $radius & "_bycount.pdf", # "k-d tree interpolation with radius: " & $radius & " pixels") # treeDist.correctEdgeCutoff(radius.float) # treeDist.plot2dTensor("/tmp/background_radius_byenergy_" & $radius & "_bycount_corrected.pdf", # "k-d tree interpolation with radius: " & $radius & " pixels") # treeDist.compDistance(kd, radius.float) # treeDist.plot2dTensor("/tmp/background_radius_byenergy_" & $radius & ".pdf", # "k-d tree interpolation with radius: " & $radius & " pixels") # treeDist.correctEdgeCutoff(radius.float) # treeDist.plot2dTensor("/tmp/background_radius_byenergy_" & $radius & "_corrected.pdf", # "k-d tree interpolation with radius: " & $radius & " pixels") # now plot interpolation based on energy echo "3d???\n\n" block kdTree3D: let tTree = stack([df["centerX", float].map_inline(toIdx(x).float), df["centerY", float].map_inline(toIdx(x).float), df["Energy", float].map_inline(x)], axis = 1) #df["Energy", float].map_inline(x * 25.6)], axis = 1) let kd = kdTree(tTree, leafSize = 16, balancedTree = true) when false: kd.plotSingleEnergySlice(1.0.keV) when false: let Es = @[1.0, 2.0, 4.0, 5.0] #linspace(0.0, 12.0, 10) #for (radius, sigma, eSigma) in [(100.0, 33.3333, 0.3), # (100.0, 15.0, 0.3), # (75.0, 75.0 / 3.0, 0.3), # (50.0, 50.0 / 3.0, 0.3), # (33.333, 11.1111, 0.3), # (25.0, 25.0 / 3.0, 0.3), # (100.0, 33.3, 0.5), # (50.0, 50.0 / 3.0, 0.5)]: for (radius, sigma, eSigma) in [(33.0, 11.111, 0.3), (33.0, 11.111, 0.5), (25.0, 25.0/3.0, 0.3), (25.0, 25.0/3.0, 0.5), (20.0, 20.0/3.0, 0.3), (20.0, 20.0/3.0, 0.5), (15.0, 15.0/3.0, 0.3), (15.0, 15.0/3.0, 0.5)]: Radius = radius Sigma = sigma EnergyRange = eSigma.keV let path = "/tmp/plots/" let suffix = &"radius_{radius:.0f}_sigma_{sigma:.0f}_energyRange_{eSigma:.1f}" let suffixTitle = &"Radius: {radius:.0f}, σ: {sigma:.0f}, ΔE: {eSigma:.1f}" echo "Generating plots for: ", suffixTitle for E in Es: kd.plotSingleEnergySlice(E.keV, outfile = path / &"back_interp_energy_{E}_{suffix}.pdf", title = &"Background interp, energy = {E} keV, {suffixTitle}") kd.plotGoldRegionBackgroundRate(outfile = path / &"background_gold_from_interp_{suffix}.pdf", title = &"Interp based gold background rate: {suffixTitle}") if true: quit() var treeDist = zeros[float]([256, 256, 10]) echo "Start computationssss" for radius in [Radius]: #arange(10, 100, 10): let Es = linspace(0.0, 12.0, 10) echo "comp 3d dist" treeDist.compDistance3D(Es, kd, radius.float, byCount = false, metric = CustomMetric) echo "correct edges" treeDist.correctEdgeCutoff3D(radius.float) # treeDist[_, _, E].plot2dTensor( # &"/tmp/background_radius_byenergy_{E}_{radius}.pdf", # &"k-d tree interpolation with radius: {radius} pixels, energy {E}") #treeDist.correctEdgeCutoff3D(radius.float) echo "plot 3d" #treeDist.plot3DTensor("/tmp/background_3d_radius_byenergy_" & $radius & ".pdf", # "k-d tree interpolation with radius: " & $radius & " pixels") treeDist.normalizeTensor(10, radius) treeDist.plot3DTensor("/tmp/background_3d_radius_byenergy_" & $radius & "_normalized.pdf", "k-d tree interpolation with radius: " & $radius & " pixels, normalized") #treeDist.correctEdgeCutoff3D(radius.float) #treeDist.plot3DTensor("/tmp/background_radius_byenergy_correccted_" & $radius & ".pdf", # "k-d tree interpolation with radius: " & $radius & " pixels corrected by edges") # now plot interpolation based on energy #block kdTreeJustMoreStuff: # let tTree = stack([df["centerX", float].map_inline(toIdx(x).float), # df["centerY", float].map_inline(toIdx(x).float)], axis = 1) # let kd = kdTree(tTree, leafSize = 16, balancedTree = true) # var treeDist = zeros[float]([256, 256, 10]) # let radius = 30 # let Es = linspace(0.0, 12.0, 10) # for E in 0 ..< Es.high: # let df = df.filter(f{`Energy` >= Es[E] and `Energy` < Es[E+1]}) # let tTree = stack([df["centerX", float].map_inline(toIdx(x).float), # df["centerY", float].map_inline(toIdx(x).float)], axis = 1) # let kd = kdTree(tTree, leafSize = 16, balancedTree = true) # for y in 0 ..< 256: # for x in 0 ..< 256: # let tup = kd.query_ball_point([x.float, y.float].toTensor, radius.float, metric = CustomMetric)#, metric = CustomMetric) # let val = compValue(tup, byCount = true) # treeDist[y, x, E] = val # # treeDist.correctEdgeCutoff(radius.float) # # treeDist[_, _, E].plot2dTensor( # # &"/tmp/background_radius_byenergy_{E}_{radius}.pdf", # # &"k-d tree interpolation with radius: {radius} pixels, energy {E}") # #treeDist.correctEdgeCutoff(radius.float) # treeDist.plot3DTensor("/tmp/background_3d_radius_byenergy_notreally_" & $radius & ".pdf", # "k-d tree interpolation with radius: " & $radius & " pixels") # #treeDist.correctEdgeCutoff(radius.float) # #treeDist.plot3DTensor("/tmp/background_radius_byenergy_correccted_" & $radius & ".pdf", # # "k-d tree interpolation with radius: " & $radius & " pixels corrected by edges") # # # # now plot interpolation based on energy if true: quit() block kdTreeEnergy: df.showBrowser() discard df.toKDE(toPlot = true, outname = "/tmp/all_data.pdf") df.plotDf("all clusters", "/tmp/all_clusters.pdf") const radius = 50 for (l, h) in [(0, 2), (2, 10)]: #0 ..< 10: let energy = l let df = df.filter(f{`Energy` >= energy.float and `Energy` < (energy + h).float}) discard df.toKDE(toPlot = true,outname = &"/tmp/range_{energy}.pdf") df.plotDf(&"clusters in energy {energy}-{energy+h} keV", &"/tmp/clusters_{energy}.pdf") echo "Events left : ", df.len let tTree = stack([df["centerX", float].map_inline(toIdx(x).float), df["centerY", float].map_inline(toIdx(x).float)], axis = 1) let kd = kdTree(tTree, leafSize = 16, balancedTree = true) var treeDist = zeros[float]([256, 256]) treeDist.compDistance(kd, radius.float, byCount = true) treeDist.plot2dTensor( &"/tmp/background_energy_{energy}_radius_{radius}_bycount.pdf", &"k-d tree interp, radius: {radius} pixels, energy: {energy} - {energy+h} keV. # cluster: {df.len}") treeDist.correctEdgeCutoff(radius.float) treeDist.plot2dTensor( &"/tmp/background_energy_{energy}_radius_{radius}_bycount_corrected.pdf", &"k-d tree interp, radius: {radius} pixels, energy: {energy} - {energy+h} keV. # cluster: {df.len}, edge corrected") treeDist.compDistance(kd, radius.float) treeDist.plot2dTensor( &"/tmp/background_energy_{energy}_radius_{radius}.pdf", &"k-d tree interp, radius: {radius} pixels, energy: {energy} - {energy+h} keV. # cluster: {df.len}") treeDist.correctEdgeCutoff(radius.float) treeDist.plot2dTensor( &"/tmp/background_energy_{energy}_radius_{radius}_corrected.pdf", &"k-d tree interp, radius: {radius} pixels, energy: {energy} - {energy+h} keV. # cluster: {df.len}, edge corrected") if true: quit()
The likelihood function used in the nature paper includes the background as a function independent of the position \(\vec{x}\). For larger detectors than ours, this is a suitable approach, as one can simply cut to an area large enough to encompass the full signal.
In the analysis of the 2014/15 data, the notion of the gold region was introduced as that described a region in the detector with the lowest background and a seemingly constant background across the whole region.
In the limit calculation of the 2017/18 dataset however, the axion image computed by the raytracing code yields an image that has signal outside of the gold region. This is of course highly undesirable, because we throw out signal information for now reason (the background should be similarly low in the extended region).
The choice of the specific likelihood method has one big advantage: including areas with large backgrounds does not matter in general, if there is no expected signal in these regions. The likelihood value will be constant for all points without a signal sensitivity. It will increase the computational cost (instead of ~30 candidates one may have 300 of course), but it doesn't worsen the result to include these areas.
With that in mind, the region to be considered will be extended to the whole center chip. This has the implication that the background now is not constant over the whole chip.
So \(B(E)\) becomes \(B(E, \vec{x})\). In order to evaluate the background at an arbitrary candidate cluster position \(\vec{x}\), we need an interpolated background rate.
To achieve this we use the background clusters left after the likelihood cut including the septem veto, i.e. fig. 414.
The interpolation that will be used will be a nearest neighbor approach. Essentially, each point will be given the interpolated value based on how many neighbors it has within a fixed search radius.
For this a k-d tree is used to efficiently compute the distance between many points.
Multiple resulting backgrounds (after filtering out the noisy pixels!) are found in ./../Figs/statusAndProgress/background_interpolation/.
The input data files for this are HDF5 files after running the likelihood cut with the following parameters:
- full chip
- septem veto
- clustering: DBSCAN, ε = 65
found in:
/home/basti/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_all_chip_septem_dbscan.h5/home/basti/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2018_all_chip_septem_dbscan.h5
The interpolation with a small search radius of 10 pixels is found in fig. 415, one with a very large radius of 100 pixels in fig. 416 and a possibly reasonable choice between homogeneity and a still good enough background rate in the center with 30 pixels in 417.
- Mathematical idea of the background interpolation
Assume the background interpolation had the idea to interpolate at an arbitrary point on the chip, but disregarding all energy information.
Then we could, in the very simplest way, associate a background rate value for each point, based on the total number of clusters within a certain radius around the point we want to describe the background rate at. That means in math:
\[ b(\vec{x}) = \frac{I}{W} = \frac{ \sum_{c_i ∈ \{ | \vec{x} - \vec{x_i} | \leq R \} } 1 }{π R² · t} \]
where
\[ c_i ∈ \{ | \vec{x} - \vec{x_i} | \leq R \} \]
describes all cluster centers \(c_i\) with position \(\vec{x_i}\), which are within the radius \(R\) around the point \(\vec{x}\). The sum over \(1\) simply means we count the number of clusters.
By normalizing by the area of a circle and the total background time \(t\) (again, ignoring the energy), we get back a background rate in counts per \(\si{cm^{-2}.s^{-1}}\) (divide by, say, \SI{10}{keV} as well, if all your clusters are only those between \(\SIrange{0}{10}{keV}\) for a background rate in standard units.
From here it is simply an equivalent extension where we do not just give each cluster the weight \(1\), but rather a weight which depends on the distance it is from the point we look at \(\vec{x}\). This weight is the "measure" \(\mathcal{M}\) in the paper and the expression of \(c_i\) is the metric (well, it uses the euclidean metric in 2D).
The final piece to understand is that the \(πR²\) comes from integrating over the metric we consider (a circle with radius \(R\)) with a fixed measure of 1.
So following the excerpt about the interpolation method from the paper:
- Background interpolation with custom metric and measure
For the background we construct a background interpolation based on all clusters found in the background dataset. This is done by defining \(b_i\) as a function of candidate position and energy using
\[ b_i(x_i, y_i, E_i) = \frac{I(x_i, y_i, E_i)}{W(x_i, y_i, E_i)} \]
where \(I\) is an intensity defined over clusters within a range \(R\) and a normalization weight \(W\). The intensity is given by
\[ I(x, y, E) = \sum_{b ∈ \{ \mathcal{D}(\vec{x}_b, \vec{x}) \leq R \}}\mathcal{M}(\vec{x}_b, E_b) = \sum_{b ∈ \{ \mathcal{D}(\vec{x}_b, \vec{x}) \leq R \} } \exp \left[ -\frac{1}{2} \mathcal{D}² / σ² \right] \text{ for clarity w/o arguments}, \]
where we introduce \(\mathcal{M}\) to refer to the measure we use and \(\mathcal{D}\) to our metric:
\begin{equation*} \mathcal{D}( (\vec{x}_1, E_1), (\vec{x}_2, E_2)) = \begin{cases} (\vec{x}_1 - \vec{x}_2)² \text{ if } |E_1 - E_2| \leq R \\ ∞ \text{ if } (\vec{x}_1 - \vec{x}_2)² > R² \\ ∞ \text{ if } |E_1 - E_2| > R \end{cases} \end{equation*}Finally, the normalization weight is the 'volume' of our measure within the boundaries set by our metric \(\mathcal{D}\):
\[ W(x', y', E') = ∫_{\mathcal{D}(\vec{x'}, \vec{x}) \leq R} ∫_{E' - E_c}^{E' + E_c} \mathcal{M}(x', y')\, \mathrm{d}x\, \mathrm{d}y\, \mathrm{d} E \]
This yields a smooth and continuous interpolation of the background over the entire chip.
The axion flux and detector efficiency are shown in fig. [BROKEN LINK: fig:detection_efficiency], with the axion image in fig. [BROKEN LINK: fig:axion_image]. Fig. [BROKEN LINK: fig:background_interpolation] shows an example of the background interpolation, with original background clusters used as a basis for the interpolation as crosses.
- Background interpolation with custom metric and measure
- Notes an limit calculation using background interpolation
Currently using the following parameters for the interpolation:
Radius = 33.3 Sigma = 11.1 EnergyRange = 0.3.keV
The discussions about the background rate below apply for these. Larger values of course do not suffer from the same b = 0 problem.
First of all we changed the implementation of the likelihood function in code to the one described in section [BROKEN LINK: sec:determine_limit_maths], eq. [BROKEN LINK: eq:likelihood_1_plus_s_over_b_form].
That means instead of the naive
\[ \mathcal{L} \supset Σ_i \ln(s_i + b_i) \] we now use: \[ \mathcal{L} \supset Σ_i \ln(1.0 + \frac{s_i}{b_i}) \] instead. This is because in code otherwise the resulting sum becomes very large, given that it's still a log. So taking the \(\exp\) becomes impossible (\(\exp^{1400}\) says hi).
In addition for the background interpolation we now create a grid of (currently, to be updated):
- 10 by 10 cells in x/y
- 20 intervals in energy
to cover the full interpolation. An example for 0.3 keV (the first interval center) using the full 3300h of background data (hence the large numbers) normalized to the expected number of counts in that cube of size 10/256×10/256×12/20 pixel²•keV.
Figure 418: Example of a grid (first layer at 0.3 keV) of the background interpolation normalized to expected number of counts in each volume over the full ~3300 h of background time. 10 intervals in x/y and 20 in E. Using this approach with the old log likelihood implementation yielded problems.
- sometimes the signal and background is 0, meaning the result is log(0) = inf
- the sum of all terms becomes so large (if the log(0) case is dealt with somehow) that exp(arg) explodes
The approach of s/b is saner, as it "renormalizes itself" to an extent (the sum might still get big).
I added some counters to see how many signal cases are 0, background are 0 and signal + background are 0.
We get output like this:
ExpRate -10.01057897300981 b == 0 : (energy: 5.02006 KiloElectronVolt, pos: (x: 6.331890951689641, y: 5.714509212104123)) b == 0 : (energy: 6.24657 KiloElectronVolt, pos: (x: 5.007987099772659, y: 4.263161399120039)) b == 0 : (energy: 6.7407 KiloElectronVolt, pos: (x: 5.529037723808754, y: 5.073390621671234)) b == 0 : (energy: 6.68518 KiloElectronVolt, pos: (x: 5.244443309844992, y: 5.129942459518077)) b == 0 : (energy: 6.99333 KiloElectronVolt, pos: (x: 5.910306924047042, y: 6.451715430422019)) b == 0 : (energy: 6.67893 KiloElectronVolt, pos: (x: 8.273825230476792, y: 7.642340990510401)) b == 0 : (energy: 8.07189 KiloElectronVolt, pos: (x: 5.686307215545322, y: 6.759320281042876)) b == 0 : (energy: 8.23012 KiloElectronVolt, pos: (x: 6.89443837734045, y: 6.505802303614423)) b == 0 : (energy: 8.27733 KiloElectronVolt, pos: (x: 7.672479685804561, y: 7.240523496285816)) b == 0 : (energy: 8.9925 KiloElectronVolt, pos: (x: 4.976138830265345, y: 4.469258935069182)) b == 0 : (energy: 8.51804 KiloElectronVolt, pos: (x: 4.874677319014947, y: 4.530622674784526)) b == 0 : (energy: 8.89131 KiloElectronVolt, pos: (x: 5.473732300114362, y: 5.165760540087661)) b == 0 : (energy: 8.70646 KiloElectronVolt, pos: (x: 6.413046099332512, y: 6.23861241570493)) b == 0 : (energy: 9.11151 KiloElectronVolt, pos: (x: 4.51902259412155, y: 4.997881338963736)) b == 0 : (energy: 9.08194 KiloElectronVolt, pos: (x: 6.003012703715648, y: 6.127977970262161)) b == 0 : (energy: 9.02828 KiloElectronVolt, pos: (x: 5.717591991796073, y: 5.672806447271474)) b == 0 : (energy: 9.00585 KiloElectronVolt, pos: (x: 7.003494272398496, y: 7.951011750307492)) b == 0 : (energy: 9.09002 KiloElectronVolt, pos: (x: 7.10388024075134, y: 7.211015005655566)) b == 0 : (energy: 9.28271 KiloElectronVolt, pos: (x: 7.776885657372958, y: 8.330602402567015)) b == 0 : (energy: 9.14102 KiloElectronVolt, pos: (x: 8.666514451356207, y: 9.468513928388699)) b == 0 : (energy: 9.72436 KiloElectronVolt, pos: (x: 6.724480803045357, y: 6.040910394022069)) b == 0 : (energy: 9.69753 KiloElectronVolt, pos: (x: 7.828530024091032, y: 7.033851473444065)) b == 0 : (energy: 9.7249 KiloElectronVolt, pos: (x: 7.375859886039128, y: 7.114695915632516)) ================================================================================ g_aγ² = 9.999999999999999e-25 g_ae² = 1e-20 Number of candidates: 471 Number of zero signal candidates: 350 Number of zero background candidates: 23 Number of zero sig & back candidates: 37
Meaning: we get actual 0 in the background. How does that happen? The gridded cube gives us non zero elements. So we draw candidates there. But then we smear them inside of the volume and at the points shown here, we actually have exactly 0 background using our parameters.
An example of such a background rate of the first point printed in the snippet above:
Figure 419: Background rate at the energy a candidate saw b = 0 exactly. We can see that given our parameters in use (R = 33, σ = 11.1, ΔE = 0.3.keV) we really do have a background of 0 at some points (color bar bottoms at 0). Therefore, I'll now try to change the parameters to use a smaller grid.
In theory though a smaller grid can never quite get rid of this problem, as the grid is unlikely to be small enough to cover exactly the same non-zero points…
As expected, increasing the grid number from 10×10 cells to 20×20 cells, gives us this:
ExpRate -10.01057897300981 b == 0 : (energy: 6.51676 KiloElectronVolt, pos: (x: 4.670892311835505, y: 4.733753169712938)) b == 0 : (energy: 6.37838 KiloElectronVolt, pos: (x: 4.585841770111567, y: 4.250060941450658)) b == 0 : (energy: 6.67148 KiloElectronVolt, pos: (x: 8.419141697886676, y: 9.019150563914415)) b == 0 : (energy: 8.23022 KiloElectronVolt, pos: (x: 4.544408750623171, y: 4.877312399395358)) b == 0 : (energy: 8.17686 KiloElectronVolt, pos: (x: 5.410146813956097, y: 5.57445057817234)) b == 0 : (energy: 8.15377 KiloElectronVolt, pos: (x: 5.21652255584068, y: 5.473809085604193)) b == 0 : (energy: 8.61204 KiloElectronVolt, pos: (x: 5.38658361312396, y: 5.513525234867119)) b == 0 : (energy: 8.53658 KiloElectronVolt, pos: (x: 5.618590960698463, y: 6.129311333767217)) b == 0 : (energy: 9.55803 KiloElectronVolt, pos: (x: 4.855358217134393, y: 4.542859468482585)) b == 0 : (energy: 9.0901 KiloElectronVolt, pos: (x: 5.494320129337995, y: 5.073960439486082)) b == 0 : (energy: 9.43972 KiloElectronVolt, pos: (x: 5.209610033935212, y: 5.582783930340959)) b == 0 : (energy: 9.12122 KiloElectronVolt, pos: (x: 5.57947712663182, y: 5.419490376516271)) b == 0 : (energy: 9.21062 KiloElectronVolt, pos: (x: 7.772429285353669, y: 8.220859222253237)) b == 0 : (energy: 9.60304 KiloElectronVolt, pos: (x: 4.937146004994323, y: 5.389417494291933)) ================================================================================ g_aγ² = 9.999999999999999e-25 g_ae² = 1e-20 Number of candidates: 503 Number of zero signal candidates: 377 Number of zero background candidates: 14 Number of zero sig & back candidates: 38
So the number has gone down, but of course not disappeared.
Update: 40 still has some left…. Even at pixel size we have some left, likely because of the energy range. A range of 12 / 20 = 0.6 is technically larger than the gaussian 2 * 0.3 keV window of the interpolation itself. Depending on where these points end up, it may be at a higher background than points in between.
Update 2: The parameters:
let radius = 40.0#33.3 let σ = radius / 3.0 let energyRange = 0.6.keV #0.3.keV let nxy = 20 let nE = 20
don't produce any issues during the initial scan, but may still later. And it's not verified that these actually produce a good interpolation!
Update 3: Indeed, running it more often causes exactly the same issue. As expected it's just more rare.
- TODO Verify parameters!
Make a plot of these. Gold region background rate as well as the interpolation slices!
- TODO NOTE: write a small script that (after reading input from HDF5 once)
one can call, giving it the above parameters and it just generates the correct plot!
- TODO Compute uncertainty on background interpolation point
Essentially try to estimate a "minimum" background for a single data point "volume" for the interpolation.
Look at the uncertainty studies we did. From it we can ask: what is the lowest background that one can detect where the chance of not getting a single entry is larger than say 50%, 75%, 90% ?
The number that comes out as a result can be used as a fallback in a sense for the b = 0 case.
In German to Klaus:
Da aber vorhin die konstante Methode schon überall quasi nur 3.05e-21 oder 3.1e-21 geworfen hat, werde ich morgen erstmal schauen, was der Grund dafür ist, dass ich gerade keine anderen Ergebnisse mehr bekomme Ich denke wir sollten doch eigentlich in der Lage sein, dass über die Unsicherheit auszugleichen. Wir wissen ja quasi was für eine Unsicherheit wir auf die Background Hypothese haben und wir können eine Aussage darüber treffen, wie "wenig" Untergrund wir überhaupt statistisch signifikant innerhalb der Zeit, die wir Background Daten genommen haben, hätten erreichen können. Das sollte doch quasi der unterste Wert sein, auf den die Interpolation gesetzt werden müsste. Oder irgendwie sowas. Ich bin langsam was müde 😅
Simple calc: 3300 h ~152 · π area 0.6 keV
1 count
import unchained let num = 1 # 1 cluster let t = 3300.h let rad = (15.0 / (256.0 / 14.0)).mm let area = rad * rad * π let ΔE = 0.6.keV let rate = num / (t.to(Second) * area * ΔE) echo rate echo "In 190h : ", rate * 190.h.to(Second) * area
So naively in a cylinder of radius 15 pixels with a single count we'd have a background rate of 6.6e-8.
That level would mean 1 ± 1 counts expected. Thus, there's a good chance not a single one would be expected. So, anything significantly below that as a background hypothesis doesn't make any sense.
- STARTED Homogeneous background for energies > 2 keV
TODO: finish the final explanation part!!
The issue with energies larger than 2 keV and performing the interpolation of all events larger than 2 in a reasonably large radius, has one specific problem.
The plot in fig. 420 shows what the interpolation for > 2 keV looks like for a radius of 80 pixels.
It is very evident that the background appears higher in the center area than in the edges / corners of the chip.
The reason for this is pretty obvious once one thinks about it deeper. Namely, an event with a significant energy that went through decent amounts of diffusion, cannot have its cluster center (given that it's X-ray like here) actually close to the edge / corner of the detector. On average its center will be half the diffusion radius away from the edges. If we then interpolate based on the cluster center information, we end up at a typical boundary problem, i.e. they are underrepresented.
Figure 420: Background interpolation for 2017/18 X-ray like data for all clusters above \SI{2}{keV} using a radius of 80 pixels. It is evident that the background in the center appears higher than at the edges, despite expecting either the opposite or constant background. Reason is cutoff at edges, so no contributions can come from outside + diffusion causing cluster centers to always be a distance away from the edges. Now, what is a good solution for this problem?
In principle we can just say "background is constant over the chip at this energy above 2 keV" and neglect the whole interpolation here, i.e. set it constant.
If we wish to keep an interpolation around, we will have to modify the data that we use to create the actual 2D interpolator.
Of course the same issue is present in the < 2 keV dataset to an extent. The question there is: does it matter? Essentially, the statement about having less background there is factually true. But only to the extent of diffusion putting the centers away from the edges, not from just picking up nothing from the area within the search radius that lies outside the chip (where thus no data can be found).
Ideally, we correct for this by scaling all points that contain data outside the chip by the fraction of area that is within the radius divided by the total area. That way we pretend that there is an 'equal' amount of background found in this area in the full radius around the point.
How?
Trigonometry for that isn't fully trivial, but also not super hard.
Keep in mind the area of a circle segment: \[ A = r² / 2 * (ϑ - sin(ϑ)) \] where \(r\) is the radius of the circle and ϑ the angle that cuts off the circle.
However, in the general case we need to know the area of a circle that is cut off from 2 sides. By subtracting the corresponding areas of circle segments for each of the lines that cut something off, we remove too much. So we need to add back:
- another circle segment, of the angle between the two angles given by the twice counted area
- the area of the triangle with the two sides given by \(R - r'\) in length, where \(r'\) is the distance that is cut off from the circle.
In combination the area remaining for a circle cut off from two (orthogonal, fortunately) lines is:
\[ E = F - A - B + C + D \] where:
- \(F\): the total area of the circle
- \(A\): the area of the first circle segment
- \(B\): the area of the second circle segment
- \(C\): the area of the triangle built by the two line cutoffs: \[ C = \frac{r' r''}{2} \] with \(r'\) as defined above for cutoff A and \(r''\) for cutoff B.
- \(D\): the area of the circle segment given by the angles between the two cutoff lines touching the circle edge: \[ D = r² / 2 * (α - sin(α)) \] where \(α\) is: \[ α = π/2 - ϑ_1 - ϑ_2 \] where \(ϑ_{1,2}\) are the related to the angles \(ϑ\) needed to compute each circle segment, by: \[ ϑ' = (π - ϑ) / 2 \] denoted as \(ϑ'\) here.
Implemented this as a prototype in: ./../Misc/circle_segments.nim UPDATE: which now also lives in TPA in the
NimUtil/helpersdirectory!Next step: incorporate this into the interpolation to re-weight the interpolation near the corners.
- Normalization of gaussian weighted k-d tree background interpolation
The background interpolation described above includes multiple steps required to finalize it.
As mentioned, we start by building a k-d tree on the data using a custom metric:
proc distance(metric: typedesc[CustomMetric], v, w: Tensor[float]): float = doAssert v.squeeze.rank == 1 doAssert w.squeeze.rank == 1 let xyDist = pow(abs(v[0] - w[0]), 2.0) + pow(abs(v[1] - w[1]), 2.0) let zDist = pow(abs(v[2] - w[2]), 2.0) if zDist <= Radius * Radius: result = xyDist else: result = zDist #if xyDist > zDist: # result = xyDist #elif xyDist < zDist and zDist <= Radius: # result = xyDist #else: # result = zDist
or in pure math:
Let \(R\) be a cutoff value.
\begin{equation} \mathcal{D}( (\vec{x}_1, E_1), (\vec{x}_2, E_2)) = \begin{cases} (\vec{x}_1 - \vec{x}_2)² \text{ if } |E_1 - E_2| \leq R \\ |E_1 - E_2| \end{cases} \end{equation}where we make sure to scale the energies such that a value for the radius in Euclidean space of the x / y geometry covers the same range as it does in energy.
This creates essentially a cylinder. In words it means we use the distance in x and y as the actual distance, unless the distance in energy is larger than the allowed cutoff, in which case we return it.
This simply assures that:
- if two clusters are close in energy, but further in Euclidean distance than the allowed cutoff, they will be removed later
- if two clusters are too far away in energy they will be removed, despite possibly being close in x/y
- otherwise the distance in energy is irrelevant.
The next step is to compute the actual background value associated with each \((x, y, E)\) point.
In the most naive approach (as presented in the first few plots in the section above), we can associate to each point the number of clusters found within a certain radius (including or excluding the energy dimension).
For obvious reasons treating each point independent of the distance as a single count (pure nearest neighbor) is problematic, as the distance matters of course. Thus, our choice is a weighted nearest neighbor. Indeed, we weigh the distance by normal distribution centered around the location at which we want to compute the background.
So, in code our total weight for an individual point is:
template compValue(tup: untyped, byCount = false, energyConst = false): untyped = if byCount: tup.idx.size.float # for the pure nearest neighbor case else: # weigh by distance using gaussian of radius being 3 sigma let dists = tup[0] var val = 0.0 for d in items(dists): # default, gaussian an energy val += smath.gauss(d, mean = 0.0, sigma = radius / 3.0) val
where
tupcontains the distances to all neighbors found within the desired radius.In math this means we first modify our distance measure \(\mathcal{D}\) from above to:
\begin{equation} \mathcal{D'}( (\vec{x}_1, E_1), (\vec{x}_2, E_2)) = \begin{cases} (\vec{x}_1 - \vec{x}_2)² \text{ if } |E_1 - E_2| \leq R \\ 0 \text{ if } (\vec{x}_1 - \vec{x}_2)² > R² \\ 0 \text{ if } |E_1 - E_2| > R \end{cases} \end{equation}to incorporate the nearest neighbor properties of dropping everything outside of the radius either in x/y or in (scaled) energy.
\begin{align*} I(\vec{x}_e, E_e) &= Σ_i \exp \left[ -\frac{1}{2} \left( \mathcal{D'}((\vec{x}_e, E_e), (\vec{x}_i, E_i)) \right)² / σ² \right] \\ I(\vec{x}_e, E_e) &= Σ_i \exp \left[ -\frac{1}{2} \mathcal{D'}² / σ² \right] \text{ for clarity w/o arguments}\\ I(\vec{x}_e, E_e) &= Σ_i \mathcal{M}(\vec{x}_i, E_i) \\ \text{where we introduce }&\mathcal{M}\text{ to refer to the measure we use.} \end{align*}where
iruns over all clusters (\(\mathcal{D'}\) takes care of only allowing points in the radius to contribute) and the subscriptestands for the evaluation point. \(σ\) is the sigma of the (non-normalized!) Gaussian distribution for the weights, which is set to \(σ = \frac{R}{3}\).This gives us a valid interpolated value for each possible value of position and energy pairs. However, these are still not normalized, nor corrected for the cutoff of the radius once it's not fully "on" the chip anymore. The normalization is done via the area of circle segments, as described in the previous section 29.1.3.3.
The normalization will be described next. For the case of unweighted points (taking every cluster in the 'cylinder'), it would simply be done by dividing by the:
- background data taking time
- energy range of interest
- volume of the cylinder
But for a weighted distance measure \(\mathcal{D'}\), we need to perform the integration over the measure (which we do implicitly for the non-weighted case by taking the area! Each point simply contributes with 1, resulting in the area of the circle).
The necessary integration over the energy can be reduced to simply dividing by the energy range (the 'cylinder height' part if one will), as everything is constant in the energy direction, i.e. no weighting in that axis.
Let's look at what happens in the trivial case for an understanding of what we are actually doing when normalizing by area of a non-weighted thing.
The measure in the unweighted case is thus: \[ \mathcal{M}(x, y) = 1 \]
Now, we need to integrate this measure over the region of interest around a point (i.e from a point x over the full radius that we consider):
\begin{align*} W &= \int_{x² + y² < R²} \mathcal{M}(x', y')\, \mathrm{d}x \mathrm{d}y \\ &= \int_{x² + y² < R²} 1\, \mathrm{d}x \mathrm{d}y \\ &= \int_0^R \int_0^{2 π} r\, \mathrm{d}r \mathrm{d}φ \\ &= \int_0^{2 π} \frac{1}{2} R² \, \mathrm{d}φ \\ &= 2 π\frac{1}{2} R² \\ &= π R² \end{align*}where the additional \(r\) after transformation from cartesian coordinates to polar coordinates is from the Jacobi determinant (ref: https://en.wikipedia.org/wiki/Jacobian_matrix_and_determinant#Example_2:_polar-Cartesian_transformation as a reminder). For this reason it is important that we start our assumption in cartesian coordinates, as otherwise we miss out that crucial factor! Unexpectedly, the result is simply the area of a circle with radius \(R\), as we intuitively assumed to be for a trivial measure.
For our actual measure we use: \[ \mathcal{M}(\vec{x}_i, E_i) = \exp \left[ - \frac{1}{2} \mathcal{D'}²((\vec{x}_e, E_e), (\vec{x}_i, E_i)) / σ² \right] \] the procedure follows in the exact same fashion (we leave out the arguments to \(\mathcal{D}\) in the further part:
\begin{align*} W &= \int_{x² + y² < R²} \mathcal{M}(x', y')\, \mathrm{d}x \mathrm{d}y \\ &= \int_{x² + y² < R²} \exp \left[ - \frac{1}{2} \mathcal{D'}² / σ² \right] \, \mathrm{d}x \mathrm{d}y \\ &= \int_0^R \int_0^{2 π} r \exp \left[ - \frac{1}{2} \mathcal{D'}² / σ² \right]\, \mathrm{d}r \mathrm{d}φ \end{align*}which can be integrated using standard procedures or just using SageMath, …:
sage: r = var('r') # for radial variable we integrate over sage: σ = var('σ') # for constant sigma sage: φ = var('φ') # for angle variable we integrate over sage: R = var('R') # for the radius to which we integrate sage: assume(R > 0) # required for sensible integration sage: f = exp(-r ** 2 / (sqrt(2) * σ) ** 2) * r sage: result = integrate(integrate(f, r, 0, R), φ, 0, 2 * pi) sage: result -2*pi*(σ^2*e^(-1/2*R^2/σ^2) - σ^2) sage: result(R = 100, σ = 33.33333).n() 6903.76027055093 - Error propagation of background interpolation
For obvious reasons the background interpolation suffers from statistical uncertainties. Ideally, we compute the resulting error from the statistical uncertainty for the points by propagating the errors through the whole computation. That is from the nearest neighbor lookup, through the sum of the weighted distance calculation and then the normalization.
We'll use https://github.com/SciNim/Measuremancer.
import datamancer, measuremancer, unchained, seqmathStart by importing some data taken from running the main program. These are the distances at some energy at pixel (127, 127) to the nearest neighbors.
when isMainModule: const data = """ dists 32.14 31.89 29.41 29.12 27.86 21.38 16.16 16.03 """
Parse and look at it:
when isMainModule: var df = parseCsvString(data) echo df
Now import the required transformations of the code, straight from the limit code (we will remove all unnecessary bits). First get the radius and sigma that we used here:
when isMainModule: let Radius = 33.3 let Sigma = Radius / 3.0 let EnergyRange = 0.3.keV
and now the functions:
template compValue(tup: untyped, byCount = false): untyped = if byCount: tup.size.float else: # weigh by distance using gaussian of radius being 3 sigma let dists = tup # `NOTE:` not a tuple here anymore var val = 0.0 for d in items(dists): val += smath.gauss(d, mean = 0.0, sigma = Sigma) val defUnit(cm²) proc normalizeValue*[T](x: T, radius, σ: float, energyRange: keV, byCount = false): auto = let pixelSizeRatio = 65536 / (1.4 * 1.4).cm² var area: float if byCount: # case for regular circle with weights 1 area = π * radius * radius # area in pixel else: area = -2*Pi*(σ*σ * exp(-1/2 * radius*radius / (σ*σ)) - (σ*σ)) let energyRange = energyRange * 2.0 #radius / 6.0 / 256.0 * 12.0 * 2.0 # fraction of full 12 keV range # we look at (factor 2 for radius) let backgroundTime = 3300.h.to(Second) let factor = area / pixelSizeRatio * # area in cm² energyRange * backgroundTime result = x / factor
compValuecomputes the weighted (or unweighted) distance measure andnormalizeValuecomputes the correct normalization based on the radius. The associated area is obtained using the integration shown in the previous section (using sagemath).Let's check if we can run the computation and see what we get
when isMainModule: let dists = df["dists", float] echo "Weighted value : ", compValue(dists) echo "Normalized value : ", compValue(dists).normalizeValue(Radius, Sigma, EnergyRange)
values that seem reasonable.
To compute the associated errors, we need to promote the functions we use above to work with
Measurement[T]objects.normalizeValuewe can just make generic (DONE). ForcompValuewe still need a Gaussian implementation (note: we don't have errors associated with \(μ\) and \(σ\) for now. We might add that.).The logic for the error calculation / getting an uncertainty from the set of clusters in the search radius is somewhat subtle.
Consider the unweighted case: If we have \(N\) clusters, we associate an uncertainty to these number of clusters to \(ΔN = √N\). Why is that? Because: \[ N = Σ_i (1 ± 1) =: f \] leads to precisely that result using linear error propagation! Each value has an uncertainty of \(√1\). Computing the uncertainty of a single value just yields \(√((∂(N)/∂N)² ΔN²) = ΔN\). Doing the same of the sum of elements, just means \[ ΔN = √( Σ_i (∂f/∂N_i)²(ΔN_i)² ) = √( Σ_i 1² ) = √N \] precisely what we expect.
We can then just treat the gaussian in the same way, namely: \[ f = Σ_i (1 ± 1) * \text{gauss}(\vec{x} - \vec{x_i}, μ = 0, σ) \] and transform it the same way. This has the effect that points that are further away contribute less than those closer!
This is implemented here (thanks to
Measuremancer, damn):proc gauss*[T](x: T, μ, σ: float): T = let arg = (x - μ) / σ res = exp(-0.5 * arg * arg) result = res proc compMeasureValue*[T](tup: Tensor[T], σ: float, byCount: bool = false): auto = if byCount: let dists = tup # only a tuple in real interp code let num = tup.size.float var val = 0.0 ± 0.0 for d in items(dists): val = val + (1.0 ± 1.0) * 1.0 # last * 1.0 represents the weight that is one !this holds! doAssert val == (num ± sqrt(num)) # sanity check that our math works out val else: # weigh by distance using gaussian of radius being 3 sigma let dists = tup # `NOTE:` not a tuple here anymore var val = 0.0 ± 0.0 for d in items(dists): let gv = (1.0 ± 1.0) * gauss(d, μ = 0.0, σ = σ) # equivalent to unweighted but with gaussian weights val = val + gv val
Time to take our data and plug it into the two procedures:
when isMainModule: let dists = df["dists", float] echo "Weighted values (byCount) : ", compMeasureValue(dists, σ = Sigma, byCount = true) echo "Normalized value (byCount) : ", compMeasureValue(dists, σ = Sigma, byCount = true) .normalizeValue(Radius, Sigma, EnergyRange, byCount = true) echo "Weighted values (gauss) : ", compMeasureValue(dists, σ = Sigma, byCount = false) echo "Normalized value (gauss) : ", compMeasureValue(dists, σ = Sigma, byCount = false) .normalizeValue(Radius, Sigma, EnergyRange)
The result mostly makes sense: Namely, in the case of the gaussian, we essentially have "less" statistics, because we weigh the events further away less. The result is a larger error on the weighted case with less statistics.
Note: In this particular case the computed background rate is significantly lower (but almost within 1σ!) than in the non weighted case. This is expected and also essentially proving the correctness of the uncertainty. The distances of the points in the input data is simply quite far away for all values.
- Random sampling to simulate background uncertainty
We'll do a simple Monte Carlo experiment to assess the uncertainties from a statistical point of view and compare with the results obtained in the section above.
First do the sampling of backgrounds part:
import std / [random, math, strformat, strutils] const outDir = "/home/basti/org/Figs/statusAndProgress/background_interpolation/uncertainty" import ./sampling_helpers proc sampleBackgroundClusters(rng: var Rand, num: int, sampleFn: (proc(x: float): float) ): seq[tuple[x, y: int]] = ## Samples a number `num` of background clusters distributed over the whole chip. result = newSeq[tuple[x, y: int]](num) # sample in `y` from function let ySamples = sampleFrom(sampleFn, 0.0, 255.0, num) for i in 0 ..< num: result[i] = (x: rng.rand(255), y: ySamples[i].round.int) import ggplotnim, sequtils proc plotClusters(s: seq[tuple[x, y: int]], suffix: string) = let df = toDf({"x" : s.mapIt(it.x), "y" : s.mapIt(it.y)}) let outname = &"{outDir}/clusters{suffix}.pdf" ggplot(df, aes("x", "y")) + geom_point(size = some(1.0)) + ggtitle(&"Sampling bias: {suffix}. Num clusters: {s.len}") + ggsave(outname) import unchained defUnit(keV⁻¹•cm⁻²•s⁻¹) proc computeNumClusters(backgroundRate: keV⁻¹•cm⁻²•s⁻¹, energyRange: keV): float = ## computes the number of clusters we need to simulate a certain background level let goldArea = 5.mm * 5.mm let area = 1.4.cm * 1.4.cm let time = 3300.h # let clusters = 10000 # about 10000 clusters in total chip background result = backgroundRate * area * time.to(Second) * energyRange import arraymancer, measuremancer import ./background_interpolation_error_propagation import numericalnim proc compClusters(fn: (proc(x: float): float), numClusters: int): float = proc hFn(x: float, ctx: NumContext[float, float]): float = (numClusters / (256.0 * fn(127.0))) * fn(x) result = simpson(hfn, 0.0, 256.0) doAssert almostEqual(hFn(127.0, newNumContext[float, float]()), numClusters / 256.0) proc computeToy(rng: var Rand, numClusters: int, radius, σ: float, energyRange: keV, sampleFn: (proc(x: float): float), correctNumClusters = false, verbose = false, suffix = ""): tuple[m: Measurement[keV⁻¹•cm⁻²•s⁻¹], num: int] = var numClusters = numClusters if correctNumClusters: numClusters = compClusters(sampleFn, numClusters).round.int let clusters = rng.sampleBackgroundClusters(numClusters.int, sampleFn) if verbose: plotClusters(clusters, suffix) # generate a kd tree based on the data let tTree = stack([clusters.mapIt(it.x.float).toTensor, clusters.mapIt(it.y.float).toTensor], axis = 1) let kd = kdTree(tTree, leafSize = 16, balancedTree = true) let tup = kd.queryBallPoint([127.float, 127.float].toTensor, radius) let m = compMeasureValue(tup[0], σ = radius / 3.0, byCount = false) .normalizeValue(radius, σ, energyRange) let num = tup[0].len if verbose: echo "Normalized value (gauss) : ", m, " based on ", num, " clusters in radius" result = (m: m, num: num) let radius = 33.3 let σ = radius / 3.0 let energyRange = 0.3.keV let num = computeNumClusters(5e-6.keV⁻¹•cm⁻²•s⁻¹, energyRange * 2.0).round.int var rng = initRand(1337) import sugar # first look at / generate some clusters to see sampling works discard rng.computeToy(num, radius, σ, energyRange, sampleFn = (x => 1.0), verbose = true, suffix = "_constant_gold_region_rate") # should be the same number of clusters! discard rng.computeToy(num, radius, σ, energyRange, sampleFn = (x => 1.0), correctNumClusters = true, verbose = true, suffix = "_constant_gold_region_rate_corrected") # now again with more statistics discard rng.computeToy(100 * num, radius, σ, energyRange, sampleFn = (x => 1.0), verbose = true, suffix = "_constant") # should be the same number of clusters! discard rng.computeToy(100 * num, radius, σ, energyRange, sampleFn = (x => 1.0), correctNumClusters = true, verbose = true, suffix = "_constant_corrected") # linear discard rng.computeToy(100 * num, radius, σ, energyRange, sampleFn = (x => x), verbose = true, suffix = "_linear") # should be the same number of clusters! discard rng.computeToy(100 * num, radius, σ, energyRange, sampleFn = (x => x), correctNumClusters = true, verbose = true, suffix = "_linear_corrected") # square discard rng.computeToy(100 * num, radius, σ, energyRange, sampleFn = (x => x*x), verbose = true, suffix = "_square") # number of clusters should differ! discard rng.computeToy(100 * num, radius, σ, energyRange, sampleFn = (x => x*x), correctNumClusters = true, verbose = true, suffix = "_square_corrected") # exp discard rng.computeToy(100 * num, radius, σ, energyRange, sampleFn = (x => exp(x/64.0)), verbose = true, suffix = "_exp64") # number of clusters should differ! discard rng.computeToy(100 * num, radius, σ, energyRange, sampleFn = (x => exp(x/64.0)), correctNumClusters = true, verbose = true, suffix = "_exp64_corrected") proc performToys(nmc: int, numClusters: int, sampleFn: (proc(x: float): float), suffix: string, correctNumClusters = true): DataFrame = var numClusters = numClusters if correctNumClusters: echo "Old number of clusters: ", numClusters numClusters = compClusters(sampleFn, numClusters).round.int echo "Corrected number of clusters: ", numClusters var data = newSeq[Measurement[keV⁻¹•cm⁻²•s⁻¹]](nmc) var clustersInRadius = newSeq[int](nmc) for i in 0 ..< nmc: if i mod 500 == 0: echo "Iteration: ", i let (m, numInRadius) = rng.computeToy(numClusters, radius, σ, energyRange, sampleFn = sampleFn) data[i] = m clustersInRadius[i] = numInRadius let df = toDf({ "values" : data.mapIt(it.value.float), "errors" : data.mapIt(it.error.float), "numInRadius" : clustersInRadius }) ggplot(df, aes("values")) + geom_histogram(bins = 500) + ggsave(&"{outDir}/background_uncertainty_mc_{suffix}.pdf") ggplot(df, aes("errors")) + geom_histogram(bins = 500) + ggsave(&"{outDir}/background_uncertainty_mc_errors_{suffix}.pdf") if numClusters < 500: ggplot(df, aes("numInRadius")) + geom_bar() + ggsave(&"{outDir}/background_uncertainty_mc_numInRadius_{suffix}.pdf") else: ggplot(df, aes("numInRadius")) + geom_histogram(bins = clustersInRadius.max) + ggsave(&"{outDir}/background_uncertainty_mc_numInRadius_{suffix}.pdf") let dfG = df.gather(["values", "errors"], key = "Type", value = "Value") ggplot(dfG, aes("Value", fill = "Type")) + geom_histogram(bins = 500, position = "identity", hdKind = hdOutline, alpha = some(0.5)) + ggtitle("Sampling bias: {suffix}. NMC = {nmc}, numClusters = {int}") + ggsave(&"{outDir}/background_uncertainty_mc_combined_{suffix}.pdf") result = dfG result["sampling"] = suffix proc performAllToys(nmc, numClusters: int, suffix = "", correctNumClusters = true) = var df = newDataFrame() df.add performToys(nmc, numClusters, (x => 1.0), "constant", correctNumClusters) df.add performToys(nmc, numClusters, (x => x), "linear", correctNumClusters) df.add performToys(nmc, numClusters, (x => x*x), "square", correctNumClusters) df.add performToys(nmc, numClusters, (x => exp(x/64.0)), "exp_x_div_64", correctNumClusters) #df = if numClusters < 100: df.filter(f{`Value` < 2e-5}) else: df let suffixClean = suffix.strip(chars = {'_'}) let pltVals = ggplot(df, aes("Value", fill = "sampling")) + facet_wrap("Type") + geom_histogram(bins = 500, position = "identity", hdKind = hdOutline, alpha = some(0.5)) + prefer_rows() + ggtitle(&"Comp diff. sampling biases, {suffixClean}. NMC = {nmc}, numClusters = {numClusters}") #ggsave(&"{outDir}/background_uncertainty_mc_all_samplers{suffix}.pdf", height = 600, width = 800) # stacked version of number in radius let width = if numClusters < 100: 800.0 else: 1000.0 # stacked version ggplot(df.filter(f{`Type` == "values"}), aes("numInRadius", fill = "sampling")) + geom_bar(position = "stack") + scale_x_discrete() + xlab("# cluster in radius") + ggtitle(&"# clusters in interp radius, {suffixClean}. NMC = {nmc}, numClusters = {numClusters}") + ggsave(&"{outDir}/background_uncertainty_mc_all_samplers_numInRadius_stacked{suffix}.pdf", height = 600, width = width) # ridgeline version ggplot(df.filter(f{`Type` == "values"}), aes("numInRadius", fill = "sampling")) + ggridges("sampling", overlap = 1.3) + geom_bar(position = "identity") + scale_x_discrete() + xlab("# cluster in radius") + ggtitle(&"# clusters in interp radius, {suffixClean}. NMC = {nmc}, numClusters = {numClusters}") + ggsave(&"{outDir}/background_uncertainty_mc_all_samplers_numInRadius_ridges{suffix}.pdf", height = 600, width = width) var pltNum: GgPlot # non stacked bar/histogram with alpha if numClusters < 100: pltNum = ggplot(df.filter(f{`Type` == "values"}), aes("numInRadius", fill = "sampling")) + geom_bar(position = "identity", alpha = some(0.5)) + scale_x_discrete() + ggtitle(&"# clusters in interp radius, {suffixClean}. NMC = {nmc}, numClusters = {numClusters}") else: let binEdges = toSeq(0 .. df["numInRadius", int].max + 1).mapIt(it.float - 0.5) pltNum = ggplot(df.filter(f{`Type` == "values"}), aes("numInRadius", fill = "sampling")) + geom_histogram(breaks = binEdges, hdKind = hdOutline, position = "identity", alpha = some(0.5)) + ggtitle(&"# clusters in interp radius, {suffixClean}. NMC = {nmc}, numClusters = {numClusters}")# + ggmulti([pltVals, pltNum], fname = &"{outDir}/background_uncertainty_mc_all_samplers{suffix}.pdf", widths = @[800], heights = @[600, 300]) # first regular MC const nmc = 100_000 performAllToys(nmc, num, suffix = "_uncorrected", correctNumClusters = false) # and now the artificial increased toy example performAllToys(nmc div 10, 10 * num, "_uncorrected_artificial_statistics", correctNumClusters = false) ## and now with cluster correction performAllToys(nmc, num, suffix = "_corrected", correctNumClusters = true) # and now the artificial increased toy example performAllToys(nmc div 10, 10 * num, "_corrected_artificial_statistics", correctNumClusters = true)
import random, seqmath, sequtils, algorithm proc cdf[T](data: T): T = result = data.cumSum() result.applyIt(it / result[^1]) proc sampleFromCdf[T](data, cdf: seq[T]): T = # sample an index based on this CDF let idx = cdf.lowerBound(rand(1.0)) result = data[idx] proc sampleFrom*[T](data: seq[T], start, stop: T, numSamples: int): seq[T] = # get the normalized (to 1) CDF for this radius let points = linspace(start, stop, data.len) let cdfD = cdf(data) result = newSeq[T](numSamples) for i in 0 ..< numSamples: # sample an index based on this CDF let idx = cdfD.lowerBound(rand(1.0)) result[i] = points[idx] proc sampleFrom*[T](fn: (proc(x: T): T), start, stop: T, numSamples: int, numInterp = 10_000): seq[T] = # get the normalized (to 1) CDF for this radius let points = linspace(start, stop, numInterp) let data = points.mapIt(fn(it)) let cdfD = cdf(data) result = newSeq[T](numSamples) for i in 0 ..< numSamples: # sample an index based on this CDF let idx = cdfD.lowerBound(rand(1.0)) result[i] = points[idx]
So, from these Monte Carlo toy experiments, we can gleam quite some insight.
We have implemented unbiased clusters as well as biased clusters.
First one example for the four different cluster samplers, with the condition each time that the number of total clusters is the same as in the constant background rate case:
Figure 421: Example of an unbiased cluster sampling. Sampled 100 times (for better visibility of the distribution) as many clusters as predicted for our background data taking. Figure 422: Example of a linearly biased cluster sampling. Sampled 100 times (for better visibility of the distribution) as many clusters as predicted for our background data taking. Figure 423: Example of a squarely biased cluster sampling. Sampled 100 times (for better visibility of the distribution) as many clusters as predicted for our background data taking. Figure 424: Example of a \(\exp(x/64)\) biased cluster sampling. Sampled 100 times (for better visibility of the distribution) as many clusters as predicted for our background data taking. With these in place, we performed two sets of Monte Carlo experiments to compute the value & uncertainty of the center point
(127, 127)using the gaussian weighted nearest neighbor interpolation from the previous section.This is done for all four different samplers and the obtained values and their errors (propagated via
Measuremancer) plotted as a histogramOnce for the number of expected clusters (based on the gold region background rate), fig. [BROKEN LINK: background_uncertainty_mc_all_samplers] and once for a lower statistics, but much 10 times higher number of clusters, fig. [BROKEN LINK: background_uncertainty_mc_all_samplers_artificial_statistics]
Figure 425: Comparison of four different samplers (unbiased + 3 biased), showing the result of \num{100000} MC toy experiments based on the expected number of clusters if the same background rate of the gold region covered the whole chip. Below a bar chart of the number of clusters found inside the radius. The number of clusters corresponds to about 5e-6 keV⁻¹•cm⁻²•s⁻¹over the whole chip.Figure 426: Comparison of four different samplers (unbiased + 3 biased), showing the result of \num{10000} MC toy experiments based on the 10 times the expected number of clusters if the same background rate of the gold region covered the whole chip. Below a histogram of the number of clusters found inside the radius. The number of clusters corresponds to about 5e-5 keV⁻¹•cm⁻²•s⁻¹over the whole chip.First of all there is some visible structure in the low statistics figure (fig. 425). The meaning of it, is not entirely clear to me. Initially, we thought it might be an integer effect of 0, 1, 2, … clusters within the radius and the additional slope being from the distance these clusters are away from the center. Further away, less weight, less background rate. But looking at the number of clusters in the radius (lowest plot in the figure), this explanation alone does not really seem to explain it.
For the high statistics case, we can see that the mean of the distribution shifts lower and lower, the more extreme the bias is. This is likely, because the bias causes a larger and larger number of clusters to land near the top corner of the chip, meaning that there are less and less clusters found within the point of interpolation. Comparing the number of clusters in radius figure for this case shows that indeed, the square and exponential bias case show a peak at lower energies.
Therefore, I also computed a correction function to compute a biased distribution that matches the background rate exactly at the center of the chip, but therefore allows for a larger number of sampled clusters in total.
We know that (projecting onto the y axis alone), there are:
\[ ∫_0^{256} f(x) dx = N \]
where \(N\) is the total number of clusters we draw and \(f(x)\) the function we use to sample. For the constant case, this means that we have a rate of \(N / 256\) clusters per pixel along the y axis (i.e. per row).
So in order to correct for this and compute the new required number of clusters in total that gives us the same rate of \(N / 256\) in the center, we can:
\[ ∫_0^{256} \frac{N}{256 · f(127)} f(x) dx = N' \]
where the point \(f(127)\) is simply the value of the "background rate" the function we currently use produces as is in the center of the chip.
Given our definition of the functions (essentially as primitive
f(x)= x,f(x) = x * x, etc. we expect the linear function to match the required background rate of the constant case exactly in the middle, i.e. at 127. And this is indeed the case (as can be seen in the new linear plot below, fig. 428).This correction has been implemented. The equivalent figures to the cluster distributions from further above are:
Figure 427: Example of an unbiased cluster sampling with the applied correction. Sampled 100 times (for better visibility of the distribution) as many clusters as predicted for our background data taking. As expected the number of clusters is still the same number as above. Figure 428: Example of a linearly biased cluster sampling with the applied correction. Sampled 100 times (for better visibility of the distribution) as many clusters as predicted for our background data taking. Figure 429: Example of a squarely biased cluster sampling with the applied correction. Sampled 100 times (for better visibility of the distribution) as many clusters as predicted for our background data taking. The correction means that the total number of clusters is now almost 2500 more than in the uncorrected case. Figure 430: Example of a \(\exp(x/64)\) biased cluster sampling with the applied correction. Sampled 100 times (for better visibility of the distribution) as many clusters as predicted for our background data taking. The correction means that the total number of clusters is now almost double the amount in the uncorrected case. The correction works nicely. It is visible that in the center the density seems to be the same as in the constant case.
From here we can again look at the same plots as above, i.e. the corrected monte carlo plots:
Figure 431: Comparison of four different samplers (unbiased + 3 biased), showing the result of \num{100000} MC toy experiments based on the expected number of clusters such that the background is biased and produces the same background rate as in the gold region in the constant case. Below a bar chart of the number of clusters found inside the radius. The number of clusters corresponds to about 5e-6 keV⁻¹•cm⁻²•s⁻¹over the whole chip.Figure 432: Comparison of four different samplers (unbiased + 3 biased), showing the result of \num{10000} MC toy experiments based on the 10 times the expected number of clusters such that the background is biased and produces the same background rate as in the gold region in the constant case. Below a histogram of the number of clusters found inside the radius. The number of clusters corresponds to about 5e-5 keV⁻¹•cm⁻²•s⁻¹over the whole chip.It can be nicely seen that the mean of the value is now again at the same place for all samplers! This is reassuring, because it implies that any systematic uncertainty due to such a bias in our real data is probably negligible, as the effects will never be as strong as simulated here.
Secondly, we can nicely see that the computed uncertainty for a single element seems to follow nicely the actual width of the distribution.
In particular this is visible in the artificial high statistics case, where the mean value of the error is comparable to the width of the
valuehistogram. - TODO Explain "ragged" structure in low statistics case
The "integer number elements in radius" hypothesis is somewhat out. What is the reason for the substructure in fig. 425.
- Random sampling to simulate background uncertainty
- Sample from background interpolation for MC
import nimhdf5, ggplotnim, os, sequtils, seqmath, unchained, strformat import ingrid / tos_helpers import arraymancer except linspace proc flatten(dfs: seq[DataFrame]): DataFrame = ## flatten a seq of DFs, which are identical by stacking them for df in dfs: result.add df.clone proc readFiles(path: string, s: seq[string]): DataFrame = var h5fs = newSeq[H5FileObj]() echo path echo s for fs in s: h5fs.add H5open(path / fs, "r") result = h5fs.mapIt( it.readDsets(likelihoodBase(), some((chip: 3, dsets: @["energyFromCharge", "centerX", "centerY"]))) .rename(f{"Energy" <- "energyFromCharge"})).flatten if result.isNil: quit("what the fuck") result = result.filter(f{`Energy` < 15.0}) for h in h5fs: discard h.close() import ./background_interpolation let path = "/home/basti/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/" let backFiles = @["lhood_2017_all_chip_septem_dbscan.h5", "lhood_2018_all_chip_septem_dbscan.h5"] var df = readFiles(path, backFiles) let kd = toNearestNeighborTree(df) Radius = 33.3 Sigma = 11.1 EnergyRange = 0.3.keV defUnit(keV⁻¹•cm⁻²•s⁻¹) let nxy = 10 let nE = 20 ## Need an offset to not start on edge, but rather within ## and stop half a step before let xyOffset = 14.0/(nxy).float / 2.0 ## XXX: fix this for real number ``within`` the chip let eOffset = 12.0/(nE).float / 2.0 let dist = (xyOffset * 2.0).mm let area = dist * dist # area of considered area echo area let ΔE = (eOffset * 2.0).keV echo ΔE let volume = area * ΔE let time = 3300.h defUnit(cm²•keV) echo volume var t = newTensor[float]([nxy, nxy, nE]) let coords = linspace(0.0 + xyOffset, 14.0 - xyOffset, nxy) let energies = linspace(0.0 + eOffset, 12.0 - eOffset, nE) echo coords echo energies #if true: quit() var xs = newSeq[float]() var ys = newSeq[float]() var cs = newSeq[float]() for yIdx in 0 ..< nxy: for xIdx in 0 ..< nxy: for iE, E in energies: let y = coords[yIdx] let x = coords[xIdx] let tup = kd.queryBallPoint([x.toIdx.float, y.toIdx.float, E].toTensor, Radius, metric = CustomMetric) let val = compValue(tup) .correctEdgeCutoff(Radius, x.toIdx, y.toIdx) .normalizeValue(Radius, EnergyRange) let valCount = val * volume * time.to(Second) echo val, " as counts: ", valCount, " at ", x, " / ", y, " E = ", E t[yIdx, xIdx, iE] = valCount echo t.sum() proc computeNumClusters(backgroundRate: keV⁻¹•cm⁻²•s⁻¹, energyRange: keV): float = ## computes the number of clusters we need to simulate a certain background level let goldArea = 5.mm * 5.mm let area = 1.4.cm * 1.4.cm let time = 3300.h # let clusters = 10000 # about 10000 clusters in total chip background result = backgroundRate * area * time.to(Second) * energyRange let num = computeNumClusters(1e-5.keV⁻¹•cm⁻²•s⁻¹, EnergyRange) #echo num / (5.mm * 5.mm) * (14.mm * 14.mm) echo num #plotSingleEnergySlice(kd, 1.0) proc plot3DTensor(t: Tensor[float], size: int, energyIdx: int, outname = "/tmp/test_tensor.pdf", title = "") = var xs = newSeq[float](size * size) var ys = newSeq[float](size * size) var cs = newSeq[float](size * size) var idx = 0 for y in 0 ..< t.shape[0]: for x in 0 ..< t.shape[1]: xs[idx] = x.float ys[idx] = y.float cs[idx] = t[y, x, energyIdx] inc idx let df = toDf(xs, ys, cs) ggplot(df, aes(f{"xs" ~ `xs` / 10.0 * 14.0}, # convert indices to coordinates f{"ys" ~ `ys` / 10.0 * 14.0}, fill = "cs")) + geom_raster() + #scale_fill_continuous(scale = (low: 0.0, high: 10.0)) + margin(top = 2) + ggtitle(title) + ggsave(outname) plot3DTensor(t, nxy, 0, outname = "/home/basti/org/Figs/statusAndProgress/background_interpolation/interpolation_gridded_first_energy_interval.pdf", title = &"First interval of gridded background interp, {energies[0]} keV center, 20 intervals in E, 10 in x/y") type Candidate = object energy: keV pos: tuple[x, y: float] import random / mersenne import alea / [core, rng, gauss, poisson] var rnd = wrap(initMersenneTwister(299792458)) proc drawCandidates(#ctx: Context, coords: seq[float], # will become field of ctx energies: seq[float], # become field of ctx eOffset, xyOffset: float, rnd: var Random, posOverride = none(tuple[x, y: float]), toPlot = false, Correct: static bool = false ): seq[Candidate] {.noinit.} = ## draws a number of random candidates from the background sample ## using the ratio of tracking to background ~19.5 # 1. iterate over every position of the background tensor # 2. draw form a poisson with mean = the value at that tensor position (is normalized to expected counts) # 3. the resulting number of candidates will be created # 3a. for each candidate, smear the position & energy within the volume of the grid cell var pois = poisson(0.0) ## Will be adjusted for each grid point var uniXY = uniform(0.0, 0.0) ## Will be adjusted for each grid point var uniE = uniform(0.0, 0.0) result = newSeqOfCap[Candidate](10000) for iE in 0 ..< energies.len: for ix in 0 ..< coords.len: for iy in 0 ..< coords.len: pois.l = t[iy, ix, iE] for _ in 0 ..< rnd.sample(pois).int: uniE.a = energies[iE] - eOffset uniE.b = energies[iE] + eOffset uniXY.a = coords[ix] - xyOffset uniXY.b = coords[ix] + xyOffset result.add Candidate(energy: rnd.sample(uniE).keV, pos: (x: rnd.sample(uniXY), y: rnd.sample(uniXY))) let cands = drawCandidates(coords, energies, eOffset, xyOffset, rnd) echo "Drew : ", cands.len, " number of candidates." for c in cands: echo c when false: ## INVESTIGATE THIS FURTHER when false: var r {.volatile.} = 0.0 when Correct: for iy in 0 ..< coords.len: for ix in 0 ..< coords.len: for iE in 0 ..< energies.len: pois.l = t[iy, ix, iE] r += pois.l # XXX: smear the position for each #let count = rnd.sample(pois) #echo "COUNT : ", count, " from ", t[iy, ix, iE] #for _ in 0 ..< rnd.sample(pois).int: # result.add Candidate(energy: energies[iE].keV, pos: (x: coords[ix], y: coords[iy])) doAssert r > 0.0 else: for iE in 0 ..< energies.len: for ix in 0 ..< coords.len: for iy in 0 ..< coords.len: pois.l = t[iy, ix, iE] r += pois.l doAssert r > 0.0 import times let t0 = epochTime() for _ in 0 ..< 1000000: discard drawCandidates(coords, energies, rnd, Correct = true) let t1 = epochTime() echo "Took ", t1 - t0, " s" let t2 = epochTime() for _ in 0 ..< 1000000: discard drawCandidates(coords, energies, rnd, Correct = false) echo "Took ", epochTime() - t2, " s"
- DONE Investigate if construction of k-d tree suffers from energy range
In our current measure that actually ignores the distance in energy for two points, don't we end up building a broken k-d tree (in terms of efficiency at least), as there is no sane way to separate along the 3rd axis in distance…
Investigate what this means.
UPDATE: When I wrote this I had completely forgotten that the construction of the kd tree doesn't even take into account the metric. It always uses a simple euclidean distance along each dimension separately.
- TODO Correct edge cutoff does not take into account weighting
This leads to overestimating areas that are far away from the center, no?
- TODO Would weighing the distance in energy make sense?
This might be interesting in the sense that it would help with the possible worsening of the background rate in energy (i.e. by spreading the lines visible in the rate over a larger energy range). By making the energy count in the weight as well, it further things would contribute less. However, an alternative is to simply reduce the "width" in energy i.e. make it smaller than the corresponding radius in x/y, i.e. 100 pixels in radius correspond to a 100/256th of the width of the radius, but we may only use 1/10th of the energy or something like this.
- Energy dependence of background rate based on position
One question still unclear is whether there is a dependency of the background rate in energy based on the position.
If there is not we can use the above interpolation as the "truth" for the position dependence and can keep the shape of the energy dependence. The absolute values then just have to be scaled accordingly.
If there is a dependence on the position for the energy, we need to do an interpolation taking into account the energy.
UPDATE: : As it turns out, the energy dependence is stronger than thought. The summary is:
- the vast majority of background still left over the whole chip is from low energy clusters. These are mainly located in the corners. This raises the question of why even though it may not be super important for the limit.
- everything above 2 keV seems to be more or less evenly distributed, with a ~same background level everywhere
- a kd tree interpolation radius of 30 pixels is not enough given the statistics of clusters in an energy range of ΔE = 1 keV (any above 2 keV)
Generated by the same files as the previous section 29.1.3, a
29.1.4. Bayes integral AFTER MAIL FROM IGOR ON :
Essentially saying that we simply integrate over and demand:
0.95 = ∫_-∞^∞ L(gae²) Π(gae²) / L0 d(gae²)
where L is the likelihood function (not the ln L!), Π is the prior that is used to exclude the unphysical region of the likelihood phase space, i.e. it is:
Π(gae²) = { 0 if gae² < 0, 1 if gae² >= 0 }
And L0 is simply a normalization constant to make sure the integral is normalized to 1.
Thus, the integral reduces to the physical range:
0.95 = ∫0^∞ L(gae²) / L0 d(gae²)
where the 0.95 is, due to normalization, simply the requirement of a 95% confidence limit.
With this out of the way I implemented this into the limit calculation
code as the lkBayesScan limit.
The resulting likelihood function in the physical region (for a single toy experiment) can be seen in fig. 455, whereas its CDF is shown in fig. 433 which is used to determine the 95% level.
After doing 1000 toy MC experiments, we get the distribution shown in fig. 434.
To verify whether the substructure in the Bayesian limits comes from a fixed integer number of candidates being found in the axion sensitive region, we will now create a plot similar to fig. 434 for artificial sets of candidates with 0 to N number of candidates in the center of the chip (x, y) = (7.0 / 7.0). A total number of 30 candidates is used. The 30 - N candidates are placed at the border of the chip to not account for axion signals, but for the background we expect.
This gives us the plot shown in fig. 435. This implementation that uses the code for the Bayesian limit (with fixed coupling stepping for the CDF) results in multiple 0 values as the limit for sets of candidates with typically more elements in the axion sensitive region. Needs to be investigated!
UPDATE: It turned out that the issue was that candidate sets that produced a ln L maximum in the physical range where incorrectly handled. The correct plot is now shown in fig. 436.
29.1.5. TODO compute sum over all backgrounds.
What number is it? not total number of expected background counts, no?
29.1.6. Verification of the usage of efficiencies
29.1.7. TODO Debug NaN values when varying σ_s in Bayesian limit
While implementing the behavior of the limit over varying systematic uncertainties, I stumbled on another issue.
The code currently () has checks that the result
of the likelihood computation using systematics does not result in a
NaN. I was sure this wouldn't happen anymore, but apparently it
still does.
The candidates saved in
./../resources/bad_candidates_bayes_limit_large_σs.csv reproduce
this, if the added checks (for decreasing behavior) in the Bayesian
limit while loop are not checked for.
The candidates here are
so they look pretty much normal.
And the \(θ_s\) phase space is at g_ae² = 1e-20:
also looks fine.
The scan of the logL however shows a problem:
so we can see that the logL space behaves normally at first, but at some point instead of continuing the downward trend it goes back up again!
Code to reproduce this. Insert this into the limit calculation proc (adjust the file we read to the resources up there!)
when true: ## debug NaN in uncertain signal let df = readCsv("/tmp/bad_candidates.txt") var cnds = newSeq[Candidate]() for row in df: cnds.add Candidate(energy: row["E"].toFloat.keV, pos: (x: row["x"].toFloat, y: row["y"].toFloat)) plotCandidates(cnds) ctx.σs_sig = 0.225 ctx.plotLikelihoodCurves(cnds) # looks fine discard ctx.computeLimit(cnds, limitKind, toPlot = true) if true: quit()
We added a check for increasing L again after it decreased and stop
the while loop in the bayesLimit proc now. This results in a
decent limit.
It made me wonder: The \(θ_s\) space depends on the coupling constant! So what does it look like in the range where the L goes crazy?
I set the g_ae² to 5e-20 (which is where we stop execution after
1000 iterations by default), way into the crazy range, and this is
what it looks like:
So at least we can see that the distribution shifts further and further to negative values, implying a lower signal (negative \(θ_s\) decreases the signal) is a better fit, which makes sense, given that a huge coupling constant causes way too much signal.
NOTE: I still don't understand why L goes back up again fully!
29.1.8. "Sudden" decrease in expected limits
While many things were changed between the initial expected limit calculations, including for the scans of different σ systematics (for signal and background), there was a sudden decrease in the expected limit when computing the final expected limit for the CAST collaboration meeting.
While using the correct inputs of background rate, raytracing etc. all played a role, the biggest effect as it turned out was the algorithm used to determine the 95% point.
We went from a fixed cutoff value to an adaptive strategy (before even considering MCMC!). This caused a significant change in the expected limits.
The old logic yielding "good" limits:
proc bayesLimit(ctx: Context, cands: seq[Candidate], toPlot: static bool = false): float = # {.gcsafe.} = var ctx = ctx const nPoints = 10000 var Ls = newSeqOfCap[float](nPoints) var cdfs = newSeqOfCap[float](nPoints) var couplings = newSeqOfCap[float](nPoints) var coupling = 0.0 let couplingStep = 1e-22 var idx = 0 # 2. compute starting values and add them when true: let L0 = ctx.evalAt(cands, 0.0) cdfs.add L0 Ls.add L0 couplings.add coupling var curL = L0 echo "Cur L ", curL #echo "L0 = ", L0, " and curL = ", curL, " abs = ", abs(ln(L0) / ln(curL)), " is nan ?? ", abs(ln(L0) / ln(curL)).isNan #if true: quit() # 3. walk from g_ae² = 0 until the ratio of the `ln` values is 0.9. Gives us good margin for CDF # calculation (i.e. make sure the CDF will have plateaued var lastL = curL var cdfVal = lastL var decreasing = false var maxVal = curL var stopVal = if curL < 5e-3: curL / 200.0 else: 5e-3 while curL > stopVal: # and idx < 1000: #ln(curL) >= 0.0: echo "Limit step ", idx, " at curL ", curL, " at g_ae²: ", ctx.g_ae², " decreasing ? ", decreasing, " curL < lastL? ", curL < lastL coupling += couplingStep curL = ctx.evalAt(cands, coupling) maxVal = max(curL, maxVal) cdfVal += curL cdfs.add cdfVal Ls.add curL couplings.add coupling if decreasing and # already decreasing curL > lastL: # rising again! Need to stop! echo "Breaking early!" #break if lastL != curL and curL < lastL: # decreasing now! decreasing = true lastL = curL inc idx let cdfsNorm = toCdf(cdfs, isCumSum = true) # 5. now find cdf @ 0.95 let idxLimit = cdfsNorm.lowerBound(0.95) # 6. coupling at this value is limit result = couplings[idxLimit]
The new adaptive logic yielding correct, but worse limits:
proc bayesLimit(ctx: Context, cands: seq[Candidate], toPlot: static bool = false): float = # {.gcsafe.} = ## compute the limit based on integrating the posterior probability according to ## Bayes theorem using a prior that is zero in the unphysical range and constant in ## the physical # 1. init needed variables var ctx = ctx var couplings = linspace(0.0, 2e-20, 10) var lh = initLimitHelper(ctx, cands, couplings) let ε = 0.005 #1e-3 # with in place, compute derivatives & insert until diff small enough var diff = Inf var at = 0 #echo lh.deriv genplot(lh, title = "MC Index: " & $ctx.mcIdx) plotSecond(lh) #echo lh.derivativesLarger(0.5) var count = 0 while diff > ε and lh.derivativesLarger(0.5): computeCouplings(lh) lh.cdf = lh.computeCdf() lh.deriv = lh.computeDeriv() at = lh.cdf.lowerBound(0.95) diff = lh.cdf[at] - 0.95 genplot(lh, title = "MC Index: " & $ctx.mcIdx) plotSecond(lh) inc count let Ls = lh.likelihoods() couplings = lh.couplings() let cdfsNorm = lh.cdf result = couplings[at]
This code requires some logic that is found in the file for the limit calc!
Note: the former method can be improved by changing the logic to at least an adaptive stopping criterion:
var stopVal = curL / 500.0 while curL > stopVal: coupling += couplingStep curL = ctx.evalAt(cands, coupling) stopVal = maxVal / 500.0 maxVal = max(curL, maxVal)
where we simply decrease the minimum value to 1/500 and also adjust based on the Ls we see in the space. If there is a maximum there is no need to go to a fixed super small value relative to the beginning, as the area will be dominated by the peak.
This latter modification yields numbers more in line with the adaptive method, while also being faster (the original method is extremely slow for certain candidates in which the L values change extremely slowly).
Comparison of different σ values based on (only 50!) toy MC samples:
The following all use the exact same input data & times:
- old method fixed cutoff:
- adaptive method
And applying the fixed stopping criteria yields:
29.1.9. Notes on MCMC parameters (chain length, starting chain etc.)
There are of course a wide variety of different parameters to be considered for a MCMC.
It seems like even in the no signal case, there is a slight possibility of going into a very unlikely area of the phase space (where probabilities are 0), if we already start from 0 (because then every new step will be accepted and thus we may walk further into unlikely territory. This is a pure random walk at that point of course. There are multiple things to keep in mind here: Firstly, ideally our likelihood function would never become absolute zero, such that we always have a "gradient" towards the more likely space. However, given our numbers that is unfortunately not the case, as the drop off is too sharp (in principle this is a good thing, as it guarantees good convergence after all). Secondly, we can restrict the space to a more narrow region in principle (where we can be sure it's 0, there is no point in allowing it to go further into that direction).
Finally,
In one (very "rare") case we started from the following parameters:
let start = @[4.94646801701552e-21, -0.2315730763663281, 0.209776111514859, 0.4953485987571491, 0.2557091758280117]
with random seed:
var rnd = wrap(initMersenneTwister(299792458 + 2))
and after a specific number of draws from the RNG:
- 5 for a first set of start parameters
- 50,000 iterations of a first chain
- 5 for a second set of start parameters
- start of the second chain, that produces ~15,000 chain steps in a region of 0 likelihood
where the RNG draws within the chain computation aren't mentioned (don't feel like checking how many it really is).
This results in the following chain (where nothing was truncated for the plot):



As such the "limit" deduced from this chain as shown in the following histogram:
is in the 1e-19 range.
The (not particularly useful code) to reproduce the RNG state in the current code is:
var totalChain = newSeq[seq[float]]() block: let start = @[rndS.rand(0.0 .. 5.0) * 1e-21, # g_ae² rndS.rand(-0.4 .. 0.4), rndS.rand(-0.4 .. 0.4), # θs, θb rndS.rand(-0.5 .. 0.5), rndS.rand(-0.5 .. 0.5)] # θs, θb let (chain, acceptanceRate) = build_MH_chain(start, @[3e-21, 0.025, 0.025, 0.05, 0.05], 50_000, fn) let start2 = @[rndS.rand(0.0 .. 5.0) * 1e-21, # g_ae² rndS.rand(-0.4 .. 0.4), rndS.rand(-0.4 .. 0.4), # θs, θb rndS.rand(-0.5 .. 0.5), rndS.rand(-0.5 .. 0.5)] # θs, θb echo start, " and ", start2 # start the problematic chain block: let start = @[4.94646801701552e-21, -0.2315730763663281, 0.209776111514859, 0.4953485987571491, 0.2557091758280117] echo "\t\tInitial chain state: ", start let (chain, acceptanceRate) = build_MH_chain(start, @[3e-21, 0.025, 0.025, 0.05, 0.05], 200_000, fn) echo "Acceptance rate: ", acceptanceRate echo "Last ten states of chain: ", chain[^10 .. ^1] totalChain.add chain result = ctx.plotChain(candidates, totalChain, computeIntegral = false)
We could in theory include only those points in the limit calculation that satisfy likelihood values > 0, but that defeats the point of using MCMC somewhat. And this is precisely the point of "burn in" of the chain after all.
With that in mind, we will change the chain approach to the following:
[X]restrict the allowed space for the parameters to a narrower region (-1 to 1 for all nuisance parameters) -> restricting the space already reduces the impact of this chain's behavior to < 10,000 problematic steps[X]drop the first 20,000 steps as "burn in"[ ]think about an "adaptive burn in" that takes into account first time the likelihood went above 0- found a case in which the chain to 50,000 steps to converge!
[X]use 5 chains[X]use more than 50,000 elements per chain? -> Yes, we finally use 3 chains with 150,000 elements & 50,000 burn in
UPDATE: By now we actually use 3 chains, with 150,000 elements and 50,000 burn in.
Truncating the first 20,000 elements thus results in the following chain state:



And while working on this I found a juicy bug, see sec. 29.1.10 below. I also changed from a binned to an unbinned CDF method now for the MCMC case (as we have a large number of inputs and don't need to bin them!).
29.1.10. TODO CDF bug found in limit calculation
The CDF implementations in the limit calculation:
template toCDF(data: seq[float], isCumSum = false): untyped = var dataCdf = data if not isCumSum: seqmath.cumSum(dataCdf) let integral = dataCdf[^1] let baseline = dataCdf[0] dataCdf.mapIt((it - baseline) / (integral - baseline))
and
proc cdfUnequal(y, x: seq[float]): seq[float] = let cumS = cumSumUnequal(y, x) let integral = cumS[^1] let baseline = cumS[0] doAssert integral != baseline, "what? " & $cumS result = cumS.mapIt((it - baseline) / (integral - baseline))
contain a serious bug. For the purpose of our limit calculation (and any other???) the baseline must never be subtracted from the cumulative sum! That just removes the whole impact the first "bin" has on the data!
[ ]investigate the impact of this in rest of our code somewhere?
baseline set to 0 running:
basti at void in ~/org/Misc ツ ./mcmclimittesting –σp 0.0 –limitKind lkBayesScan –nmc 50 Dataframe with 3 columns and 6 rows: Idx σs σb expLimits dtype: float float float 0 0.05 0.05 5.4861e-21 1 0.1 0.1 4.8611e-21 2 0.15 0.15 5.5556e-21 3 0.2 0.2 6.5278e-21 4 0.25 0.25 7.2222e-21 5 0.3 0.3 7.7778e-21
baseline set to data[0]:
basti at void in ~/org/Misc ツ ./mcmclimittesting –σp 0.0 –limitKind lkBayesScan –nmc 50 Dataframe with 3 columns and 6 rows: Idx σs σb expLimits dtype: float float float 0 0.05 0.05 5.5556e-21 1 0.1 0.1 5.2778e-21 2 0.15 0.15 5.8333e-21 3 0.2 0.2 6.6667e-21 4 0.25 0.25 7.7778e-21 5 0.3 0.3 8.8889e-21
It certainly has a very serious effect! Especially given the limited
statistics as well (nmc = 50)
29.1.11. Limits for different cases
These all use the "real" inputs, as far as I can tell:
- 169h of effective tracking time
- 3318 - 169h of effective background time
- background as given by CDL mapping fixed code
- "wider" raytracing
- No systematics, adaptive Bayes scan
Expected limit: 5.416666666666666e-21
- No systematics, non-adaptive Bayes scan
Expected limit: 5.500000000000005e-21
- No systematics, MCMC
Expected limit: 5.560323376271403e-21
- Systematics as "needed" with MCMC parameters as determined above
Running:
basti at void in ~/org/Misc ツ ./mcmc_limit_testing \ -f lhood_2017_all_vetoes_dbscan_cdl_mapping_fixed.h5 \ -f lhood_2018_all_vetoes_dbscan_cdl_mapping_fixed.h5 \ --σ_p 0.05 --limitKind lkMCMC --computeLimit --nmc 50
so extremely low statistics!
gae² = 5.663551538352475e-21 yielding gae gaγ = 7.525e-23 GeV⁻¹ with gaγ = 1e-12 GeV⁻¹
which is for 3 MCMC chains with 50,000 burn in and 150,000 samples each (so effectively 300,000 samples).
So given the statistics there is a high chance the number may change significantly from here.
Need more performance for more statistics.
NOTE: running with a modified position uncertainty (i.e.
x + θ * 7instead ofx * (1 + θ)) yields: 5.705224563009264e-21 -> 7.55e-23 for the exact same arguments and data.- Multiprocessing with correct parameters
After we switched over to
procpoolnow and running 100,000 toys over night, we get the following expected limit:/mcmc_limit_testing -f lhood_2017_all_vetoes_dbscan_cdl_mapping_fixed.h5 \ -f lhood_2018_all_vetoes_dbscan_cdl_mapping_fixed.h5 \ --σ_p 0.05 --limitKind lkMCMC --nmc 100000
gae² = 5.589011482509822e-21 gae · gaγ = 7.476e-23 GeV⁻¹
The histogram of all toy limits is:
and the data of all limits: ./../resources/mc_limit_lkMCMC_skInterpBackground_nmc_100000_uncertainty_ukUncertain_σs_0.0469_σb_0.0028_posUncertain_puUncertain_σp_0.0500.csv
Now, whether this is our "final" limit, who knows…
Still have to check 3 things:
[ ]does our data really contain the solar tracking data as well as the background data as it is now?[ ]is the nuisance parameter for the position fine as it is? i.e. usingx + θ_x * 7.0instead ofx * (1 + θ_x)(which behaves weirdly for values away from the center!)[ ]is the normalization of the axion signal without the strongback correct? Or should we normalize the axion signal by placing the strongback on top and then normalizing? Probably require the latter!
- Multiprocessing with correct parameters
- Different sets of vetoes (test run for automation)
[/]
Over the night of we ran the limit calculation (now in TPA) for the different sets of likelihood outputs as generated by ./../../CastData/ExternCode/TimepixAnalysis/Analysis/createAllLikelihoodCombinations.nim in particular in ./../resources/lhood_limits_automation_testing/lhood_outputs_adaptive_fadc using MCMC with the help of ./../../CastData/ExternCode/TimepixAnalysis/Analysis/runLimits.nim
The outputs are in file:///home/basti/org/resources/lhood_limits_automation_testing/lhood_outputs_adaptive_fadc_limits
IMPORTANT: These are not in any way final, because they do not take into account the dead times associated with the septem & line veto, as well as the efficiency of the FADC veto! Nor are the tracking times necessarily correct!
The code was run as a test bed to check the automation of the limit method for different inputs and to get an idea for the expected limits.
Let's output the different expected limits from each output CSV file.
import os, datamancer, strutils, sugar const path = "/home/basti/org/resources/lhood_limits_automation_testing/lhood_outputs_adaptive_fadc_limits/" const prefix = "/t/lhood_outputs_adaptive_fadc_limits/mc_limit_lkMCMC_skInterpBackground_nmc_2000_uncertainty_ukUncertain_σs_0.0469_σb_0.0028_posUncertain_puUncertain_σp_0.0500" var df = newDataFrame() for f in walkFiles(path / "mc_limit_lkMCMC_skInterpBackground_nmc_2000_*.csv"): let name = f.dup(removePrefix(prefix)).dup(removeSuffix(".csv")) let dfLoc = readCsv(f) let limit = sqrt(dfLoc["limits", float].percentile(50)) df.add toDf(limit, name) echo df.toOrgTable()
These numbers are the \(g_{ae}\) number only and need to be combined with \(g_{aγ} = \SI{1e-12}{GeV⁻¹}\) and compared with the current best limit of \(\SI{8.1e-23}{GeV⁻¹}\).
: Rerunning today with latest version of
createAllLikelihoodCombinationsutilizing different veto percentiles for the FADC.mcmc_limit_calculationis now running.Let's rerun the same on the nmc = 1000 data with the different FADC veto percentiles:
[ ]TURN THIS INTO A TABLE THAT ALREADY CONTAINS THE VETO SETUPS ETC IN A NICER FORMAT
import os, datamancer, strutils, sugar const path = "/home/basti/org/resources/lhood_limits_automation_preliminary/lhood_outputs_adaptive_fadc_limits/" const prefix = "mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0469_σb_0.0028_posUncertain_puUncertain_σp_0.0500" var df = newDataFrame() for f in walkFiles(path / "mc_limit_lkMCMC_skInterpBackground_nmc_1000_*.csv"): let name = f.extractFilename.dup(removePrefix(prefix)).dup(removeSuffix(".csv")) let dfLoc = readCsv(f) let limit = sqrt(dfLoc["limits", float].percentile(50)) * 1e-12 df.add toDf(limit, name) echo df.toOrgTable()
: The above was still with the wrong duration used in background / tracking as well as using all data including tracking data.
: Note: see the journal for continuing along these lines with more features and other cases we look at (what is mentioned in the meeting notes).
Based on the new H5 output we can do the above in a neater way, without relying on any of the file name logic etc! And we can generate the full table automatically as well.
import os, nimhdf5, datamancer, strutils, sugar type VetoKind = enum fkScinti, fkFadc, fkSeptem, fkLineVeto LimitData = object expectedLimit: float limitNoSignal: float vetoes: set[VetoKind] eff: Efficiency Efficiency = object totalEff: float # total efficiency multiplier based on signal efficiency of lnL cut, FADC & veto random coinc rate signalEff: float # the lnL cut signal efficiency used in the inputs vetoPercentile: float # if FADC veto used, the percentile used to generate the cuts septemVetoRandomCoinc: float # random coincidence rate of septem veto lineVetoRandomCoinc: float # random coincidence rate of line veto septemLineVetoRandomCoinc: float # random coincidence rate of septem + line veto proc expLimit(limits: seq[float]): float = result = sqrt(limits.percentile(50)) * 1e-12 proc readVetoes(h5f: H5File): set[VetoKind] = let flags = h5f["/ctx/logLFlags", string] for f in flags: result.incl parseEnum[VetoKind](f) proc fromH5[T: SomeNumber](h5f: H5File, res: var T, name, path: string) = ## Reads the attribute `name` from `path` into `res` let grp = h5f[path.grp_str] res = grp.attrs[name, T] proc readEfficiencies(h5f: H5File): Efficiency = for field, val in fieldPairs(result): h5f.fromH5(val, field, "/ctx/eff") proc readLimit(fname: string): LimitData = var h5f = H5open(fname, "r") let limits = h5f["/limits", float] let noCands = h5f.attrs["limitNoSignal", float] let vetoes = readVetoes(h5f) let effs = readEfficiencies(h5f) result = LimitData(expectedLimit: expLimit(limits), limitNoSignal: noCands, vetoes: vetoes, eff: effs) proc asDf(limit: LimitData): DataFrame = result = toDf({ "ε_lnL" : limit.eff.signalEff, "Scinti" : fkScinti in limit.vetoes, "FADC" : fkFadc in limit.vetoes, "ε_FADC" : 1.0 - (1.0 - limit.eff.vetoPercentile) * 2.0, "Septem" : fkSeptem in limit.vetoes, "Line" : fkLineVeto in limit.vetoes, "ε_Septem" : limit.eff.septemVetoRandomCoinc, "ε_Line" : limit.eff.lineVetoRandomCoinc, "ε_SeptemLine" : limit.eff.septemLineVetoRandomCoinc, "Total eff." : limit.eff.totalEff, "Limit no signal" : limit.limitNoSignal, "Expected Limit" : limit.expectedLimit }) const path = "/t/lhood_outputs_adaptive_fadc_limits/" const prefix = "mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0469_σb_0.0028_posUncertain_puUncertain_σp_0.0500" var df = newDataFrame() for f in walkFiles(path / "mc_limit_lkMCMC_skInterpBackground_nmc_1000_*.h5"): let name = f.extractFilename.dup(removePrefix(prefix)).dup(removeSuffix(".csv")) let limit = readLimit(f) #echo name, " = ", limit df.add asDf(limit) echo df.toOrgTable()
UPDATE: As the limits above still used the wrong background time, we will now run all of it again and regenerate the above table. The old table containing the correct input files with some fields missing, but using the old background time is below in sec. [BROKEN LINK: sec:limits:exp_limits_wrong_time].
Expected limits from ./../resources/lhood_limits_automation_with_nn_support/ which should be more or less correct now (but they lack the eccentricity line veto cut value, so it's 0 in all columns!
cd $TPA/Tools/generateExpectedLimitsTable ./generateExpectedLimitsTable --path ~/org/resources/lhood_limits_automation_with_nn_support/limits
NOTE: These have different rows for different ε line veto cutoffs, but the table does not highlight that fact! 0.8602 corresponds to ε = 1.0, i.e. disable the cutoff.
εlnL Scinti FADC εFADC Septem Line eccLineCut εSeptem εLine εSeptemLine Total eff. Limit no signal Expected Limit 0.9 true true 0.98 false true 0 0.7841 0.8602 0.7325 0.7587 3.7853e-21 7.9443e-23 0.9 true false 0.98 false true 0 0.7841 0.8602 0.7325 0.7742 3.6886e-21 8.0335e-23 0.9 true true 0.98 false true 0 0.7841 0.8794 0.7415 0.7757 3.6079e-21 8.1694e-23 0.8 true true 0.98 false true 0 0.7841 0.8602 0.7325 0.6744 4.0556e-21 8.1916e-23 0.8 true true 0.98 false true 0 0.7841 0.8602 0.7325 0.6744 4.0556e-21 8.1916e-23 0.9 true true 0.98 false true 0 0.7841 0.8946 0.7482 0.7891 3.5829e-21 8.3198e-23 0.8 true true 0.98 false true 0 0.7841 0.8794 0.7415 0.6895 3.9764e-21 8.3545e-23 0.8 true true 0.9 false true 0 0.7841 0.8602 0.7325 0.6193 4.4551e-21 8.4936e-23 0.9 true true 0.98 false true 0 0.7841 0.9076 0.754 0.8005 3.6208e-21 8.5169e-23 0.8 true true 0.98 false true 0 0.7841 0.8946 0.7482 0.7014 3.9491e-21 8.6022e-23 0.8 true true 0.98 false true 0 0.7841 0.9076 0.754 0.7115 3.9686e-21 8.6462e-23 0.9 true false 0.98 true true 0 0.7841 0.8602 0.7325 0.6593 4.2012e-21 8.6684e-23 0.7 true true 0.98 false true 0 0.7841 0.8602 0.7325 0.5901 4.7365e-21 8.67e-23 0.9 true true 0.98 true true 0 0.7841 0.8602 0.7325 0.6461 4.3995e-21 8.6766e-23 0.7 true false 0.98 false true 0 0.7841 0.8602 0.7325 0.6021 4.7491e-21 8.7482e-23 0.8 true true 0.98 true true 0 0.7841 0.8602 0.7325 0.5743 4.9249e-21 8.7699e-23 0.8 true true 0.98 false false 0 0.7841 0.8602 0.7325 0.784 3.6101e-21 8.8059e-23 0.8 true true 0.8 false true 0 0.7841 0.8602 0.7325 0.5505 5.1433e-21 8.855e-23 0.7 true true 0.98 false true 0 0.7841 0.8794 0.7415 0.6033 4.4939e-21 8.8649e-23 0.8 true true 0.98 true false 0 0.7841 0.8602 0.7325 0.6147 4.5808e-21 8.8894e-23 0.9 true false 0.98 true false 0 0.7841 0.8602 0.7325 0.7057 3.9383e-21 8.9504e-23 0.7 true true 0.98 false true 0 0.7841 0.8946 0.7482 0.6137 4.5694e-21 8.9715e-23 0.8 true true 0.9 true true 0 0.7841 0.8602 0.7325 0.5274 5.3406e-21 8.9906e-23 0.9 true true 0.9 true true 0 0.7841 0.8602 0.7325 0.5933 4.854e-21 9e-23 0.8 false false 0.98 false false 0 0.7841 0.8602 0.7325 0.8 3.5128e-21 9.0456e-23 0.8 true false 0.98 false false 0 0.7841 0.8602 0.7325 0.8 3.5573e-21 9.0594e-23 0.7 true true 0.98 false true 0 0.7841 0.9076 0.754 0.6226 4.5968e-21 9.0843e-23 0.7 true true 0.98 true true 0 0.7841 0.8602 0.7325 0.5025 5.627e-21 9.1029e-23 0.8 true true 0.9 false false 0 0.7841 0.8602 0.7325 0.72 3.8694e-21 9.1117e-23 0.8 true true 0.9 true false 0 0.7841 0.8602 0.7325 0.5646 4.909e-21 9.2119e-23 0.7 true false 0.98 true true 0 0.7841 0.8602 0.7325 0.5128 5.5669e-21 9.3016e-23 0.7 true false 0.98 true false 0 0.7841 0.8602 0.7325 0.5489 5.3018e-21 9.3255e-23 0.7 true true 0.9 true true 0 0.7841 0.8602 0.7325 0.4615 6.1471e-21 9.4509e-23 0.8 true true 0.8 false false 0 0.7841 0.8602 0.7325 0.64 4.5472e-21 9.5113e-23 0.8 true true 0.8 true true 0 0.7841 0.8602 0.7325 0.4688 5.8579e-21 9.5468e-23 0.8 true true 0.8 true false 0 0.7841 0.8602 0.7325 0.5018 5.6441e-21 9.5653e-23 - Expected limits July 2023
See
journal.orgfor more details around the calculation around this time!./generateExpectedLimitsTable \ --path ~/org/resources/lhood_lnL_04_07_23/limits/ \ --prefix mc_limit_lkMCMC_skInterpBackground_nmc_1000 \ --path ~/org/resources/lhood_MLP_06_07_23/limits/ \ --prefix mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty
- Expected limits July 2023 with more statistics
The table above used only 1000 toy limits to compute the expected limit. To have a prettier plot for the presentation on as well as to get a more certain result, we generated more samples (30k for the best case and 15k for the next few).
More details are found in
journal.orgon the dates before .For the best case:
./generateExpectedLimitsTable --path ~/org/resources/lhood_MLP_06_07_23/limits/ --prefix mc_limit_lkMCMC_skInterpBackground_nmc_30000_
ε Type Scinti FADC εFADC Septem Line eccLineCut εSeptem εLine εSeptemLine Total eff. Limit no signal [GeV⁻¹] Expected limit [GeV⁻¹] Exp. limit variance [GeV⁻¹] Exp. limit σ [GeV⁻¹] 0.9107 MLP true true 0.98 false true 1 0.7841 0.8602 0.7325 0.7677 5.9559e-23 7.5824e-23 6.0632e-51 7.7866e-26 For the next worse cases:
./generateExpectedLimitsTable \ --path ~/org/resources/lhood_MLP_06_07_23/limits/ \ --prefix mc_limit_lkMCMC_skInterpBackground_nmc_15000_
ε Type Scinti FADC εFADC Septem Line eccLineCut εSeptem εLine εSeptemLine Total eff. Limit no signal [GeV⁻¹] Expected limit [GeV⁻¹] Exp. limit variance [GeV⁻¹] Exp. limit σ [GeV⁻¹] 0.9718 MLP true true 0.98 false true 1 0.7841 0.8602 0.7325 0.8192 5.8374e-23 7.6252e-23 1.6405e-50 1.2808e-25 0.8474 MLP true true 0.98 false true 1 0.7841 0.8602 0.7325 0.7143 6.1381e-23 7.6698e-23 1.4081e-50 1.1866e-25 0.7926 MLP true true 0.98 false true 1 0.7841 0.8602 0.7325 0.6681 6.2843e-23 7.8222e-23 1.3589e-50 1.1657e-25 0.7398 MLP true true 0.98 false true 1 0.7841 0.8602 0.7325 0.6237 6.5704e-23 7.9913e-23 1.6073e-50 1.2678e-25 - Expected limits with too long time
The files to generate these numbers are: ./../resources/lhood_limits_automation_correct_duration/
εlnL Scinti FADC εFADC Septem Line εSeptem εLine εSeptemLine Total eff. Limit no signal Expected Limit 0.8 true true 0.98 false true 0.7841 0.8602 0.7325 0.6744 3.9615e-21 7.958e-23 0.8 true false 0.98 false true 0.7841 0.8602 0.7325 0.6881 3.8739e-21 8.1849e-23 0.8 true true 0.9 false true 0.7841 0.8602 0.7325 0.6193 4.3163e-21 8.3183e-23 0.8 true true 0.98 true true 0.7841 0.8602 0.7325 0.5743 4.6195e-21 8.5274e-23 0.8 true true 0.8 false true 0.7841 0.8602 0.7325 0.5505 4.7792e-21 8.5958e-23 0.8 true true 0.98 false false 0.7841 0.8602 0.7325 0.784 3.4501e-21 8.6618e-23 0.8 true true 0.98 true false 0.7841 0.8602 0.7325 0.6147 4.3118e-21 8.6887e-23 0.8 true false 0.98 false false 0.7841 0.8602 0.7325 0.8 3.3996e-21 8.8007e-23 0.8 true false 0.98 true true 0.7841 0.8602 0.7325 0.586 4.561e-21 8.8266e-23 0.8 true false 0.98 true false 0.7841 0.8602 0.7325 0.6273 4.2011e-21 8.8528e-23 0.8 false false 0.98 false false 0.7841 0.8602 0.7325 0.8 3.3243e-21 8.8648e-23 0.8 true true 0.9 true false 0.7841 0.8602 0.7325 0.5646 4.6701e-21 8.8912e-23 0.8 true true 0.9 true true 0.7841 0.8602 0.7325 0.5274 5.1526e-21 8.8965e-23 0.8 true true 0.9 false false 0.7841 0.8602 0.7325 0.72 3.6485e-21 8.917e-23 0.9 true true 0.8 true true 0.7841 0.8602 0.7325 0.5274 4.8752e-21 9.0607e-23 0.8 true true 0.8 true false 0.7841 0.8602 0.7325 0.5018 5.2437e-21 9.312e-23 0.8 true true 0.8 true true 0.7841 0.8602 0.7325 0.4688 5.58e-21 9.3149e-23 0.8 true true 0.8 false false 0.7841 0.8602 0.7325 0.64 4.1102e-21 9.539e-23 0.7 true true 0.8 true true 0.7841 0.8602 0.7325 0.4102 6.1919e-21 9.716e-23
29.2. Limit method mathematical explanation from scratch
This section will cover explaining the basic ideas of the limit calculation methods and the math involved. At the moment I need to write this for the talk about the limit method ./../Talks/LimitMethod/limit_method.html but it will also be useful both for the thesis as well as other people (I hope).
- Context and terminology
An experiment tries to detect a new phenomenon of the kind where you expect very little signal compared to background sources. We have a dataset in which the experiment is sensitive to the phenomenon, another dataset in which it is not sensitive and finally a theoretical model of our expected signal.
Any data entry (after cleaning) in the sensitive dataset is a candidate. Each candidate is drawn from a distribution of the present background plus the expected signal contribution (c = s + b). Any entry in the non sensitive dataset is background only.
- Goal
- compute the value of a parameter (coupling constant) such that there is 95% confidence that the combined hypothesis of signal and background sources are compatible with the background only hypothesis.
- Condition
Our experiment should be such that the data in some "channels" of our choice can be modeled by a Poisson distribution
\[ P_{\text{Pois}}(k; λ) = \frac{λ^k e^{-λ}}{k!}. \]
Each such channel with an expected mean of \(λ\) counts has probability \(P_{\text{Pois}}(k; λ)\) to measure \(k\) counts. Because the poisson distribution (as written here) is a probability density function, multiple different channels can be combined to a "likelihood" for an experiment outcome by taking the product of each channel's poisson probability
\[ \mathcal{L}(λ) = \prod_i P_{i, \text{Pois}}(k; λ) = \prod_i \frac{λ_i^{k_i} e^{-λ_i}}{k_i!} \]
i.e. given a set of \(k_i\) recorded counts for all different channels \(i\) with expected means \(λ_i\) the "likelihood" gives us the literal likelihood to record exactly that experimental outcome. Note that the parameter of the likelihood function is the mean \(λ\) and not the recorded data \(k\)! The likelihood function describes the likelihood for a fixed set of data (our real measured counts) for different parameters (our signal & background models - where background model is constant as well).
In addition the method described below is valid under the assumption that our experiment did not have a statistically significant detection in the signal sensitive dataset compared to the background dataset!
- Implementation
The likelihood function as described in the previous section is not helpful to compute a limit for the usage with different datasets as described before. For that case we want to have some kind of a "test statistic" that relates the sensitive dataset with its seen candidates to the background dataset. For practical purposes we prefer to define such a statistic which is monotonically increasing in the number of candidates (see T. Junk's 1999 paper for details or read sec. 17.4.1.1). There are different choices possible, but the one we use is: \[ Q(s, b) = \prod_i \frac{P_{\text{pois}}(c_i; s_i + b_i)}{P_{\text{pois}}(c_i; b_i)} \] so the ratio of the signal plus background over the pure background hypothesis. The number \(c_i\) is the real number of measured candidates. So the numerator gives the probability to measure \(c_i\) counts in each channel \(i\) given the signal plus background hypothesis. On the other hand the denominator measures the probability to measure \(c_i\) counts in each channel \(i\) assuming only the background hypothesis.
For each channel \(i\) the ratio of probabilities itself is not strictly speaking a probability density function, because the integral
\[ \int_{-∞}^{∞}Q\, \mathrm{d}x = N \neq 1 \]
where \(N\) can be interpreted as a hypothetical number of total number of counts measured in the experiment. A PDF requires this integral to be 1.
As a result the full construct \(Q\) of the product of these ratios is technically not a likelihood function either. It is usually referred to as an "extended likelihood function".
For all practical purposes though we will continue to treat is as a likelihood function and call it \(L\) as usual.
Note the important fact that \(Q\) really is only a function of our signal hypothesis \(s\) and our background model \(b\). Each experimental outcome has its own \(Q\). This is precisely why the likelihood function describes everything about an experimental outcome (at least if the signal and background models are reasonably understood) and thus different experiments can be combined by combining them in "likelihood space" (multiplying their \(Q\) or adding \(\ln Q\) values) to get a combined likelihood to compute a limit for.
- Deriving a practical version of \(Q\)
The version of \(Q\) presented above is still quite impractical to use and the ratio of exponentials of the Poisson distributions can be simplified significantly:
\begin{align*} Q &= \prod_i \frac{P(c_i, s_i + b_i)}{P(c_i, b_i)} = \prod_i \frac{ \frac{(s_i + b_i)^{n_i}}{n_i!} e^{-(s_i + b_i)} }{ \frac{b_i^{n_i}}{n_i!} e^{-b_i}} \\ &= \prod_i \frac{e^{-s_i} (s_i + b_i)^{c_i}}{b_i^{c_i}} = e^{-s_\text{tot}} \prod_i \frac{(s_i + b_i)^{c_i}}{b_i^{c_i}} \\ &= e^{-s_\text{tot}} \prod_i \left(1 + \frac{s_i}{b_i} \right)^{c_i} \end{align*}This really is the heart of computing a limit with a number of \(s_{\text{tot}}\) expected events from the signal hypothesis (depending on the parameter to be studied, the coupling constant), \(c_i\) measured counts in each channel and \(s_i\) expected signal events and \(b_i\) expected background events in that channel.
As mentioned previously though the choice of what a channel is, is completely up to us! One such choice might be binning the candidates in energy. However, there is one choice that is particularly simple and is often referred to as the "unbinned likelihood". Namely, we create channels in time such that each "time bin" is so short as to either have 0 entries (most channels) or 1 entry. This means we have a large number of channels, but because of our definition of \(Q\) this does not matter. All channels with 0 candidates do not contribute to \(Q\) (they are \((1 + \frac{s_i}{b_i})^0 = 1\)). As a result our expression of \(Q\) simplifies further to:
\[ Q = e^{-s_\text{tot}} \prod_i \left(1 + \frac{s_i}{b_i}\right) \]
where \(i\) is now all channels where a candidate is contained (\(c_i = 1\)).
- How to explicitly compute \(Q\)
Our simplified version of \(Q\) using very short time bins now allows to explicitly compute the likelihood for a set of parameters. Let's now look at each of the constituents \(s_{\text{tot}}\), \(s_i\) and \(b_i\) and discuss how they are computed. We will focus on the explicit case of an X-ray detector behind a telescope at CAST.
Here it is important to note that the signal hypothesis depends on the coupling constant we wish to compute a limit for, we will just call it \(g\) in the remainder (it may be \(g_{aγ}\) or \(g_{ae}\) or any other coupling constant). And this is the actual parameter of \(Q\), but more on that in the next section on how a limit is actually computed later.
First of all the signal contribution in each channel \(s_i\). It is effectively a number of counts that one would expect within the time window of the channel \(i\). While this seems tricky given that we have not explicitly defined such a window we can:
- either assume our time interval to be infinitesimally small and give a signal rate (i.e. per second)
- or make use of the neat property that our expression only contains the ratio of \(s_i\) and \(b_i\). What this means is that we can choose our units ourselves, as long as we use the same units for \(s_i\) as for \(b_i\)!
We will use the second case and scale each candidate's signal and background contribution to the total tracking time (signal sensitive dataset length). Each parameter with a subscript \(i\) is the corresponding value that the candidate has we are currently looking at (e.g. \(E_i\) is the energy of the recorded candidate \(i\) used to compute the expected signal).
\begin{equation} \label{eq:limit_method_signal_si} s_i(g) = f(g, E_i) · A · t · P_{a \rightarrow γ}(g_{aγ}) · ε(E_i) · r(x_i, y_i) \end{equation}where:
- \(f(g, E_i)\) is the axion flux at energy \(E_i\) in units of \(\si{keV^{-1}.cm^{-2}.s^{-1}}\) as a function of \(g\).
- \(A\) is the area of the magnet bore in \(\si{cm²}\)
- \(t\) is the tracking time in \(\si{s}\)
- \(P_{a \rightarrow γ}\) is the conversion probability of the axion converting into a photon computed via \[ P_{a \rightarrow γ}(g_{aγ}) = \left( \frac{g_{aγ} B L}{2} \right)² \] written in natural units (meaning if we wish to use the equation as written here we need to convert \(B = \SI{9}{T}\) and \(L = \SI{9.26}{m}\) into values expressed in powers of electronvolt \(\si{eV}\)).
- \(ε(E_i)\) is the combined detection efficiency, i.e. the combination of X-ray telescope effective area, the transparency of the detector window and the absorption probability of an X-ray in the gas.
- \(r(x_i, y_i)\) is the expected amount of flux from the solar axion flux after it is focused by the X-ray telescope in the readout plane of the detector at the candidate's position \((x_i, y_i)\) (this requires a raytracing model). It should be expressed as a fractional value in units of \(\si{cm^{-2}}\).
As a result the units of \(s_i\) are then given in \(\si{keV^{-1}.cm^{-2}}\) with the tracking time integrated out. If one computes a limit for \(g_{aγ}\) then \(f\) and \(P\) both depend on the coupling of interest. In case of e.g. an axion-electron coupling limit \(g_{ae}\) the conversion probability can be treated as constant (with a fixed \(g_{aγ}\)).
Secondly the background hypothesis \(b_i\) for each channel. Its value depends on whether we assume a constant background model, an energy dependent one or even an energy plus position dependent model. In either case the main point is to evaluate that background model at the (potentially) position \((x_i, y_i)\) of the candidate and energy \(E_i\) of the candidate. The value should then be scaled to the same units of as \(s_i\), namely \(\si{keV^{-1}.cm^{-2}}\). Depending on how the model is defined this might just be a multiplication by the total tracking time in seconds.
The final piece is the total signal \(s_{\text{tot}}\), corresponding to the total number of counts expected from our signal hypothesis for the given dataset. This is nothing else as the integration of \(s_i\) over the entire energy range and detection area. However, because \(s_i\) implies the signal for candidate \(i\), we write \(s(E, x, y)\) to mean the equivalent signal as if we had a candidate at \((E, x, y)\) \[ s_{\text{tot}} = ∫_0^{E_{\text{max}}} ∫_A s(E, x, y) \mathrm{d}E \mathrm{d}x \mathrm{d}y \] where \(A\) simply implies integrating the full area in which \((x, y)\) is defined (the axion flux is bounded within a region much smaller than the active detection area and hence all contributions outside are 0).
- Computing a limit from \(Q\)
With the above we are now able to evaluate \(Q\) for a set of candidates \({c_i(E_i, x_i, y_i)}\). As mentioned before it is important to realize that \(Q\) is a function of the coupling constant \(g\), \(Q(g)\) with all other parameters effectively constant in the context of "one experiment".
With this in mind the "limit" is defined as the 95-th percentile of \(Q(g)\) within the physical region of \(g\) (the region \(g < 0\) is explicitly ignored, as a coupling constant cannot be negative! This can be "rigorously" justified in Bayesian statistics by saying the prior \(π(g)\) is 0 for \(g < 0\).).
So we can define it implicitly as:
\begin{equation} \label{eq:limit_method:limit_def} 0.95 = \frac{∫_0^{g'} Q(g) \mathrm{d}g}{∫_0^∞ Q(g) \mathrm{d}g} \end{equation}In practice the integral cannot be evaluated until infinity. Fortunately, our choice of \(Q\) in the first place means that the function converges to \(0\) quickly for large values of \(g\). Therefore, we only need to compute values to a "large enough" value of \(g\) to get the shape of \(Q(g)\). From there we can use any numerical approach (via an empirical CDF for example) to determine the coupling constant \(g'\) that corresponds to the 95-th percentile of \(Q(g)\).
In an intuitive sense the limit means the following: \(\SI{95}{\percent}\) of all coupling constants that reproduce the data we measured - given our signal and background hypotheses - are smaller than \(g'\).
Fig. 437 shows the likelihood as a function of \(g_{ae}²\). The blue area is the lower \SI{95}{\%} of the parameter space and the red area is everything below. Therefore, the limit in this particular set of toy candidates is at the intersection of the two colors.
Figure 437: Example likelihood as a function of \(g_{ae}²\) for a set of toy candidates. Blue is the lower 95-th percentile of the integral over the likelihood function and red the upper 5-th. The limit is at the intersection. - Drawing toy candidates and computing an expected limit
Assuming a constant background over some chip area with only an energy dependence, the background hypothesis can be used to draw toy candidates that can be used in place for the real candidates to compute limits.
In this situation the background hypothesis can be modeled as follows:
\[ B = \{ P_{\text{Pois}}(k; λ = b_i) \: | \: \text{for all energy bins } E_i \}, \]
that is the background is the set of all energy bins \(E_i\), where each bin content is described by a Poisson distribution with a mean and expectation value of \(λ = b_i\) counts.
To compute a set of toy candidates then, we simply iterate over all energy bins and draw a number from each Poisson distribution. This is the number of candidates in that bin for the toy. Given that we assumed a constant background over the chip area, we finally need to draw the \((x_i, y_i)\) positions for each toy candidate from a uniform distribution.
These sets of toy candidates can be used to compute an "expected limit". The term expected limit is usually understood to mean the median of sets of representative toy candidates. If \(L_{t_i}\) is the limit of the toy candidate set \(t_i\), the expected limit \(\langle L \rangle\) is defined as
\[ \langle L \rangle = \mathrm{median}( \{ L_{t_i} \} ) \]
If the number of toy candidate sets is large enough the expected limit should prove accurate. The real limit will then be below or above with \(\SI{50}{\%}\) chance each.
- Including systematics
The aforementioned likelihood ratio assumes perfect knowledge of the inputs for the signal and background hypotheses. In practice neither of these is known perfectly though. Each input typically has associated a small systematic uncertainty (e.g. the width of the detector window is only known up to N nanometers, the pressure in the chamber only up to M milli bar, magnet length only up to A centimeter etc.). These all affect the "real" numbers one should actually calculate with. So how does one treat these uncertainties?
The basic starting point is realizing that the values we use are our "best guess" of the real value. Usually it is a reasonable approximation that the real value will likely be within some standard deviation around that best guess, following a normal distribution. Further, it is a good idea to identify all systematic uncertainties and classify them by which aspect of \(s_i\) or \(b_i\) they affect (amount of signal or background or the position { in some other type of likelihood function possibly other } ). Another reasonable assumption is to combine different uncertainties of the same type by the square root of their squared sum, i.e. computing the euclidean radius N dimensions (for N uncertainties of the same type).
For example assuming we had these systematics (expressed as relative numbers from the best guess):
- signal uncertainties:
- magnet length: \SI{0.2}{\%}
- magnet bore diameter: \SI{2.3}{\%}
- window thickness: \SI{0.6}{\%}
- position uncertainty (of where the axion image is projected):
- detector alignment: \SI{5}{\%}
- background uncertainty:
- A: \SI{0.5}{\%} (whatever it may be, all real ones of mine are very specific)
From here we compute 3 combined systematics:
- \(σ_s = \sqrt{ 0.2² + 2.3² + 0.6²} = \SI{2.38}{\%}\)
- \(σ_p = \SI{5}{\%}\)
- \(σ_b = \SI{0.5}{\%}\)
The previous explanation and assumptions already tells us everything about how to encode these uncertainties into the limit calculation. For a value corresponding to our "best guess" we want to recover the likelihood function \(Q\) from before. The further we get away from our "best guess" the more the likelihood function should be "penalized", i.e. the actual likelihood of that parameter given our data should be lower. We will see in a minute what is meant by "being at the 'best guess'" or "away from it".
We encode this by multiplying the initial likelihood \(Q\) with additional normal distributions, one for each uncertainty (4 in total in our case, signal, background, and two position uncertainties). Each adds an additional parameter, a "nuisance parameter".
To illustrate the details, let's look at the case of adding a single nuisance parameter. In particular we'll look at the nuisance parameter for the signal as it requires more care.
The idea is to express our uncertainty of a parameter - in this case the signal - by introducing an additional parameter \(s_i'\). In contrast to \(s_i\) it describes a possible other value of \(s_i\) due to our systematic uncertainty. For simplicity we rewrite our likelihood \(Q\) as \(Q'(s_i, s_i', b_i)\):
\[ Q' = e^{-s'_\text{tot}} \prod_i (1 + \frac{s_i'}{b_i}) · \exp\left[-\frac{1}{2} \left(\frac{s_i' - s_i}{σ_s'}\right)² \right] \]
where \(s_i'\) takes the place of the \(s_i\). The added gaussian then provides a penalty for any deviation from \(s_i\). The standard deviation of the gaussian \(σ_s'\) is the actual systematic uncertainty on our parameter \(s_i\) in units of \(s_i\) (so not in percent as we showed examples further up, but as an effective number of counts { or whatever unit \(s_i\) is expressed in } ).
This form of adding a secondary parameter \(s_i'\) of the same units as \(s_i\) is not the most practical, but maybe provides the best explanation as to how the name 'penalty term' arises for the added gaussian. If \(s_i = s_i'\) then the exponential term is \(1\) meaning the likelihood remains unchanged. For any other value the exponential is \(< 1\) decreasing the likelihood \(Q'\).
By a change of variables we can replace the "unitful" parameter \(s_i'\) by a unitless number \(ϑ_s\). We would like the exponential to be \(\exp(-ϑ_s²/(2 σ_s²))\) to only express deviation from our best guess \(s_i\). \(ϑ_s = 0\) means no deviation and \(|ϑ_s| = 1\) implies \(s_i = -s_i'\). Note that the standard deviation of this is now \(σ_s\) and not \(σ_s'\) as seen in the expression above. This \(σ_s\) corresponds to our systematic uncertainty on the signal as a percentage.
To arrive at this expression:
\begin{align*} \frac{s_i' - s_i}{σ_s'} &= \frac{ϑ_s}{σ_s} \\ \Rightarrow s_i' &= \frac{σ_s'}{σ_s} ϑ_s + s_i \\ \text{with } s_i &= \frac{σ_s'}{σ_s} \\ s_i' &= s_i + s_i ϑ_s \\ \Rightarrow s_i' &= s_i (1 + ϑ_s) \\ \end{align*}where we made use of the fact that the two standard deviations are related by the signal \(s_i\) (which can be seen by defining \(ϑ_s\) as the normalized difference \(ϑ_s = \frac{s'_i - s_i}{s_i}\)).
This results in the following final (single nuisance parameter) likelihood \(Q'\):
\[ Q' = e^{-s'_\text{tot}} \prod_i (1 + \frac{s_i'}{b_i}) · \exp\left[-\frac{1}{2} \left(\frac{ϑ_s}{σ_s}\right)² \right] \]
where \(s_i' = s_i (1 + ϑ_s)\) and similarly \(s_{\text{tot}}' = s_{\text{tot}} ( 1 + ϑ_s )\) (the latter just follows because \(1 + ϑ_s\) is a constant under the different channels \(i\), see the appendix below).
The same approach is used to encode the background systematic uncertainty. The position uncertainty is generally handled the same, but the \(x\) and \(y\) coordinates are treated separately.
As shown in eq. \eqref{eq:limit_method_signal_si} the signal \(s_i\) actually depends on the positions \((x_i, y_i)\) of each candidate via the raytracing image \(r\).
With this we can introduce the nuisance parameters by replacing \(r\) by an \(r'\) such that \[ r' ↦ r(x_i - x'_i, y_i - y'_i) \] which effectively moves the center position by \((x'_i, y'_i)\). In addition we need to add penalty terms for each of these introduced parameters:
\[ \mathcal{L}' = \exp[-s] \cdot \prod_i \left(1 + \frac{s'_i}{b_i}\right) \cdot \exp\left[-\left(\frac{x_i - x'_i}{\sqrt{2}σ} \right)² \right] \cdot \exp\left[-\left(\frac{x_i - x'_i}{\sqrt{2}σ} \right)² \right] \]
where \(s'_i\) is now the modification from above using \(r'\) instead of \(r\). Now we perform the same substitution as we do for \(θ_b\) and \(θ_s\) to arrive at:
\[ \mathcal{L}' = \exp[-s] \cdot \prod_i \left(1 + \frac{s'_i}{b_i}\right) \cdot \exp\left[-\left(\frac{θ_x}{\sqrt{2}σ_x} \right)² \right] \cdot \exp\left[-\left(\frac{θ_y}{\sqrt{2}σ_y} \right)² \right] \]
The substitution for \(r'\) means the following for the parameters: \[ r' = r\left(x (1 + θ_x), y (1 + θ_y)\right) \] where essentially a deviation of \(|θ| = 1\) means we move the spot to the edge of the chip.
Putting all these four nuisance parameters together we have
\begin{align} \label{eq:limit_method:likelihood_function_def} Q' &= \left(\prod_i \frac{P_{\text{pois}}(n_i; s_i + b_i)}{P_{\text{pois}}(n_i; b_i)}\right) \cdot \mathcal{N}(θ_s, σ_s) \cdot \mathcal{N}(θ_b, σ_b) \cdot \mathcal{N}(θ_x, σ_x) \cdot \mathcal{N}(θ_y, σ_y) \\ Q'(g, ϑ_s, ϑ_b, ϑ_x, ϑ_y) &= e^{s'_\text{tot}} \prod_i (1 + \frac{s_i''}{b_i'}) · \exp\left[-\frac{1}{2} \left(\frac{ϑ_s}{σ_s}\right)² -\frac{1}{2} \left(\frac{ϑ_b}{σ_b}\right)² -\frac{1}{2} \left(\frac{ϑ_x}{σ_x}\right)² -\frac{1}{2} \left(\frac{ϑ_y}{σ_y}\right)² \right] \end{align}where here the doubly primed \(s_i''\) refers to modification for the signal nuisance parameter as well as for the position uncertainty via \(r'\).
- signal uncertainties:
- Computing a limit with nuisance parameters
The likelihood function we started with \(Q\) was only a function of the coupling constant \(g\) we want to compute a limit for. With the inclusion of the four nuisance parameters however, \(Q'\) is now a function of 5 parameters, \(Q'(g, ϑ_s, ϑ_b, ϑ_x, ϑ_y)\). Following our definition of a limit via a fixed percentile of the integral over the coupling constant, eq. \eqref{eq:limit_method:limit_def}, leads to problem for \(Q'\). If anything one could define a contour describing the 95-th percentile of the "integral volume", but this would lead to infinitely many values of \(g\) that describe said contour.
As a result to still define a sane limit value, the concept of the marginal likelihood function \(Q'_M\) is introduced. The idea is to integrate out the nuisance parameters
\[ Q'_M(g) = \iiiint_{-∞}^∞ Q'(g, ϑ_s, ϑ_b, ϑ_x, ϑ_y)\,\mathrm{d}ϑ_s\mathrm{d}ϑ_b\mathrm{d}ϑ_x\mathrm{d}ϑ_y \]
Depending on the exact definition of \(Q'\) in use these integrals may be analytically computable. In many cases however they are not and numerical techniques to evaluate the integral must be utilized.
Aside from the technical aspects about how to evaluate \(Q'_M(g)\) at a specific \(g\), the limit calculation continues exactly as for the case without nuisance parameters once \(Q'_M(g)\) is defined as such.
- Practical calculation of \(Q'_M(g)\) in our case
In case of our explicit likelihood function eq. \eqref{eq:limit_method:likelihood_function_def} there is already one particular case that makes the marginal likelihood not analytically integrable because the \(b_i' = b_i(1 + ϑ_b)\) term introduces a singularity for \(ϑ_b = -1\). For practical purposes this is not too relevant, as values approaching \(ϑ_b = -1\) would imply having zero background and within a reasonable systematic uncertainty the penalty term makes contributions in this limit so small such that this area does not physically contribute to the integral.
Using standard numerical integration routines (simpson, adaptive Gauss-Kronecker etc.) are all too expensive to compute such a four-fold integration under the context of computing many limits for an expected limit. For this reason Monte Carlo approaches are used, in particular the Metropolis-Hastings (MH) Markov Chain Monte Carlo (MCMC) is used. The basic idea of general Monte Carlo integration routines is to evaluate the function at random points and computing the integral based on the function evaluation at these points (by scaling the evaluations correctly). Unless the function is very 'spiky' in the integration space, Monte Carlo approaches provide good accuracy at a fraction of the computational effort as normal numerical algorithms even in higher dimensions. However, we can do better than relying on fully random points in the integration space. The Metropolis-Hastings algorithm tries to evaluate the function more often in those points where the contributions are large. The basic idea is the following:
[ ]REWRITE THIS
Pick a random point in the integration space as a starting point \(p_0\). Next, pick another random point \(p_1\) within the vicinity of \(p_0\). If the function evaluates to a larger value at \(p_1\) accept it as the new current position. If it is smaller, accept it with a probability of \(1 - \frac{f(p_{i-1})}{f(p_i)}\) (i.e. if the new value is close to the old one we accept it with a high probability and if the new one is much lower accept it rarely). This guarantees to pick values inching closer to the most contributing areas of the integral in the integration space, while still allowing to get out of local maxima due to the random acceptance of "worse" positions.
If a new point is accepted and becomes the current position, the "chain" of points is extended (hence "Markov Chain"). By creating a chain of reasonable length the integration space is taken into account well enough. Because the initial point is completely random (up to some possible prior) the first \(N\) links of the chain are in a region of low interest (and depending on the interpretation of the chain "wrong"). For that reason one defines a cutoff \(N_b\) of the first elements that are thrown away as "burn-in" before using the chain to evaluate the integral or parameters.
In addition it can be valuable to not only start a single Markov Chain from one random point, but instead start multiple chains from different points in the integration space. This increases the chance to cover different regions of interest even in the presence of multiple peaks separated too far away to likely "jump over" via the probabilistic acceptance.
Furthermore, outside of using Metropolis-Hastings we still have to make sure the evaluation of \(Q'(g, ϑ_s, ϑ_b, ϑ_x, ϑ_y)\) is fast. We will discuss this in the next section about the evaluation of \(Q'\).
[ ]Check sagemath calculations for x and y systematics
- Evaluate \(Q'\) in our case
[ ]background position dependent
[ ]use k-d tree to store background cluster information of (x, y, E) per cluster. Interpolation using custom metric with gaussian weighting in (x, y) but constant weight in E[ ]towards corners need to correct for loss of area
template computeBackground(): untyped {.dirty.} = let px = c.pos.x.toIdx let py = c.pos.y.toIdx interp.kd.query_ball_point([px.float, py.float, c.energy.float].toTensor, radius = interp.radius, metric = CustomMetric) .compValue() .correctEdgeCutoff(interp.radius, px, py) # this should be correct .normalizeValue(interp.radius, interp.energyRange, interp.backgroundTime) .toIntegrated(interp.trackingTime)
[ ]background values cached, to avoid recomputing values if same candidate is asked for
[ ]Signal[ ]detection efficiency, window (w/o strongback) + gas + telescope efficiency (energy dependent)[ ]axion flux, rescale by gae²[ ]conversion prob[ ]raytracing result (telescope focusing) + window strongback
[ ]candidate sampling[ ]handled using a grid of NxNxM volumes (x, y, E)[ ]sample in each volume & assign uniform positions in volume
29.2.1. Note about likelihood integral
The likelihood is a product of probability density functions. However, it is important to note that the likelihood is a function of the parameter and not the data. As such integrating over all parameters does not necessarily equate to 1!
[ ]If one takes the inverse and assumes an \(L(k) = Π_i P(k_i; λ_i)\) instead, does that integrate to 1 if integrated over all \(k\)?
29.2.2. \(s'\) is equivalent to \(s_i'\) ?
so indeed, this is perfectly valid.
29.2.3. Derivation of short form of Q
This uses the logarithm form, but the non log form is even easier actually.
\begin{align*} \ln \mathcal{Q} &= \ln \prod_i \frac{ \frac{(s_i + b_i)^{n_i}}{n_i!} e^{-(s_i + b_i)} }{ \frac{b_i^{n_i}}{n_i!} e^{-b_i} } \\ &= \sum_i \ln \frac{ \frac{(s_i + b_i)^{n_i}}{n_i!} e^{-(s_i + b_i)} }{ \frac{b_i^{n_i}}{n_i!} e^{-b_i} } \\ &= \sum_i \ln \frac{(s_i + b_i)^{n_i}}{n_i!} e^{-(s_i + b_i)} - \ln \frac{b_i^{n_i}}{n_i!} e^{-b_i} \\ &= \sum_i n_i \ln (s_i + b_i) - \ln n_i! - (s_i + b_i) - (n_i \ln b_i - \ln n_i! -b_i) \\ &= \sum_i n_i \ln (s_i + b_i) - (s_i + b_i) - n_i \ln b_i + b_i \\ &= \sum_i n_i \ln (s_i + b_i) - (s_i + b_i - b_i) - n_i \ln b_i \\ &= \sum_i n_i \ln \left(\frac{s_i + b_i}{b_i}\right) - s_i \\ &= -s_{\text{tot}} + \sum_i n_i \ln \left(\frac{s_i + b_i}{b_i} \right) \\ &\text{or alternatively} \\ &= -s_{\text{tot}} + \sum_i n_i \ln \left(1 + \frac{s_i}{b_i} \right) \\ \end{align*}29.2.4. Implementing a basic limit calculation method
Simplest implementation:
- single channel
- no detection efficiencies etc., just a flux that scales with \(g²\)
- constant background (due to single channel)
- no telescope, i.e. area for signal flux is the same as for background (due to no focusing)
import unchained, math ## Assumptions: const totalTime = 100.0.h # 100 of "tracking time" const totalArea = 10.cm² # assume 10 cm² area (magnet bore and chip! This case has no telescope) defUnit(cm⁻²•s⁻¹) proc flux(g: float): cm⁻²•s⁻¹ = ## Dummy flux. Just the coupling constant squared · 1e-6 result = 1e-6 * (g*g).cm⁻²•s⁻¹ proc totalFlux(g: float): float = ## Flux integrated to total time and area result = flux(g) * totalTime.to(Second) * totalArea ## Assume signal and background in counts of the single channel! ## (Yes, `signal` is the same as `totalFlux` in this case) proc signal(g: float): float = flux(g) * totalTime * totalArea ## Signal only depends on coupling in this simple model proc background(): float = 1e-6.cm⁻²•s⁻¹ * totalTime * totalArea ## Single channel, i.e. constant background proc likelihood(g: float, cs: int): float = ## `cs` = number of candidates in the single channel result = exp(-totalFlux(g)) # `e^{-s_tot}` result *= pow(1 + signal(g) / background(), cs.float) proc poisson(k: int, λ: float): float = λ^k * exp(-λ) / (fac(k)) echo "Background counts = ", background(), ". Probabilty to measure 4 counts given background: ", poisson(4, background()) echo "equal to signal counts at g = 1: ", signal(1.0) echo "Likelihood at g = 1 for 4 candidates = ", likelihood(1.0, 4) ## Let's plot it from 0 to 3 assuming 4 candidates import ggplotnim let xs = linspace(0.0, 3.0, 100) let ys = xs.map_inline(likelihood(x, 4)) ggplot(toDf(xs, ys), aes("xs", "ys")) + geom_line() + ggsave("/tmp/simple_likelihood.pdf") ## Compute limit, CDF@95% import algorithm let yCumSum = ys.cumSum() # cumulative sum let yMax = yCumSum.max # maximum of the cumulative sum let yCdf = yCumSum.map_inline(x / yMax) # normalize to get (empirical) CDF let limitIdx = yCdf.toSeq1D.lowerBound(0.95) # limit at 95% of the CDF echo "Limit at : ", xs[limitIdx]
Background counts = 3.6. Probabilty to measure 4 counts given background: 0.1912223391751322 equal to signal counts at g = 1: 3.6 Likelihood at g = 1 for 4 candidates = 0.4371795591566811 Limit at : 1.12121212121212
More realistic implementation, above plus:
- real solar axion flux
- TODO: (detection efficiency) (could just use fixed efficiency)
- X-ray telescope without usage of local flux information
- multiple channels in energy
import unchained, math, seqmath, sequtils, algorithm ## Assumptions: const totalTime = 100.0.h # 100 of "tracking time" const areaBore = π * (2.15 * 2.15).cm² const chipArea = 5.mm * 5.mm # assume all flux is focused into an area of 5x5 mm² # on the detector. Relevant area for background! defUnit(GeV⁻¹) defUnit(cm⁻²•s⁻¹) defUnit(keV⁻¹) defUnit(keV⁻¹•cm⁻²•s⁻¹) ## Constants defining the channels and background info const Energies = @[0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5, 9.5].mapIt(it.keV) Background = @[0.5e-5, 2.5e-5, 4.5e-5, 4.0e-5, 1.0e-5, 0.75e-5, 0.8e-5, 3e-5, 3.5e-5, 2.0e-5] .mapIt(it.keV⁻¹•cm⁻²•s⁻¹) # convert to a rate ## A possible set of candidates from `Background · chipArea · totalTime · 1 keV` ## (1e-5 · 5x5mm² · 100h = 0.9 counts) Candidates = @[0, 2, 7, 3, 1, 0, 1, 4, 3, 2] proc solarAxionFlux(ω: keV, g_aγ: GeV⁻¹): keV⁻¹•cm⁻²•s⁻¹ = # axion flux produced by the Primakoff effect in solar core # in units of keV⁻¹•m⁻²•yr⁻¹ let flux = 2.0 * 1e18.keV⁻¹•m⁻²•yr⁻¹ * (g_aγ / 1e-12.GeV⁻¹)^2 * pow(ω / 1.keV, 2.450) * exp(-0.829 * ω / 1.keV) # convert flux to correct units result = flux.to(keV⁻¹•cm⁻²•s⁻¹) func conversionProbability(g_aγ: GeV⁻¹): UnitLess = ## the conversion probability in the CAST magnet (depends on g_aγ) ## simplified vacuum conversion prob. for small masses let B = 9.0.T let L = 9.26.m result = pow( (g_aγ * B.toNaturalUnit * L.toNaturalUnit / 2.0), 2.0 ) from numericalnim import simpson # simpson numerical integration routine proc totalFlux(g_aγ: GeV⁻¹): float = ## Flux integrated to total time, energy and area # 1. integrate the solar flux ## NOTE: in practice this integration must not be done in this proc! Only perform once! let xs = linspace(0.0, 10.0, 100) let fl = xs.mapIt(solarAxionFlux(it.keV, g_aγ)) let integral = simpson(fl.mapIt(it.float), # convert units to float for compatibility xs).cm⁻²•s⁻¹ # convert back to units (integrated out `keV⁻¹`!) # 2. compute final flux by "integrating" out the time and area result = integral * totalTime * areaBore * conversionProbability(g_aγ) ## NOTE: only important that signal and background have the same units! proc signal(E: keV, g_aγ: GeV⁻¹): keV⁻¹ = ## Returns the axion flux based on `g` and energy `E` result = solarAxionFlux(E, g_aγ) * totalTime.to(Second) * areaBore * conversionProbability(g_aγ) proc background(E: keV): keV⁻¹ = ## Compute an interpolation of energies and background ## NOTE: For simplicity we only evaluate at the channel energies anyway. In practice ## one likely wants interpolation to handle all energies in the allowed range correctly! let idx = Energies.lowerBound(E) # get idx of this energy ## Note: area of interest is the region on the chip, in which the signal is focused! ## This also allows us to see that the "closer" we cut to the expected axion signal on the ## detector, the less background we have compared to the *fixed* signal flux! result = (Background[idx] * totalTime * chipArea).to(keV⁻¹) proc likelihood(g_aγ: GeV⁻¹, energies: seq[keV], cs: seq[int]): float = ## `energies` = energies corresponding to each channel ## `cs` = each element is number of counts in that energy channel result = exp(-totalFlux(g_aγ)) # `e^{-s_tot}` for i in 0 ..< cs.len: let c = cs[i] # number of candidates in this channel let E = energies[i] # energy of this channel let s = signal(E, g_aγ) let b = background(E) result *= pow(1 + signal(E, g_aγ) / background(E), c.float) ## Let's plot it from 0 to 3 assuming 4 candidates import ggplotnim # define coupling constants let xs = logspace(-13, -10, 300).mapIt(it.GeV⁻¹) # logspace 1e-13 GeV⁻¹ to 1e-8 GeV⁻¹ let ys = xs.mapIt(likelihood(it, Energies, Candidates)) let df = toDf({"xs" : xs.mapIt(it.float), ys}) ggplot(df, aes("xs", "ys")) + geom_line() + ggsave("/tmp/energy_bins_likelihood.pdf") ## Compute limit, CDF@95% import algorithm # limit needs non logspace x & y data! (at least if computed in this simple way) let xLin = linspace(0.0, 1e-10, 1000).mapIt(it.GeV⁻¹) let yLin = xLin.mapIt(likelihood(it, Energies, Candidates)) let yCumSum = yLin.cumSum() # cumulative sum let yMax = yCumSum.max # maximum of the cumulative sum let yCdf = yCumSum.mapIt(it / yMax) # normalize to get (empirical) CDF let limitIdx = yCdf.lowerBound(0.95) # limit at 95% of the CDF echo "Limit at : ", xLin[limitIdx] # Code outputs: # Limit at : 6.44645e-11 GeV⁻¹
Limit at : 6.44645e-11 GeV⁻¹
"Realistic" implementation:
[ ]FINISH
import unchained, datamancer type Candidate = tuple[x, y: mm, E: keV] GUnit = float ## Unit of the coupling constant we study. Might be `float` or `GeV⁻¹` etc ## Define some compound units we use! defUnit(keV⁻¹•cm⁻²•s⁻¹) defUnit(keV⁻¹•m⁻²•yr⁻¹) defUnit(cm⁻²) defUnit(keV⁻¹•cm⁻²) proc detectionEff() = proc calcTotalFlux(axModel: string): DataFrame = ## Just read the CSV file of the solar axion flux and convert the energy ## column from eV to keV, as well as the flux from m⁻² yr⁻¹ to cm⁻² s⁻¹. ## Use resulting DF to compute flux by integration over energy. proc convert(x: float): float = result = x.keV⁻¹•m⁻²•yr⁻¹.to(keV⁻¹•cm⁻²•s⁻¹).float let df = readCsv(axModel) let E = df["Energy / eV", float].map_inline(x.eV.to(keV).float) ## Convert eV to keV let flux = df["Flux / keV⁻¹ m⁻² yr⁻¹", float].map_inline(convert(x)) ## Convert flux # get flux after detection efficiency let effFlux = flux *. detectionEff() result = simpson(df["Flux", float].toSeq1D, df["Energy [keV]", float].toSeq1D) ## Compute the total axion flux const FluxFile = "/home/basti/CastData/ExternCode/AxionElectronLimit/axion_diff_flux_gae_1e-13_gagamma_1e-12.csv" let FluxIntegral = calcTotalFlux(FluxFile) proc totalFlux(g: GUnit): float = let areaBore = π * (2.15 * 2.15).cm² let integral = ctx.integralBase.rescale(g^2) result = integral.cm⁻²•s⁻¹ * areaBore * ctx.totalTrackingTime.to(s) * conversionProbability() proc likelihood(g: GUnit, cs: seq[Candidate]): float = ## Computes the likelihood given the candidates and coupling constant `g`. let sTotal = totalFlux(g)
29.3. Estimating variance of the expected limit
We are severely limited in the number of toy candidates we can run for the expected limit calculation. At most maybe 100k samples is possible for the best case scenario.
Generally though it would be nice if we could estimate the uncertainty on the median from the width of the distribution! Using the variance or standard deviation is problematic though, because they take into account the absolute value of the limits, which we don't want.
https://en.wikipedia.org/wiki/Median_absolute_deviation MAD - the median absolute deviation - could be interesting, but suffers from the problem that if we want to use it as a consistent estimator we need a scale factor \(k\) (see the link).
Therefore, we will simply use bootstrapping, i.e. we resample the data N times, compute our statistics and then estimate the variance of our median!
In pseudo code:
let samples = generateSamples(N) const BootStraps = M var medians = newSeq[float](M) for i in 0 ..< M: # resample var newSample = newSeq[float](N) for j in 0 ..< N: newSample[j] = samples[rnd.rand(0 ..< N)] # get an index and take its value # compute our statistics medians.add median(newSample) echo "Variance of medians = ", medians.variance
which would then give us the variance of all bootstrapped medians!
This is implemented in generateExpectedLimitsTable.
29.4. Note on limit without candidates
In the case without any candidates in the signal sensitive regions (or
none at all, red line in our plots) it's important to keep in mind
that the number we show on the plot / in the table is itself sensitive
to statistical variations. The number is also computed using lkMCMC
and therefore it's not the true number!
[ ]Write a sanity check that computes this limit e.g. 500 times and shows
the spread. (already in the TODO list above)
30. TODO write notes about expectedlimitsσ*.pdf plots
31. Neural networks for classification
In this section we will again consider neural networks for classification of clusters. From the work in my bachelor and master thesis this is certainly an extremely promising avenue to improve the background rate.
For the application of neural networks to CAST like data, there are two distinct approaches one can take.
- make use of the already computed geometrical properties of the clusters and use a simple MLP network with ~1 hidden layer.
- use the raw pixel data (treating data as "images") and use a CNN approach.
The former is more computationally efficient, but could in theory introduce certain biases (clustering, choice of variables etc.). The latter avoids that by seeing the data "as it is", but is significantly more expensive. 2-3 orders of magnitude more neurons involved (even if the network is very sparse, as most pixels are empty).
In addition to these to approaches, another possibility lies in training not one network for the whole energy range, but rather treat it like the likelihood data: train one network for each X-ray tube target instead and for prediction use the network corresponding to the energy of the cluster to classify.
We will start with the simplest approach, namely the property based MLP with a single network for the whole energy range.
The training data will be the CDL X-ray tube data, after applying the 'cleaning cuts' we also use to generate the reference spectra for the likelihood method. For the background data we will start with a single, very long, background dataset taken at CAST. Ideally one should sample from a range of different background runs to better reflect changes in the detector behavior!
This input dataset is split into half, one set for training and the other half for testing purposes (to check if we have overtraining etc.).
To compare the results, we will compare with the likelihood method by:
- computing the likelihood distributions of the CDL data & the background data and compare it with the neural network prediction for the same data.
- computing the ROC curves of the two methods (signal efficiency vs. background rejection)
- finally computing a background rate based on the neural network & comparing it to the likelihood based background rate.
To implement all this we will use Flambeau, our Nim wrapper for
libtorch (the C++ backend to PyTorch).
31.1. Single MLP with one hidden layer
Aside from data preparation etc. the code used at this moment () is as follows (this code does not compile as 'useless' things have been taken out):
import flambeau/[flambeau_nn] import flambeau / tensors import strformat import nimhdf5 import ingrid / [tos_helpers, ingrid_types] import os, strutils, sequtils, random, algorithm, options, cligen import datamancer {.experimental: "views".} let bsz = 100 # batch size defModule: type IngridNet* = object of Module hidden* = Linear(14, 500) # 14 input neurons, 500 neurons on hidden layer classifier* = Linear(500, 2) proc forward(net: IngridNet, x: RawTensor): RawTensor = var x = net.hidden.forward(x).relu() return net.classifier.forward(x).squeeze(1) let validReadDSets = XrayReferenceDsets - { igNumClusters, igFractionInHalfRadius, igRadiusDivRmsTrans, igRadius, igBalance, igLengthDivRadius, igTotalCharge } + { igCenterX, igCenterY } let validDsets = validReadDSets - { igLikelihood, igCenterX, igCenterY} proc train(model: IngridNet, optimizer: var Optimizer, input, target: RawTensor, device: Device) = let dataset_size = input.size(0) var toPlot = false for epoch in 0 .. 1000: var correct = 0 if epoch mod 25 == 0: toPlot = true var predictions = newSeqOfCap[float](dataset_size) var targets = newSeqOfCap[int](dataset_size) for batch_id in 0 ..< dataset_size div bsz: # Reset gradients. optimizer.zero_grad() # minibatch offset in the Tensor let offset = batch_id * bsz let x = input[offset ..< offset + bsz, _ ] let target = target[offset ..< offset + bsz] # Running input through the network let output = model.forward(x) let pred = output.argmax(1) if toPlot: # take 0th column predictions.add output[_, 0].toNimSeq[:float] targets.add target[_, 0].toNimSeq[:int] correct += pred.eq(target.argmax(1)).sum().item(int) # Computing the loss var loss = sigmoid_cross_entropy(output, target) # Compute the gradient (i.e. contribution of each parameter to the loss) loss.backward() # Correct the weights now that we have the gradient information optimizer.step() if toPlot: let train_loss = correct.float / dataset_size.float64() # loss.item(float) echo &"\nTrain set: Average loss: {train_loss:.4f} " & &"| Accuracy: {correct.float64() / dataset_size.float64():.3f}" ## create output plot if toPlot: plotTraining(predictions, targets) let preds = predictions.mapIt(clamp(-it, -50.0, 50.0)) rocCurve(preds, targets) toPlot = false proc test(model: IngridNet, input, target: RawTensor, device: Device): (seq[float], seq[int]) = ## returns the predictions / targets let dataset_size = input.size(0) var correct = 0 var predictions = newSeqOfCap[float](dataset_size) var targets = newSeqOfCap[int](dataset_size) no_grad_mode: for batch_id in 0 ..< dataset_size div bsz: # minibatch offset in the Tensor let offset = batch_id * bsz let x = input[offset ..< offset + bsz, _ ].to(device) let target = target[offset ..< offset + bsz].to(device) # Running input through the network let output = model.forward(x) # get the larger prediction along axis 1 (the example axis) let pred = output.argmax(1) # take 0th column predictions.add output[_, 0].toNimSeq[:float] targets.add target[_, 0].toNimSeq[:int] correct += pred.eq(target.argmax(1)).sum().item(int) # Computing the loss let test_loss = correct.float / dataset_size.float64() echo &"\nTest set: Average loss: {test_loss:.4f} " & &"| Accuracy: {correct.float64() / dataset_size.float64():.3f}" ## create output plot plotTraining(predictions, targets) let preds = predictions.mapIt(clamp(-it, -50.0, 50.0)) result = (preds, targets) proc predict(model: IngridNet, input, target: RawTensor, device: Device, cutVal: float): seq[int] = ## returns the predictions / targets let dataset_size = input.size(0) var correct = 0 var predictions = newSeq[float]() no_grad_mode: for batch_id in 0 ..< dataset_size div bsz: # minibatch offset in the Tensor let offset = batch_id * bsz let x = input[offset ..< offset + bsz, _ ].to(device) let target = target[offset ..< offset + bsz].to(device) # Running input through the network let output = model.forward(x) # get the larger prediction along axis 1 (the example axis) let pred = output.argmax(1) # take 0th column predictions.add output[_, 0].toNimSeq[:float] for i in 0 ..< bsz: if output[i, 0].item(float) > cutVal: result.add (offset + i).int # else add the index of this event that looks like signal correct += pred.eq(target.argmax(1)).sum().item(int) let test_loss = correct.float / dataset_size.float64() echo &"\nPredict set: Average loss: {test_loss:.4f} " & &"| Accuracy: {correct.float64() / dataset_size.float64():.3f}" proc main(fname: string) = let dfCdl = prepareCdl() let dfBack = prepareBackground(fname, 186).drop(["centerX", "centerY"]) var df = newDataFrame() df.add dfCdl df.add dfBack # create likelihood plot df.plotLikelihoodDist() let (logL, logLTargets) = df.logLValues() df.plotLogLRocCurve() df.drop(igLikelihood.toDset(fkTpa)) let (trainTup, testTup) = generateTrainTest(df) let (trainIn, trainTarg) = trainTup let (testIn, testTarg) = testTup Torch.manual_seed(1) var device_type: DeviceKind if Torch.cuda_is_available(): echo "CUDA available! Training on GPU." device_type = kCuda else: echo "Training on CPU." device_type = kCPU let device = Device.init(device_type) var model = IngridNet.init() model.to(device) # Stochastic Gradient Descent var optimizer = SGD.init( model.parameters(), SGDOptions.init(0.005).momentum(0.2) #learning_rate = 0.005 ) # Learning loop model.train(optimizer, trainIn.to(kFloat32).to(device), trainTarg.to(kFloat32).to(device), device) let (testPredict, testTargets) = model.test(testIn, testTarg, device) when isMainModule: dispatch main
The following network:
- MLP: Input: 14 neurons Hidden Layer: 500 neurons Output: 2 neurons
- Algorithm: Stochastic gradient descent with momentum. Learning rate: 0.005 Momentum: 0.2
- training for 1000 epochs with a batch size of 100.
used input parameters:
igHits, igSkewnessLongitudinal, igSkewnessTransverse, igRmsTransverse, igEccentricity, igHits, igKurtosisLongitudinal, igKurtosisTransverse, igLength, igWidth, igRmsLongitudinal, igRotationAngle, igEnergyFromCharge, igFractionInTransverseRms,
Background data: run 186 (11 day long run at end of first data taking period, March 2018).
First the likelihood distribution in fig. 438.
The equivalent plot for the neural network classification is shown in fig. 439.
If we restrict ourselves to the background data in the gold region only and predict all (2017 / beginning 2018) of it with the network, we get the distribution shown in fig. 440, where we have computed the global cut value for a software efficiency of ε = 80%.
We can now compare the ROC curves of the two approaches, see fig. 441.
Keep in mind that the ROC curve for the logL method here is essentially a 'mean' of the individual targets. The split curves for each target are shown in fig. 442.
Let's now split the ROC curves by the different CDL targets to compare with this figure for the MLP prediction as well. First in fig. 443 is this plot after only 10 epochs (~5 seconds of training time).
In fig. 444 we have the same plot again, but after the 1000 epochs we used for all other plots here.
And finally we can use the network to predict on the full 2017 / beginning 2018 dataset to get a background rate, see fig. 445, using a signal efficiency of ε = 80%.
Comparing to the likelihood based background rate using the same ε = 80%, as seen in fig. 446.
31.2. TODO Things to consider / do
- proper normalization of all inputs.
-> Need to get
DataLoaderto properly work, so that we don't have to perform normalization manually for all properties maybe. Or write a proc that does normalization ourselves. - apply the network to all calibration runs in order to check the efficiency in those as a sanity check!
- DONE apply the trained network to the individual CDL targets to compute the separate ROC curves! (without training individual one)
31.3. Combination of NN w/ vetoes
./../../CastData/ExternCode/TimepixAnalysis/Tools/NN_playground/predict_event.nim
./predict_event ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2017_crGold_septemveto_lineveto_dbscan.h5 \ ~/CastData/ExternCode/TimepixAnalysis/resources/LikelihoodFiles/lhood_2018_crGold_septemveto_lineveto_dbscan.h5 \ --lhood --cutVal 3.2 --totalTime 3400
Within 0 - 8 keV: 6.413398692810459e-6 keV⁻¹•cm⁻²•s⁻¹ Within 0 - 2.8 & 4.0 - 8.0 keV: 4.133025759323338e-6 keV⁻¹•cm⁻²•s⁻¹
yields
31.4. FADC data, applying a CNN [0/1]
Another avenue that might actually turn out to be helpful is to attempt to train a CNN on the raw FADC spectra. Using a 1D kernel it should be pretty successful I imagine. Especially because our manually computed observables have a high chance of being very particular to the energy of the recorded events. A CNN may learn from the particular shapes etc.
[ ]implement this!
32. Timepix3 support in TPA
To develop Timepix3 support in TPA we have a 55Fe test file in ./../../../../mnt/1TB/Uni/Tpx3/data_take_2021-04-01_10-53-34.h5 which is still only the raw data.
We now need to do the following things to get it to work:
- Since we do not want to build a raw data parser (2 x 32-bit words)
to convert them to (x, y, TOT, TOA) values, we check out the
baf2branch of the tpx3 code: https://github.com/SiLab-Bonn/tpx3-daq/tree/baf2 In theanalysis.pyfile: https://github.com/SiLab-Bonn/tpx3-daq/blob/baf2/tpx3/analysis.py the_interpret_raw_datafunction converts this to an array of the parsed data. The output of running theanalysis.py(I suppose) stores that data in the same H5 file. UPDATE: this has since been implemented by hand after all: ./../../CastData/ExternCode/TimepixAnalysis/Tools/Timepix3/readTpx3RawTest.nim (will be merged intoraw_data_manipulationsoon) - The TOT calibrations are performed and fitted with the same function as for the Timepix1. This is already performed (and there is a H5 file for the TOT calibration), which we can use. Easiest for the beginning might be to automatically add an entry to the InGrid database by handing it a TOT calibration file.
- Write a
raw_data_manipulationhandler that can take Tpx3 H5 files and read the chip information + the parsed data and hand over to write H5 function as exist. - From there handle Tpx3 data the same way as Timepix1 data. In a few cases we might handle them differently in some respects.
- (optional at the beginning) Add handling of ToA data in TPA.
UPDATE: Points 4 and 5 have also since been implemented.
The following will describe quickly the main ideas of the current Tpx3 reconstruction
32.1. Raw data reconstruction
The file listed above: ./../../CastData/ExternCode/TimepixAnalysis/Tools/Timepix3/readTpx3RawTest.nim performs the raw conversion from 32 bit words into the following datastructure:
Tpx3MetaData = object index_start: uint32 index_stop: uint32 data_length: uint32 timestamp_start: float timestamp_stop: float scan_param_id: uint32 discard_error: uint32 decode_error: uint32 trigger: float Tpx3Data* = object data_header*: uint8 header*: uint8 hit_index*: uint64 x*: uint8 y*: uint8 TOA*: uint16 TOT*: uint16 EventCounter*: uint16 HitCounter*: uint8 FTOA*: uint8 scan_param_id*: uint16 chunk_start_time*: cdouble iTOT*: uint16 TOA_Extension*: uint64 TOA_Combined*: uint64
Tpx3MetaData is used to know where the binary uint32 information
for data packets start and stop.
The raw data is read and parsed into Tpx3Data.
After that we get an HDF5 file containing the original
/configuration group (with the run & chip meta data) and a
/interpreted/hit_data_0 dataset, which contains the Tpx3Data as a
composite datatype.
The latter is done, as that is the same data structure used in the
Python code if one uses that to reconstruct.
These files can then be given to the raw_data_manipulation tool by
using the --tpx3 argument.
This performs a first clustering of the input data based on ToA data,
which happens here:
./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/private/tpx3_utils.nim
in computeTpx3RunParameters.
It uses the following basic logic for clustering:
if not ( toa == lastToa or # 0. ( toa <= lastToa + clusterTimeCutoff and toa > clusterStart ) or # 1. ( toa >= (clusterStart - clusterTimeCutoff) and toa < lastToa ) or # 2. (overflow + toa) <= (lastToa + clusterTimeCutoff)):
If this condition is true (note the not in front of the main condition), the current cluster being built is stopped and added to the result. The following variables are used:
toa: the ToA value of the current pixellastToa: The ToA value of the last pixelclusterStart: The ToA value of the first pixel in the cluster. If the condition is true and we start a new cluster, thenclusterStartwill be set totoaclusterTimeCutoff: A cutoff defined inconfig.tomlin clock cyclesoverflow: 214, as we use a 14 bit counter for the ToA clock.
We also correct overflows in a cluster. If we see pixels near 0 and near 16384, we assume an overflow happened and increase an overflow counter. For each pixel we add the current overflow counter to the result. Only 3 overflows are possible, before we raise an exception within a single cluster (which would make it a humongous cluster anyway! 65536 clock cycles long in time. That's ~1.6e6 ns or 1.6 ms.
After this is done, we treat the data as regular Timepix data, with the exception of propagating the ToA values through to the final result.
One distinction is: after cluster finding, we perform a check on
duplicate pixels here:
./../../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/private/clustering.nim
in toCluster via:
when T is PixTpx3: # in Tpx3 case filter out all pixels that appear multiple times in a # short time let p = pixels[idx] let pi = (x: p.x, y: p.y) if pi notin pixMap: pixMap.incl pi result.add p
essentially we take exactly the first pixel in a cluster. If any are activated later, they are dropped.
From there, we compute a few ToA geometry variables:
- length
- mean
- RMS
- minimum value
The minimum value because when computing these properties we subtract the minimum value to have all clusters start at a ToA value of 0. (the ToA combined data is not touched!).
32.2. Peculiarities seen in Tpx3 data
Looking at the Tpx3 data showed a few very weird things. This is only a summary. Look at the (unfortunately not public) discord discussions in the Tpx3 channel.
- Comparison plots of Fe55 run & CAST Fe55 run for the different geometric variables look mostly comparable!
- same for background data does not look comparable at all.
Things we learned about the latter:
- many "events" are extremely noisy. Have many with O(>65000) pixels active in weird patterns
- clusters appear to often have pixels active multiple times.
The plotData script was extended significantly to isolate specific
cases and create plots for e.g. different cuts on the data (but
different distributions) or extract events for certain cuts etc. etc.
An example call to plotData:
/plotData --h5file /tmp/reco_0423.h5 --runType=rtBackground \ --eventDisplay 0 --cuts '("hits", 2, 1000000)' \ --cuts '("rmsTransverse", -0.5, 0.05)' \ --applyAllCuts --head 100
Meaning input is background data, create event displays for run 0 with the two cuts defined here applied to the data. Create the first 100 events.
Isolating things like:
- number of hits
fractionInTransverseRmseither== 1 or == 0was very helpful. Equal to 0 happens for events, which are rather sparse around thecenterX/centerYdata. The ones with equal 1 are those with pixels active multiple times. In that case one often sees events with O(50) hits, but only a single pixel is actually active!- …
32.2.1. TODO add plots for:
- single pixel, multiple hits
- fraction in transverse equal 0
- ToA of all data
- ToA length
32.3. Comparison plots background
The background rate currently has a large increase at low energies. Much more so than CAST data.
To better understand, some comparison plots between Tpx3 background and CAST background.
Generated the plots with:
plotData --h5file /t/reco_tpx3_background.h5 \ --runType rtBackground --chips 0 --chips 3 \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --applyAllCuts --h5Compare ~/CastData/data/DataRuns2018_Reco.h5 --ingrid
with the following cuts and masked regions defined in the config TOML file:
[Cuts] cuts = [1] [Cuts.1] applyFile = ["reco_tpx3_background.h5"] applyDset = [] # if any given, apply this cut to all plots of the datasets in this array dset = "toaLength" # dataset the cut applies to min = 0.0 # minimum allowed value max = 20.0 # maximum allowed value [MaskRegions] # in theory this could also identify itself by chip name & run period, but that's a bit restrictive regions = [1, 2] [MaskRegions.1] applyFile = ["reco_tpx3_background.h5"] applyDset = [] x = [150, 250] # mask x range from <-> to y = [130, 162] # mask y range from <-> to [MaskRegions.2] applyFile = ["reco_tpx3_background.h5"] applyDset = [] x = [125, 135] # mask x range from <-> to y = [110, 120] # mask y range from <-> to
TODO: add plot from home!
33. Automatically generated run list
The following run list is created by the writeRunList tool:
./../../CastData/ExternCode/TimepixAnalysis/Tools/writeRunList/writeRunList.nim
based on the tracking logs.
| Run # | Type | DataType | Start | End | Length | # trackings | # frames | # FADC | Backup? | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| 76 | rtBackground | rfNewTos | 2 days 10:44 | 1 | 88249 | 19856 | y | |||
| 77 | rtBackground | rfNewTos | 1 days 00:03 | 1 | 36074 | 8016 | y | |||
| 78 | rtBackground | rfNewTos | 0 days 15:17 | 1 | 23506 | 5988 | y | |||
| 79 | rtBackground | rfNewTos | 1 days 03:22 | 1 | 40634 | 8102 | y | |||
| 80 | rtBackground | rfNewTos | 0 days 23:40 | 1 | 35147 | 6880 | y | |||
| 81 | rtBackground | rfNewTos | 1 days 00:06 | 1 | 35856 | 7283 | y | |||
| 82 | rtBackground | rfNewTos | 1 days 15:56 | 2 | 59502 | 12272 | y | |||
| 83 | rtCalibration | rfNewTos | 0 days 00:59 | 0 | 4915 | 4897 | y | |||
| 84 | rtBackground | rfNewTos | 1 days 01:11 | 1 | 37391 | 7551 | y | |||
| 85 | rtBackground | rfNewTos | 0 days 02:45 | 0 | 4104 | 899 | y | |||
| 86 | rtBackground | rfNewTos | 1 days 04:29 | 1 | 42396 | 9656 | y | |||
| 87 | rtBackground | rfNewTos | 1 days 12:11 | 2 | 54786 | 15123 | y | |||
| 88 | rtCalibration | rfNewTos | 0 days 00:59 | 0 | 4943 | 4934 | y | |||
| 89 | rtBackground | rfNewTos | 1 days 02:57 | 1 | 25209 | 6210 | y | |||
| 90 | rtBackground | rfNewTos | 1 days 01:09 | 1 | 37497 | 8122 | y | |||
| 91 | rtBackground | rfNewTos | 1 days 01:20 | 1 | 37732 | 8108 | y | |||
| 92 | rtBackground | rfNewTos | 1 days 21:32 | 1 | 67946 | 14730 | y | |||
| 93 | rtCalibration | rfNewTos | 0 days 01:00 | 0 | 4977 | 4968 | y | |||
| 94 | rtBackground | rfNewTos | 1 days 05:46 | 1 | 44344 | 9422 | y | |||
| 95 | rtBackground | rfNewTos | 4 days 08:06 | 1 | 154959 | 33112 | y | |||
| 96 | rtCalibration | rfNewTos | 0 days 07:01 | 0 | 34586 | 34496 | y | |||
| 97 | rtBackground | rfNewTos | 2 days 07:57 | 1 | 83404 | 18277 | y | |||
| 98 | rtBackground | rfNewTos | 0 days 19:36 | 1 | 29202 | 6285 | y | |||
| 99 | rtBackground | rfNewTos | 1 days 09:27 | 1 | 49921 | 10895 | y | |||
| 100 | rtBackground | rfNewTos | 0 days 23:53 | 1 | 35658 | 7841 | y | |||
| 101 | rtBackground | rfNewTos | 0 days 13:37 | 1 | 20326 | 4203 | y | |||
| 102 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9919 | 9898 | y | |||
| 103 | rtBackground | rfNewTos | 1 days 08:19 | 1 | 47381 | 7867 | y | |||
| 104 | rtBackground | rfNewTos | 1 days 00:00 | 1 | 35220 | 5866 | y | |||
| 105 | rtBackground | rfNewTos | 0 days 23:51 | 1 | 34918 | 5794 | y | |||
| 106 | rtBackground | rfNewTos | 1 days 00:14 | 1 | 35576 | 6018 | y | |||
| 107 | rtBackground | rfNewTos | 0 days 06:44 | 1 | 9883 | 1641 | y | |||
| 108 | rtCalibration | rfNewTos | 0 days 04:00 | 0 | 19503 | 19448 | y | |||
| 109 | rtBackground | rfNewTos | 0 days 17:32 | 1 | 28402 | 8217 | y | |||
| 110 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9804 | 9786 | y | |||
| 111 | rtBackground | rfNewTos | 0 days 02:53 | 0 | 4244 | 644 | y | |||
| 112 | rtBackground | rfNewTos | 3 days 15:55 | 2 | 128931 | 19607 | y | |||
| 113 | rtBackground | rfNewTos | 1 days 00:03 | 1 | 35100 | 5174 | y | |||
| 114 | rtBackground | rfNewTos | 0 days 11:43 | 1 | 17111 | 2542 | y | |||
| 115 | rtBackground | rfNewTos | 1 days 02:21 | 1 | 40574 | 9409 | y | |||
| 116 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9741 | 9724 | y | |||
| 117 | rtBackground | rfNewTos | 0 days 21:33 | 1 | 31885 | 5599 | y | |||
| 118 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9771 | 9748 | y | |||
| 119 | rtBackground | rfNewTos | 0 days 16:57 | 1 | 25434 | 4903 | y | |||
| 120 | rtCalibration | rfNewTos | 0 days 04:00 | 0 | 19308 | 19261 | y | |||
| 121 | rtBackground | rfNewTos | 0 days 22:14 | 1 | 33901 | 6947 | y | |||
| 122 | rtCalibration | rfNewTos | 0 days 05:57 | 0 | 29279 | 29208 | y | |||
| 123 | rtBackground | rfNewTos | 0 days 23:45 | 1 | 34107 | 3380 | y | |||
| 124 | rtBackground | rfNewTos | 2 days 01:50 | 2 | 71703 | 7504 | y | |||
| 125 | rtBackground | rfNewTos | 0 days 13:22 | 1 | 19262 | 1991 | y | |||
| 126 | rtCalibration | rfNewTos | 0 days 02:59 | 0 | 14729 | 14689 | y | |||
| 127 | rtBackground | rfNewTos | 2 days 04:50 | 1 | 75907 | 7663 | y | |||
| 128 | rtCalibration | rfNewTos | 0 days 09:05 | 0 | 44806 | 44709 | y | |||
| 145 | rtCalibration | rfNewTos | 0 days 03:22 | 0 | 16797 | 16796 | y | |||
| 146 | rtBackground | rfNewTos | 0 days 21:30 | 1 | 32705 | 3054 | y | |||
| 147 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 10102 | 10102 | y | |||
| 148 | rtBackground | rfNewTos | 0 days 20:37 | 1 | 31433 | 3120 | y | |||
| 149 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9975 | 9975 | y | |||
| 150 | rtBackground | rfNewTos | 0 days 21:42 | 1 | 33192 | 3546 | y | |||
| 151 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9907 | 9907 | y | |||
| 152 | rtBackground | rfNewTos | 0 days 20:10 | 1 | 30809 | 3319 | y | |||
| 153 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 10103 | 10102 | y | |||
| 154 | rtBackground | rfNewTos | 0 days 20:12 | 1 | 30891 | 3426 | y | |||
| 155 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9861 | 9861 | y | |||
| 156 | rtBackground | rfNewTos | 0 days 11:35 | 1 | 17686 | 1866 | y | |||
| 157 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9962 | 9962 | y | |||
| 158 | rtBackground | rfNewTos | 2 days 13:03 | 1 | 93205 | 9893 | y | |||
| 159 | rtCalibration | rfNewTos | 0 days 04:00 | 0 | 19879 | 19878 | y | |||
| 160 | rtBackground | rfNewTos | 2 days 19:28 | 1 | 103145 | 11415 | y | |||
| 161 | rtCalibration | rfNewTos | 0 days 04:00 | 0 | 19944 | 19943 | y | |||
| 162 | rtBackground | rfNewTos | 3 days 03:08 | 3 | 114590 | 11897 | y | |||
| 163 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 10093 | 10093 | y | |||
| 164 | rtBackground | rfNewTos | 1 days 20:18 | 2 | 67456 | 6488 | y | |||
| 165 | rtCalibration | rfNewTos | 0 days 04:00 | 0 | 19882 | 19879 | y | |||
| 166 | rtBackground | rfNewTos | 0 days 17:38 | 1 | 26859 | 2565 | y | |||
| 167 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9938 | 9938 | y | |||
| 168 | rtBackground | rfNewTos | 5 days 20:16 | 0 | 213545 | 20669 | y | |||
| 169 | rtCalibration | rfNewTos | 0 days 06:00 | 0 | 29874 | 29874 | y | |||
| 170 | rtBackground | rfNewTos | 0 days 21:42 | 1 | 33098 | 3269 | y | |||
| 171 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9999 | 9999 | y | |||
| 172 | rtBackground | rfNewTos | 0 days 18:50 | 1 | 28649 | 2773 | y | |||
| 173 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9898 | 9897 | y | |||
| 174 | rtBackground | rfNewTos | 0 days 19:48 | 1 | 30163 | 2961 | y | |||
| 175 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 10075 | 10075 | y | |||
| 176 | rtBackground | rfNewTos | 1 days 02:19 | 1 | 40084 | 3815 | y | |||
| 177 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9967 | 9966 | y | |||
| 178 | rtBackground | rfNewTos | 4 days 18:09 | 5 | 174074 | 17949 | y | |||
| 179 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9887 | 9887 | y | |||
| 180 | rtBackground | rfNewTos | 1 days 21:22 | 1 | 69224 | 7423 | y | |||
| 181 | rtCalibration | rfNewTos | 0 days 04:00 | 0 | 20037 | 20036 | y | |||
| 182 | rtBackground | rfNewTos | 1 days 19:14 | 2 | 65888 | 6694 | y | |||
| 183 | rtCalibration | rfNewTos | 0 days 03:59 | 0 | 20026 | 20026 | y | |||
| 184 | rtBackground | rfNewTos | 3 days 13:45 | 0 | 130576 | 12883 | y | |||
| 185 | rtCalibration | rfNewTos | 0 days 03:59 | 0 | 19901 | 19901 | y | |||
| 186 | rtBackground | rfNewTos | 11 days 21:00 | 0 | 434087 | 42830 | y | |||
| 187 | rtCalibration | rfNewTos | 0 days 04:00 | 0 | 19667 | 19665 | y | |||
| 188 | rtBackground | rfNewTos | 5 days 14:00 | 0 | 204281 | 20781 | y | |||
| 239 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9565 | 9518 | y | |||
| 240 | rtBackground | rfNewTos | 1 days 01:21 | 1 | 38753 | 4203 | y | |||
| 241 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9480 | 9426 | y | |||
| 242 | rtBackground | rfNewTos | 1 days 03:24 | 1 | 41933 | 4843 | y | |||
| 243 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9488 | 9429 | y | |||
| 244 | rtBackground | rfNewTos | 0 days 18:52 | 1 | 28870 | 3317 | y | |||
| 245 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9573 | 9530 | y | |||
| 246 | rtBackground | rfNewTos | 0 days 18:18 | 1 | 27970 | 2987 | y | |||
| 247 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9389 | 9334 | y | |||
| 248 | rtBackground | rfNewTos | 1 days 04:04 | 1 | 42871 | 4544 | y | |||
| 249 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9473 | 9431 | y | |||
| 250 | rtBackground | rfNewTos | 0 days 20:54 | 1 | 31961 | 3552 | y | |||
| 251 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9551 | 9503 | y | |||
| 253 | rtCalibration | rfNewTos | 0 days 02:20 | 0 | 11095 | 11028 | y | |||
| 254 | rtBackground | rfNewTos | 1 days 01:23 | 1 | 38991 | 4990 | y | |||
| 255 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9378 | 9330 | y | |||
| 256 | rtBackground | rfNewTos | 1 days 20:29 | 1 | 68315 | 8769 | y | |||
| 257 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9648 | 9592 | y | |||
| 258 | rtBackground | rfNewTos | 0 days 15:55 | 1 | 24454 | 3103 | y | |||
| 259 | rtCalibration | rfNewTos | 0 days 01:14 | 0 | 5900 | 5864 | y | |||
| 260 | rtCalibration | rfNewTos | 0 days 01:30 | 0 | 7281 | 7251 | y | |||
| 261 | rtBackground | rfNewTos | 2 days 19:43 | 3 | 103658 | 12126 | y | |||
| 262 | rtCalibration | rfNewTos | 0 days 05:59 | 0 | 28810 | 28681 | y | |||
| 263 | rtBackground | rfNewTos | 0 days 19:52 | 1 | 30428 | 3610 | y | |||
| 264 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9595 | 9544 | y | |||
| 265 | rtBackground | rfNewTos | 1 days 23:21 | 1 | 72514 | 8429 | y | |||
| 266 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9555 | 9506 | y | |||
| 267 | rtBackground | rfNewTos | 0 days 04:48 | 0 | 7393 | 929 | y | |||
| 268 | rtBackground | rfNewTos | 0 days 11:04 | 1 | 16947 | 1974 | y | |||
| 269 | rtCalibration | rfNewTos | 0 days 04:00 | 0 | 19382 | 19302 | y | |||
| 270 | rtBackground | rfNewTos | 1 days 23:34 | 2 | 72756 | 8078 | y | |||
| 271 | rtCalibration | rfNewTos | 0 days 02:43 | 0 | 13015 | 12944 | y | |||
| 272 | rtBackground | rfNewTos | 2 days 18:58 | 3 | 102360 | 11336 | y | |||
| 273 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9535 | 9471 | y | |||
| 274 | rtBackground | rfNewTos | 2 days 20:45 | 3 | 105187 | 12101 | y | |||
| 275 | rtCalibration | rfNewTos | 0 days 02:43 | 0 | 13179 | 13116 | y | |||
| 276 | rtBackground | rfNewTos | 4 days 04:17 | 2 | 153954 | 19640 | y | |||
| 277 | rtCalibration | rfNewTos | 0 days 13:48 | 0 | 66052 | 65749 | y | |||
| 278 | rtBackground | rfNewTos | 0 days 18:36 | 0 | 28164 | 3535 | y | |||
| 279 | rtBackground | rfNewTos | 2 days 04:07 | 2 | 79848 | 9677 | y | |||
| 280 | rtCalibration | rfNewTos | 0 days 04:00 | 0 | 19189 | 19112 | y | |||
| 281 | rtBackground | rfNewTos | 1 days 23:04 | 1 | 72230 | 8860 | y | |||
| 282 | rtCalibration | rfNewTos | 0 days 02:43 | 0 | 12924 | 12860 | y | |||
| 283 | rtBackground | rfNewTos | 2 days 16:07 | 3 | 98246 | 11965 | y | |||
| 284 | rtCalibration | rfNewTos | 0 days 03:59 | 0 | 19017 | 18904 | y | |||
| 285 | rtBackground | rfNewTos | 2 days 00:33 | 2 | 74405 | 8887 | y | |||
| 286 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9766 | 9715 | y | |||
| 287 | rtBackground | rfNewTos | 0 days 20:01 | 1 | 30598 | 3393 | y | |||
| 288 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9495 | 9443 | y | |||
| 289 | rtBackground | rfNewTos | 0 days 20:03 | 1 | 30629 | 3269 | y | |||
| 290 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9457 | 9394 | y | |||
| 291 | rtBackground | rfNewTos | 1 days 14:24 | 2 | 58602 | 6133 | y | |||
| 292 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9475 | 9426 | y | |||
| 293 | rtBackground | rfNewTos | 2 days 04:07 | 1 | 79677 | 8850 | y | |||
| 294 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9514 | 9467 | y | |||
| 295 | rtBackground | rfNewTos | 0 days 19:37 | 1 | 29981 | 3271 | y | |||
| 296 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9565 | 9517 | y | |||
| 297 | rtBackground | rfNewTos | 1 days 18:15 | 2 | 68124 | 12530 | y | |||
| 298 | rtBackground | rfNewTos | 1 days 12:01 | 1 | 53497 | 0 | y | |||
| 299 | rtBackground | rfNewTos | 0 days 11:29 | 1 | 17061 | 0 | y | |||
| 300 | rtCalibration | rfNewTos | 0 days 02:00 | 0 | 9466 | 9415 | y | |||
| 301 | rtBackground | rfNewTos | 1 days 16:43 | 2 | 62454 | 7751 | y | |||
| 302 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9616 | 9577 | y | |||
| 303 | rtBackground | rfNewTos | 0 days 23:48 | 1 | 36583 | 4571 | y | |||
| 304 | rtCalibration | rfNewTos | 0 days 01:59 | 0 | 9531 | 9465 | y | |||
| 306 | rtBackground | rfNewTos | 0 days 04:58 | 1 | 7546 | 495 | y |