1. Journal (day to day) extended
1.1. [1/1]
[X]arraymancer NN DSL PR
1.2. [3/5]
[X]write mail to CAST PC about talk at Patras.[ ]implement MCMC multiple chains starting & mean value of them for limit calculation[ ]fix segfault when multithreading[X]compute correct "depth" for raytracing focal spot[X]split window strongback from signal & position uncertainty Implemented the axion image w/o window & strongback separately. Still have to be implemented into limit calc.
1.3. [2/4]
[X]implement MCMC multiple chains starting & mean value of them for limit calculation[ ]fix segfault when multithreading[X]implement strongback / signal split into limit calc[ ]Timepix3 background rate!
1.4.
Questions for meeting with Klaus today:
- Did you hear something from Igor? -> Nope he hasn't either. Apparently Igor is very busy currently. But Klaus doesn't think there will be any showstoppers regarding making the data available.
- For reference distributions and logL morphing:
We morph bin wise on pre-binned data. This leads to jumps in the logL cut value.
Maybe a good idea after all not use a histogram, but a smooth KDE? Unbinned is not directly possible, because we don't have data to compute an unbinned distribution for everything outside main fluorescence lines! -> Klaus had a good idea here: We can estimate the systematic effect of our binning by moving the bin edges by half a bin width to the left / right and computing the expected limit based on these. If the effective limit changes, we know there is some systematic effect going on. More likely though, the expected limit remains unchanged (within variance) and therefore the systematic impact is smaller than the variance of the limit.
About septem veto and line veto: What to do with random coincidences? Is it honest to use those clusters? -> Klaus had an even better idea here: we can estimate the dead time by doing the following:
- read full septemboard data
- shuffle center + outer chip event data around such that we know the two are not correlated
- compute the efficiency of the septem veto.
In theory 0% of all events should trigger either the septem or the line veto. The percentage that does anyway is our random coincidence!
1.5.
All files that were in /tmp/playground (/t/playground) referenced
here and in the meeting notes are backed up in
~/development_files/07_03_2023/playground
(to not make sure we lose something / for reference to recreate some
in development behavior etc.)
Just because I'll likely shut down the computer for the first time in 26 days soon and not sure if everything was backed up from there. I believe so, but who knows.
1.6.
Let's rerun the likelihood after adding the tracking information
back to the H5 files and fixing how the total duration is calculated
from the data files.
Previously we used the total duration in every case, even when excluding tracking information and thus having less time in actuality. 'Fortunately', all background rate plots in the thesis as of today ran without any tracking info in the H5 files, meaning they include the solar tracking itself. Therefore the total duration is correct in those cases.
Run-2 testing (all vetoes):
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out /tmp/playground/test_run2.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --lineveto --scintiveto --fadcveto --septemveto \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5
Run-3 testing (all vetoes):
likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/playground/test_run3.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --lineveto --scintiveto --fadcveto --septemveto \ --calibFile ~/CastData/data/CalibrationRuns2018_Reco.h5
The likelihood outputs are here: ./resources/background_rate_test_correct_time_no_tracking/
Background:
plotBackgroundRate \ /tmp/playground/test_run2.h5 \ /tmp/playground/test_run3.h5 \ --combName 2017/18 \ --combYear 2017 \ --centerChip 3 \ --title "Background rate from CAST data, all vetoes" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_crGold_all_vetoes.pdf \ --outpath /tmp/playground/ \ --quiet
The number coming out (see title of the generated plot) is now 3158.57 h, which matches our (new :( ) expectation.
which is also in the same directory: ./resources/background_rate_test_correct_time_no_tracking/background_rate_crGold_all_vetoes.pdf
[X]RerunwriteRunListand update thestatusAndProgressandthesistables about times![ ]Update data in
thesis! Run-2:./writeRunList -b ~/CastData/data/DataRuns2017_Reco.h5 -c ~/CastData/data/CalibrationRuns2017_Reco.h5
Type: rtBackground total duration: 14 weeks, 6 days, 11 hours, 25 minutes, 59 seconds, 97 milliseconds, 615 microseconds, and 921 nanoseconds In hours: 2507.433082670833 active duration: 2238.783333333333 trackingDuration: 4 days, 10 hours, and 20 seconds In hours: 106.0055555555556 active tracking duration: 94.12276972527778 nonTrackingDuration: 14 weeks, 2 days, 1 hour, 25 minutes, 39 seconds, 97 milliseconds, 615 microseconds, and 921 nanoseconds In hours: 2401.427527115278 active background duration: 2144.666241943055
Solar tracking [h] Background [h] Active tracking [h] Active background [h] Total time [h] Active time [h] 106.006 2401.43 94.1228 2144.67 2507.43 2238.78 Type: rtCalibration total duration: 4 days, 11 hours, 25 minutes, 20 seconds, 453 milliseconds, 596 microseconds, and 104 nanoseconds In hours: 107.4223482211111 active duration: 2.601388888888889 trackingDuration: 0 nanoseconds In hours: 0.0 active tracking duration: 0.0 nonTrackingDuration: 4 days, 11 hours, 25 minutes, 20 seconds, 453 milliseconds, 596 microseconds, and 104 nanoseconds In hours: 107.4223482211111 active background duration: 2.601391883888889
Solar tracking [h] Background [h] Active tracking [h] Active background [h] Total time [h] Active time [h] 0 107.422 0 2.60139 107.422 2.60139 Run-3:
./writeRunList -b ~/CastData/data/DataRuns2018_Reco.h5 -c ~/CastData/data/CalibrationRuns2018_Reco.h5
Type: rtBackground total duration: 7 weeks, 23 hours, 13 minutes, 35 seconds, 698 milliseconds, 399 microseconds, and 775 nanoseconds In hours: 1199.226582888611 active duration: 1079.598333333333 trackingDuration: 3 days, 2 hours, 17 minutes, and 53 seconds In hours: 74.29805555555555 active tracking duration: 66.92306679361111 nonTrackingDuration: 6 weeks, 4 days, 20 hours, 55 minutes, 42 seconds, 698 milliseconds, 399 microseconds, and 775 nanoseconds In hours: 1124.928527333056 active background duration: 1012.677445774444
Solar tracking [h] Background [h] Active tracking [h] Active background [h] Total time [h] Active time [h] 74.2981 1124.93 66.9231 1012.68 1199.23 1079.6 Type: rtCalibration total duration: 3 days, 15 hours, 3 minutes, 47 seconds, 557 milliseconds, 131 microseconds, and 279 nanoseconds In hours: 87.06321031416667 active duration: 3.525555555555556 trackingDuration: 0 nanoseconds In hours: 0.0 active tracking duration: 0.0 nonTrackingDuration: 3 days, 15 hours, 3 minutes, 47 seconds, 557 milliseconds, 131 microseconds, and 279 nanoseconds In hours: 87.06321031416667 active background duration: 3.525561761944445
Solar tracking [h] Background [h] Active tracking [h] Active background [h] Total time [h] Active time [h] 0 87.0632 0 3.52556 87.0632 3.52556
[X]Rerun the
createAllLikelihoodCombinationsnow that tracking information is there. -> Currently running (Update: We could now combine the below with the one further down that excludes the FADC!)../createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crGold --regions crAll \ --vetoSets "{fkScinti, fkFadc, fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --fadcVetoPercentiles 0.9 --fadcVetoPercentiles 0.95 --fadcVetoPercentiles 0.99 \ --out /t/lhood_outputs_adaptive_fadc \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing
Found here: ./resources/lhood_limits_automation_correct_duration/
[ ]Now generate the other likelihood outputs we need for more expected limit cases from sec. [BROKEN LINK: sec:meetings:10_03_23] in StatusAndProgress:
[ ]Calculate expected limits also for the following cases:[X]Septem, line combinations without the FADC[ ]Best case (lowest row of below) with lnL efficiencies of:[ ]0.7[ ]0.9
The former (septem, line without FADC) will be done using
createAllLikelihoodCombinations:./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crGold --regions crAll \ --vetoSets "{+fkScinti, fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --out /t/lhood_outputs_adaptive_fadc \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing
For simplicity, this will regenerate some of the files already generated (i.e. the no vetoes & the scinti case)
These files are also found here: ./resources/lhood_limits_automation_correct_duration/ together with a rerun of the regular cases above.
Plot the background clusters to see if we indeed have less over the whole chip.
plotBackgroundClusters \ /t/lhood_outputs_adaptive_duplicated_fadc_stuff/likelihood_cdl2018_Run2_crAll_scinti_vetoPercentile_0.99_fadc_vetoPercentile_0.99_septem_vetoPercentile_0.99_line_vetoPercentile_0.99.h5 \ /t/lhood_outputs_adaptive_duplicated_fadc_stuff/likelihood_cdl2018_Run3_crAll_scinti_vetoPercentile_0.99_fadc_vetoPercentile_0.99_septem_vetoPercentile_0.99_line_vetoPercentile_0.99.h5 \ --zMax 5 \ --title "X-ray like clusters of CAST data after all vetoes" \ --outpath /tmp/playground/ \ --filterNoisyPixels \ --filterEnergy 12.0 \ --suffix "_all_vetoes"
Available here: resources/background_rate_test_correct_time_no_tracking/background_cluster_centers_all_vetoes.pdf
Where we can see that indeed we now have less than 10,000 clusters compared to the ~10,500 we had when using all data (including tracking).
1.7.
Continue from yesterday:
[X]Now generate the other likelihood outputs we need for more expected limit cases from sec. [BROKEN LINK: sec:meetings:10_03_23] in StatusAndProgress: -> All done, path to files below.
[X]Calculate expected limits also for the following cases:[X]Septem, line combinations without the FADC (done yesterday)[X]Best case (lowest row of below) with lnL efficiencies of:[X]0.7[X]0.9
The latter has now also been implemented as functionality in
likelihoodandcreateAllLikelihoodCombinations(adjust signal efficiency from command line and add options to runner). Now run:./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crGold --regions crAll \ --signalEfficiency 0.7 --signalEfficiency 0.9 \ --vetoSets "{+fkScinti, +fkFadc, +fkSeptem, fkLineVeto}" \ --fadcVetoPercentile 0.9 \ --out /t/lhood_outputs_adaptive_fadc \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --dryRun
to reproduce numbers for the best expected limit case together with a lnL signal efficiency of 70 and 90%.
Finally, these files are also in: ./resources/lhood_limits_automation_correct_duration/ which means now all the setups we initially care about are there.
Let's look at the background rate we get from the 70% vs the 90% case:
plotBackgroundRate \ likelihood_cdl2018_Run2_crGold_signalEff_0.7_scinti_fadc_septem_line_vetoPercentile_0.9.h5 \ likelihood_cdl2018_Run3_crGold_signalEff_0.7_scinti_fadc_septem_line_vetoPercentile_0.9.h5 \ likelihood_cdl2018_Run2_crGold_signalEff_0.9_scinti_fadc_septem_line_vetoPercentile_0.9.h5 \ likelihood_cdl2018_Run3_crGold_signalEff_0.9_scinti_fadc_septem_line_vetoPercentile_0.9.h5 \ --names "0.7" --names "0.7" \ --names "0.9" --names "0.9" \ --centerChip 3 \ --title "Background rate from CAST data, incl. all vetoes, 70vs90" \ --showNumClusters --showTotalTime \ --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_cast_all_vetoes_70p_90p.pdf \ --outpath . \ --quiet
[INFO]:Dataset: 0.7 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 4.1861e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 3.4884e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.9 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 9.3221e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 7.7684e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.7 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 8.6185e-06 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 4.3093e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.9 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 1.4775e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 7.3873e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.7 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.8116e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 4.0259e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.9 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 3.4650e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 7.7000e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.7 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 1.4423e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 5.7691e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.9 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 2.4273e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 9.7090e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.7 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 4.3972e-06 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 1.0993e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.9 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 1.1785e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 2.9461e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.7 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 2.7790e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 3.4738e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.9 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 5.3998e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 6.7497e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.7 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 1.3895e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 2.3159e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: 0.9 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 3.0956e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 5.1594e-06 keV⁻¹·cm⁻²·s⁻¹
yielded
which shows quite the incredibly change especially in the 8 keV peak!
And in the 4 to 8 keV range we even almost got to the 1e-7 range for the 70% case (however note that in this case the total efficiency is only about 40% or so!).
[X]Verify that the signal efficiency used is written to output logL file[X]If not implement -> It was not, now implemented.
[X]read signal efficiency from logL file in mcmclimit and stop using the efficiency includingεin the context. Instead merge with the calculator for veto efficiencies. -> Implemented.
From meeting notes:
[ ]Verify that those elements with lower efficiency indeed have \(R_T = 0\) at higher values! -> Just compute \(R_T = 0\) for all input files and output result, easiest.
1.8.
Old model from March 2022 ./resources/mlp_trained_march2022.pt :
Test set: Average loss: 0.9876 | Accuracy: 0.988 Cut value: 2.483305978775025 Test set: Average loss: 0.9876 | Accuracy: 0.988 Total efficiency = 0.8999892098945267 Test set: Average loss: 0.9995 | Accuracy: 0.999 Target: Ag-Ag-6kV eff = 0.9778990694345026 Test set: Average loss: 0.9916 | Accuracy: 0.992 Target: Al-Al-4kV eff = 0.9226669690441093 Test set: Average loss: 0.9402 | Accuracy: 0.940 Target: C-EPIC-0.6kV eff = 0.6790938280413843 Test set: Average loss: 0.9941 | Accuracy: 0.994 Target: Cu-EPIC-0.9kV eff = 0.8284986713906112 Test set: Average loss: 0.9871 | Accuracy: 0.987 Target: Cu-EPIC-2kV eff = 0.8687534321801208 Test set: Average loss: 1.0000 | Accuracy: 1.000 Target: Cu-Ni-15kV eff = 0.9939449541284404 Test set: Average loss: 0.9999 | Accuracy: 1.000 Target: Mn-Cr-12kV eff = 0.9938112429087158 Test set: Average loss: 1.0000 | Accuracy: 1.000 Target: Ti-Ti-9kV eff = 0.9947166683932456
New model from yesterday ./resources/mlp_trained_bsz8192_hidden_5000.pt :
Test set: Average loss: 0.9714 | Accuracy: 0.971 Cut value: 1.847297704219818 Test set: Average loss: 0.9714 | Accuracy: 0.971 Total efficiency = 0.8999892098945267 Test set: Average loss: 0.9945 | Accuracy: 0.994 Target: Ag-Ag-6kV eff = 0.9525769506084467 Test set: Average loss: 0.9804 | Accuracy: 0.980 Target: Al-Al-4kV eff = 0.9097403333711305 Test set: Average loss: 0.8990 | Accuracy: 0.899 Target: C-EPIC-0.6kV eff = 0.7640920442383161 Test set: Average loss: 0.9584 | Accuracy: 0.958 Target: Cu-EPIC-0.9kV eff = 0.8211913197519929 Test set: Average loss: 0.9636 | Accuracy: 0.964 Target: Cu-EPIC-2kV eff = 0.8543657331136738 Test set: Average loss: 0.9980 | Accuracy: 0.998 Target: Cu-Ni-15kV eff = 0.981651376146789 Test set: Average loss: 0.9978 | Accuracy: 0.998 Target: Mn-Cr-12kV eff = 0.9807117070654977 Test set: Average loss: 0.9982 | Accuracy: 0.998 Target: Ti-Ti-9kV eff = 0.9776235367243344
Calculated with determineCdlEfficiency.
To get a background rate estimate we use the simple functionality in
the NN training tool itself. Note that it needs the total time as an
input to scale the data correctly (hardcoded has a value of Run-2, but
for Run-3 need to use the totalTime argument!)
Background rate at 95% for Run-2 data:
cd ~/CastData/ExternCode/TimepixAnalysis/Tools/NN_playground
./train_ingrid -f ~/CastData/data/DataRuns2017_Reco.h5 --ε 0.95
which yields:
Background rate at 90% for Run-2 data:
cd ~/CastData/ExternCode/TimepixAnalysis/Tools/NN_playground
./train_ingrid -f ~/CastData/data/DataRuns2017_Reco.h5 --ε 0.9
Test set: Average loss: 0.9714 | Accuracy: 0.971 Cut value: 1.847297704219818
Test set: Average loss: 0.9714 | Accuracy: 0.971 Total efficiency = 0.8999892098945267
Test set: Average loss: 0.9945 | Accuracy: 0.994 Target: Ag-Ag-6kV eff = 0.9525769506084467
Test set: Average loss: 0.9804 | Accuracy: 0.980 Target: Al-Al-4kV eff = 0.9097403333711305
Test set: Average loss: 0.8990 | Accuracy: 0.899 Target: C-EPIC-0.6kV eff = 0.7640920442383161
Test set: Average loss: 0.9584 | Accuracy: 0.958 Target: Cu-EPIC-0.9kV eff = 0.8211913197519929
Test set: Average loss: 0.9636 | Accuracy: 0.964 Target: Cu-EPIC-2kV eff = 0.8543657331136738
Test set: Average loss: 0.9980 | Accuracy: 0.998 Target: Cu-Ni-15kV eff = 0.981651376146789
Test set: Average loss: 0.9978 | Accuracy: 0.998 Target: Mn-Cr-12kV eff = 0.9807117070654977
Test set: Average loss: 0.9982 | Accuracy: 0.998 Target: Ti-Ti-9kV eff = 0.9776235367243344
which yields:
Background rate at 80% for Run-2 data:
cd ~/CastData/ExternCode/TimepixAnalysis/Tools/NN_playground
./train_ingrid -f ~/CastData/data/DataRuns2017_Reco.h5 --ε 0.8
Test set: Average loss: 0.9714 | Accuracy: 0.971 Cut value: 3.556154251098633
Test set: Average loss: 0.9714 | Accuracy: 0.971 Total efficiency = 0.799991907420895
Test set: Average loss: 0.9945 | Accuracy: 0.994 Target: Ag-Ag-6kV eff = 0.856209735146743
Test set: Average loss: 0.9804 | Accuracy: 0.980 Target: Al-Al-4kV eff = 0.7955550515931511
Test set: Average loss: 0.8990 | Accuracy: 0.899 Target: C-EPIC-0.6kV eff = 0.6546557260078487
Test set: Average loss: 0.9584 | Accuracy: 0.958 Target: Cu-EPIC-0.9kV eff = 0.7030558015943313
Test set: Average loss: 0.9636 | Accuracy: 0.964 Target: Cu-EPIC-2kV eff = 0.7335529928610653
Test set: Average loss: 0.9980 | Accuracy: 0.998 Target: Cu-Ni-15kV eff = 0.9102752293577981
Test set: Average loss: 0.9978 | Accuracy: 0.998 Target: Mn-Cr-12kV eff = 0.9036616812790098
Test set: Average loss: 0.9982 | Accuracy: 0.998 Target: Ti-Ti-9kV eff = 0.8947477468144618
which yields:
Background rate at 70% for Run-2 data:
cd ~/CastData/ExternCode/TimepixAnalysis/Tools/NN_playground
./train_ingrid -f ~/CastData/data/DataRuns2017_Reco.h5 --ε 0.7
Test set: Average loss: 0.9714 | Accuracy: 0.971 Cut value: 5.098100709915161
Test set: Average loss: 0.9714 | Accuracy: 0.971 Total efficiency = 0.6999946049472634
Test set: Average loss: 0.9945 | Accuracy: 0.994 Target: Ag-Ag-6kV eff = 0.7380100214745884
Test set: Average loss: 0.9804 | Accuracy: 0.980 Target: Al-Al-4kV eff = 0.6931624900782402
Test set: Average loss: 0.8990 | Accuracy: 0.899 Target: C-EPIC-0.6kV eff = 0.5882090617195861
Test set: Average loss: 0.9584 | Accuracy: 0.958 Target: Cu-EPIC-0.9kV eff = 0.6312001771479185
Test set: Average loss: 0.9636 | Accuracy: 0.964 Target: Cu-EPIC-2kV eff = 0.6507413509060955
Test set: Average loss: 0.9980 | Accuracy: 0.998 Target: Cu-Ni-15kV eff = 0.7844036697247706
Test set: Average loss: 0.9978 | Accuracy: 0.998 Target: Mn-Cr-12kV eff = 0.7790613718411552
Test set: Average loss: 0.9982 | Accuracy: 0.998 Target: Ti-Ti-9kV eff = 0.7758209882937946
which yields:
[X]Add background rates for new model using 95%, 90%, 80%[X]And the outputs as above for the local efficiencies
[ ]Implement selection of global vs. local target efficiency[ ]Check the efficiency we get when applying the model to the 55Fe calibration data (need same efficiency!). Cross check our helper program that does this for lnL method[ ]Check the background rate from the Run-3 data. Is it compatible? Or does it break down?
Practical:
[ ]Move the model logic over tolikelihood.nimas a replacement for thefkAggressiveveto[ ]including the selection of the target efficiency
[X]Clean up veto system inlikelihoodfor better insertion of NN[X]add lnL as a form of veto
[X]add vetoes for MLP and ConvNet (in principle)[ ]move NN code to main ingrid module[ ]make MLP / ConvNet types accessible inlikelihoodif compiled on cpp backend (and with CUDA?)[ ]add path to model file[ ]adjustCutValueInterpolatorto make it work for both lnL as well as NN. Idea is the same!
Questions:
[ ]Is there still a place for something like an equivalent for the likelihood morphing? In this case based on likely just interpolating the cut values?
1.9.
TODOs from yesterday:
[X]Add background rates for new model using 95%, 90%, 80%[X]And the outputs as above for the local efficiencies
[X]Implement selection of global vs. local target efficiency[X]Check the efficiency we get when applying the model to the 55Fe calibration data (need same efficiency!). Cross check our helper program that does this for lnL method -> Wroteeffective_eff_55fe.niminNN_playground-> Efficiency in 55Fe data is abysmal! ~40-55 % at 95% ![ ]Check the background rate from the Run-3 data. Is it compatible? Or does it break down?
Practical:
[ ]Move the model logic over tolikelihood.nimas a replacement for thefkAggressiveveto[ ]including the selection of the target efficiency
[X]Clean up veto system inlikelihoodfor better insertion of NN[X]add lnL as a form of veto
[X]add vetoes for MLP and ConvNet (in principle)[ ]move NN code to main ingrid module[X]make MLP / ConvNet types accessible inlikelihoodif compiled on cpp backend (and with CUDA?)[X]add path to model file[X]adjustCutValueInterpolatorto make it work for both lnL as well as NN. Idea is the same!
Questions:
[ ]Is there still a place for something like an equivalent for the likelihood morphing? In this case based on likely just interpolating the cut values?
With the refactor of likelihood we can now do things like disable
the lnL cut itself and only use the vetoes.
NOTE: All outputs below that were placed in /t/testing can be
found in ./resources/nn_testing_outputs/.
For example look at only using the FADC (with a much harsher cut than usual):
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out /tmp/testing/test_run2_only_fadc.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --fadcveto \ --vetoPercentile 0.75 \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5 plotBackgroundRate /t/testing/test_run2_only_fadc.h5 \ --combName "onlyFadc" \ --combYear 2017 \ --centerChip 3 \ --title "Background rate from CAST data, only FADC veto" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_fadc_veto.pdf \ --outpath /t/testing/ --quiet
The plot is here:
The issue is there's a couple of runs that have no FADC / in which the
FADC was extremely noisy, hence all we really see is the background distribution of those.
But for the more interesting stuff, let's try to create the background rate using the NN veto at 95% efficiency!:
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out /tmp/testing/test_run2_only_mlp_0.95.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/mlp_trained_bsz8192_hidden_5000.pt \ --nnSignalEff 0.95 \ --nnCutKind global \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5
plotBackgroundRate /t/testing/test_run2_only_mlp_0.95.h5 \ --combName "onlyMLP" \ --combYear 2017 \ --centerChip 3 \ --title "Background rate from CAST data, only MLP @ 95%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_mlp_0.95.pdf \ --outpath /t/testing/ --quiet
At 90% global efficiency:
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out /tmp/testing/test_run2_only_mlp_0.9.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/mlp_trained_bsz8192_hidden_5000.pt \ --nnSignalEff 0.9 \ --nnCutKind global \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5
plotBackgroundRate /t/testing/test_run2_only_mlp_0.9.h5 \ --combName "onlyMLP" \ --combYear 2017 \ --centerChip 3 \ --title "Background rate from CAST data, only MLP @ 90%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_mlp_0.9.pdf \ --outpath /t/testing/ --quiet
At 80% global efficiency:
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out /tmp/testing/test_run2_only_mlp_0.8.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/mlp_trained_bsz8192_hidden_5000.pt \ --nnSignalEff 0.8 \ --nnCutKind global \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5
plotBackgroundRate /t/testing/test_run2_only_mlp_0.8.h5 \ --combName "onlyMLP" \ --combYear 2017 \ --centerChip 3 \ --title "Background rate from CAST data, only MLP @ 90%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_mlp_0.8.pdf \ --outpath /t/testing/ --quiet
At 70% global efficiency:
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out /tmp/testing/test_run2_only_mlp_0.7.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/mlp_trained_bsz8192_hidden_5000.pt \ --nnSignalEff 0.7 \ --nnCutKind global \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5
plotBackgroundRate /t/testing/test_run2_only_mlp_0.7.h5 \ --combName "onlyMLP" \ --combYear 2017 \ --centerChip 3 \ --title "Background rate from CAST data, only MLP @ 90%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_mlp_0.7.pdf \ --outpath /t/testing/ --quiet
NOTE: Make sure to set neuralNetCutKind to local in the config file!
And local 95%:
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out /tmp/testing/test_run2_only_mlp_local_0.95.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/mlp_trained_bsz8192_hidden_5000.pt \ --nnSignalEff 0.95 \ --nnCutKind local \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5
plotBackgroundRate /t/testing/test_run2_only_mlp_local_0.95.h5 \ --combName "onlyMLP" \ --combYear 2017 \ --centerChip 3 \ --title "Background rate from CAST data, only MLP @ local 95%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_mlp_local_0.95.pdf \ --outpath /t/testing/ --quiet
And local 80%:
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out /tmp/testing/test_run2_only_mlp_local_0.8.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/mlp_trained_bsz8192_hidden_5000.pt \ --nnSignalEff 0.8 \ --nnCutKind local \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5
plotBackgroundRate /t/testing/test_run2_only_mlp_local_0.8.h5 \ --combName "onlyMLP" \ --combYear 2017 \ --centerChip 3 \ --title "Background rate from CAST data, only MLP @ local 80%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_mlp_local_0.8.pdf \ --outpath /t/testing/ --quiet
1.10.
Continue on from yesterday:
[ ]Implement 55Fe calibration data into the training process. E.g. add about 1000 events per calibration run to the training data as signal target to have a wider distribution of what real X-rays should look like. Hopefully that increases our efficiency![ ]It seems like only very few events pass the cuts in
readCalibData(e.g. what we use for effective efficiency check and in mixed data training). Why is that? Especially for escape peak often less than 300 events are valid! So little statistics, really? Looking at spectra, e.g. in~/CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/out/CalibrationRuns2018_Raw_2020-04-28_15-06-54there is really this little statistics in the escape peak often. (peaks at less than per bin 50!) How do these spectra look without any cuts? Are our cuts rubbish? Quick look:plotData --h5file ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --runType rtCalibration \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --region crSilver \ --ingrid --separateRuns
seems to support that there simply isn't much more statistics available!
[X]First training with a mixed set of data, using, per run:
- min(500, total) escape peak (after cuts)
- min(500, total) photo peak (after cuts)
- 6000 background
- all CDL
-> all of these are of course shuffled and then split into training and test datasets The resulting model is in: ./resources/nn_devel_mixing/trained_mlp_mixed_data.pt
The generated plots are in: ./Figs/statusAndProgress/neuralNetworks/development/mixing_data/
Looking at these figures we can see mainly that the ROC curve is extremely 'clean', but fitting for the separation seen in the training and validation output distributions.
[ ]effective efficiencies for 55Fe[ ]efficiencies of CDL data!
[X]make loss / accuracy curves log10[ ]Implement snapshots of the model during training whenever the training and test (or only test) accuracy improves
As discussed in the meeting today (sec. [BROKEN LINK: sec:meetings:17_03_23] in notes), let's rerun all expected limits and add the new two, namely:
[ ]redo all expected limit calculations with the following new cases:- 0.9 lnL + scinti + FADC@0.98 + line
- 0.8 lnL + scinti + FADC@0.98 + line εcut:
- 1.0, 1.2, 1.4, 1.6
The standard cases (lnL 80 + all veto combinations with different FADC settings):
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crGold --regions crAll \ --vetoSets "{fkLogL, fkScinti, fkFadc, fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --fadcVetoPercentiles 0.9 --fadcVetoPercentiles 0.95 --fadcVetoPercentiles 0.99 \ --out /t/lhood_outputs_adaptive_fadc \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing
The no septem veto + different lnL efficiencies:
[X]0.9 lnL + scinti + FADC@0.98 + line
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crGold --regions crAll \ --vetoSets "{+fkLogL, +fkScinti, fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --signalEfficiency 0.7 --signalEfficiency 0.9 \ --out /t/lhood_outputs_adaptive_fadc \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing
The older case of changing lnL efficiency with different lnL efficiency
[X]add a case with less extreme FADC veto
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crGold --regions crAll \ --signalEfficiency 0.7 --signalEfficiency 0.9 \ --vetoSets "{+fkLogL, +fkScinti, +fkFadc, +fkSeptem, fkLineVeto}" \ --fadcVetoPercentile 0.95 --fadcVetoPercentile 0.99 \ --out /t/lhood_outputs_adaptive_fadc \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing
And finally different eccentricity cutoffs for the line veto:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crGold --regions crAll \ --signalEfficiency 0.7 --signalEfficiency 0.8 --signalEfficiency 0.9 \ --vetoSets "{+fkLogL, +fkScinti, +fkFadc, fkLineVeto}" \ --eccentricityCutoff 1.0 --eccentricityCutoff 1.2 --eccentricityCutoff 1.4 --eccentricityCutoff 1.6 \ --out /t/lhood_outputs_adaptive_fadc \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing
The output H5 files will be placed in: ./resources/lhood_limits_automation_with_nn_support
1.11.
With the likelihood output files generated over night in
resources/lhood_limits_automation_with_nn_support
it's now time to let the limits run.
I noticed something else was missing from these files: I forgot to re
add the actual vetoes in use to the output (because those were written
manually).
[X]addflagstoLikelihoodContextto auto serialize them[X]updatemcmclimit code to use the new serialized data as veto efficiency and veto usage[X]rerun all limits with all the different setups.[X]updaterunLimitsto be smarter about what has been done. In principle we can now quit the limit calculation and it should continue automatically on a restart (with the last file worked on!)
The script we actually ran today. This will be part of the thesis (or a variation thereof).
#!/usr/bin/zsh cd ~/CastData/ExternCode/TimepixAnalysis/Analysis/ ./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crGold --regions crAll \ --vetoSets "{fkLogL, fkScinti, fkFadc, fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --fadcVetoPercentiles 0.9 --fadcVetoPercentiles 0.95 --fadcVetoPercentiles 0.99 \ --out /t/lhood_outputs_adaptive_fadc \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 12 ./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crGold --regions crAll \ --vetoSets "{+fkLogL, +fkScinti, fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --signalEfficiency 0.7 --signalEfficiency 0.9 \ --out /t/lhood_outputs_adaptive_fadc \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 12 ./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crGold --regions crAll \ --signalEfficiency 0.7 --signalEfficiency 0.9 \ --vetoSets "{+fkLogL, +fkScinti, +fkFadc, +fkSeptem, fkLineVeto}" \ --fadcVetoPercentile 0.95 --fadcVetoPercentile 0.99 \ --out /t/lhood_outputs_adaptive_fadc \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 12 ./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crGold --regions crAll \ --signalEfficiency 0.7 --signalEfficiency 0.8 --signalEfficiency 0.9 \ --vetoSets "{+fkLogL, +fkScinti, +fkFadc, fkLineVeto}" \ --eccentricityCutoff 1.0 --eccentricityCutoff 1.2 --eccentricityCutoff 1.4 --eccentricityCutoff 1.6 \ --out /t/lhood_outputs_adaptive_fadc \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 12
(all in ./resources/lhood_limits_automation_with_nn_support/)
And currently running:
./runLimits --path ~/org/resources/lhood_limits_automation_with_nn_support --nmc 1000
Train NN:
./train_ingrid ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --ε 0.95 \ --modelOutpath /tmp/trained_mlp_mixed_data.pt
NOTE: The one thing I just realized is: the accuracy we print is of course related to the actual prediction of the network, i.e. which of the two output neurons is the maximum value. So maybe our approach of only looking at one and adjusting based on that is just dumb and the network actually is much better than we think?
The numbers we see as accuracy actually make sense.
Consider predictBackground output:
Pred set: Average loss: 0.0169 | Accuracy: 0.9956 p inds len 1137 compared to all 260431
The 1137 clusters left after cuts correspond exactly to 99.56% (this is based on using the network's real prediction and not the output + cut value). The question here is: at what signal efficiency is this? From the CDL data it would seem to be at ~99%.
The network we trained today, including checkpoints is here: ./resources/nn_devel_mixing/18_03_23/
[X]Check what efficiency we get for calibration data instead of background -> Yeah, it is also at over 99% efficiency. So we get a 1e-5 background rate at 99% efficiency. At least that's not too bad.
:
The processed expected limit files are now here:
./resources/lhood_limits_automation_with_nn_support/limits/
with the logL output files in the lhood folder.
We'll continue on later with the processed.txt file as a guide there.
1.12.
The expected limits for
resources/lhood_limits_automation_with_nn_support/limits/ are
still running because our processed continuation check was incorrect
(looking at full path & not actually skipping files!).
Back to the NN: Let's look at the output of the network for both output neurons. Are they really essentially a mirror of one another?
predictAll in train_ingrid creates a plot of the different data
kinds (55Fe, CDL, background) and each neurons output prediction.
This yields the following plot by running:
./train_ingrid ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --ε 0.95 \ --modelOutpath ~/org/resources/nn_devel_mixing/18_03_23/trained_mlp_mixed_datacheckpoint_epoch_95000_loss_0.0117_acc_0.9974.pt \ --predict
where we can see the following points:
- the two neurons are almost perfect mirrors, but not exactly
- selecting the
argmaxof the two neurons gives us that neuron, which has a positive value almost certainly, due to the mirror nature around 0. It could be different (e.g. both neurons giving a positive or a negative value), but looking at the data this does not seem to happen (if then very rarely). - a cut value of
0should reproduce pretty exactly the standard neural network prediction of picking theargmax
Question: Can the usage of both neurons be beneficial given the small but existing differences in the distributions? Not sure how if so.
An earlier checkpoint (the one before the extreme jump in the loss
value based on the loss figure; need to regenerate it, but similar to
except as log10) yields the following neuron output:
./train_ingrid ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --ε 0.95 \ --modelOutpath ~/org/resources/nn_devel_mixing/18_03_23/trained_mlp_mixed_datacheckpoint_epoch_65000_loss_0.0103_acc_0.9977.pt \ --predict
We can clearly see that at this stage in training the two types of signal data are predicted quite differently! In that sense the latest model is actually much more like what we want, i.e. same prediction for all different kinds of X-rays!
What does the case with the worst loss look like?
./train_ingrid ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --ε 0.95 \ --modelOutpath ~/org/resources/nn_devel_mixing/18_03_23/trained_mlp_mixed_datacheckpoint_epoch_70000_loss_0.9683_acc_0.9977.pt \ --predict
Interestingly essentially the same. But the accuracy is the same as before, only the loss is different. Not sure why that might be.
Training the network again after the charge bug was fixed:
./train_ingrid ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --ε 0.95 \ --modelOutpath /t/nn_training/trained_model_charge_cut_bug_fixed.pt
which are stored here:
./resources/nn_devel_mixing/19_03_23_charge_bug_fixed/
with the generated plots:
./Figs/statusAndProgress/neuralNetworks/development/charge_cut_bug_fixed
Looking at the loss plot, at around epoch 83000 the training data
started to outpace the test data (test didn't get any worse though and
test accuracy improved slightly).
Also the ~all_prediction.pdf plot showing how CDL and 55Fe data is
predicted is interesting. The CDL data is skewed significantly more
to the right than 55Fe, explaining the again prevalent difference in
55Fe efficiency for a given CDL eff:
./effective_eff_55fe \ -f ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/19_03_23_charge_bug_fixed/trained_model_charge_cut_bug_fixedcheckpoint_epoch_100000_loss_0.0102_acc_0.9978.pt \ --ε 0.95
Run: 83 for target: signal Keeping : 759 of 916 = 0.8286026200873362
Run: 88 for target: signal Keeping : 763 of 911 = 0.8375411635565313
Run: 93 for target: signal Keeping : 640 of 787 = 0.8132147395171537
Run: 96 for target: signal Keeping : 4591 of 5635 = 0.8147293700088731
Run: 102 for target: signal Keeping : 1269 of 1588 = 0.7991183879093199
Run: 108 for target: signal Keeping : 2450 of 3055 = 0.8019639934533551
Run: 110 for target: signal Keeping : 1244 of 1554 = 0.8005148005148005
Run: 116 for target: signal Keeping : 1404 of 1717 = 0.8177052999417589
Run: 118 for target: signal Keeping : 1351 of 1651 = 0.8182919442761962
Run: 120 for target: signal Keeping : 2784 of 3413 = 0.8157046586580721
Run: 122 for target: signal Keeping : 4670 of 5640 = 0.8280141843971631
Run: 126 for target: signal Keeping : 2079 of 2596 = 0.8008474576271186
Run: 128 for target: signal Keeping : 6379 of 7899 = 0.8075705785542474
Run: 145 for target: signal Keeping : 2950 of 3646 = 0.8091058694459682
Run: 147 for target: signal Keeping : 1670 of 2107 = 0.7925961082107261
Run: 149 for target: signal Keeping : 1536 of 1936 = 0.7933884297520661
Run: 151 for target: signal Keeping : 1454 of 1839 = 0.790647090810223
Run: 153 for target: signal Keeping : 1515 of 1908 = 0.7940251572327044
Run: 155 for target: signal Keeping : 1386 of 1777 = 0.7799662352279122
Run: 157 for target: signal Keeping : 1395 of 1817 = 0.7677490368739681
Run: 159 for target: signal Keeping : 2805 of 3634 = 0.7718767198679142
Run: 161 for target: signal Keeping : 2825 of 3632 = 0.7778083700440529
Run: 163 for target: signal Keeping : 1437 of 1841 = 0.7805540467137425
Run: 165 for target: signal Keeping : 3071 of 3881 = 0.7912909044060809
Run: 167 for target: signal Keeping : 1557 of 2008 = 0.775398406374502
Run: 169 for target: signal Keeping : 4644 of 5828 = 0.7968428277282087
Run: 171 for target: signal Keeping : 1561 of 1956 = 0.7980572597137015
Run: 173 for target: signal Keeping : 1468 of 1820 = 0.8065934065934066
Run: 175 for target: signal Keeping : 1602 of 2015 = 0.7950372208436725
Run: 177 for target: signal Keeping : 1557 of 1955 = 0.7964194373401534
Run: 179 for target: signal Keeping : 1301 of 1671 = 0.7785757031717534
Run: 181 for target: signal Keeping : 2685 of 3426 = 0.7837127845884413
Run: 183 for target: signal Keeping : 2821 of 3550 = 0.7946478873239436
Run: 185 for target: signal Keeping : 3063 of 3856 = 0.7943464730290456
Run: 187 for target: signal Keeping : 2891 of 3616 = 0.7995022123893806
This is for a local efficiency. So once again 95% in CDL correspond to about 80% in 55Fe. Not ideal.
Let's try to train a network that also includes the total charge, so it has some idea of the gas gain in the events.
Otherwise we leave the settings as is:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --ε 0.95 \ --modelOutpath /t/nn_training/trained_model_incl_totalCharge.pt
Interestingly, when including the total charge, the loss of the test data remains lower than of the training set! Models: ./resources/nn_devel_mixing/19_03_23_with_total_charge/ and plots: ./Figs/statusAndProgress/neuralNetworks/development/with_total_charge/ Looking at the total charge, we see the same behavior of CDL and 55Fe data essentially. The background distribution has changed a bit.
We could attempt to change our definition of our loss
function. Currently we in now way enforce that our result should be
close to our target [1, 0] and [0, 1]. Using a MSE loss for
example would make sure of that.
Now training with MSE loss. -> Couldn't get anything sensible out of MSE loss. Chatted with BingChat and it couldn't quite help me (different learning rate etc.), but it did suggest to try L1 loss (mean absolute error), which I am running with now.
L1 loss: ./resources/nn_devel_mixing/19_03_23_l1_loss/ ./Figs/statusAndProgress/neuralNetworks/development/l1_loss/ The all prediction plot is interesting. We see the same-ish behavior in this case as in the cross entropy loss. In the training dataset we can even more clearly see two distinct peaks. However, especially the effective efficiencies in the 55Fe data are all over the place:
./effective_eff_55fe \ -f ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/19_03_23_l1_loss/trained_model_incl_totalCharge_l1_losscheckpoint_epoch_100000_loss_0.0157_acc_0.9971.pt \ --ε 0.95
Run: 83 for target: signal Keeping : 781 of 916 = 0.8526200873362445
Run: 88 for target: signal Keeping : 827 of 911 = 0.9077936333699231
Run: 93 for target: signal Keeping : 612 of 787 = 0.7776365946632783
Run: 96 for target: signal Keeping : 4700 of 5635 = 0.8340727595385981
Run: 102 for target: signal Keeping : 1292 of 1588 = 0.8136020151133502
Run: 108 for target: signal Keeping : 2376 of 3055 = 0.7777414075286416
Run: 110 for target: signal Keeping : 1222 of 1554 = 0.7863577863577863
Run: 116 for target: signal Keeping : 1453 of 1717 = 0.8462434478741991
Run: 118 for target: signal Keeping : 1376 of 1651 = 0.8334342822531798
Run: 120 for target: signal Keeping : 2966 of 3413 = 0.8690301787283914
Run: 122 for target: signal Keeping : 5049 of 5640 = 0.8952127659574468
Run: 126 for target: signal Keeping : 2157 of 2596 = 0.8308936825885979
Run: 128 for target: signal Keeping : 6546 of 7899 = 0.8287124952525636
Run: 145 for target: signal Keeping : 2729 of 3646 = 0.7484914975315414
Run: 147 for target: signal Keeping : 1517 of 2107 = 0.7199810156620788
Run: 149 for target: signal Keeping : 1152 of 1936 = 0.5950413223140496
Run: 151 for target: signal Keeping : 1135 of 1839 = 0.6171832517672649
Run: 153 for target: signal Keeping : 1091 of 1908 = 0.5718029350104822
Run: 155 for target: signal Keeping : 974 of 1777 = 0.5481148002250985
Run: 157 for target: signal Keeping : 978 of 1817 = 0.5382498624105668
Run: 159 for target: signal Keeping : 2083 of 3634 = 0.5731975784259769
Run: 161 for target: signal Keeping : 2152 of 3632 = 0.5925110132158591
Run: 163 for target: signal Keeping : 1264 of 1841 = 0.6865833785985878
Run: 165 for target: signal Keeping : 2929 of 3881 = 0.7547023962896161
Run: 167 for target: signal Keeping : 1467 of 2008 = 0.7305776892430279
Run: 169 for target: signal Keeping : 4458 of 5828 = 0.7649279341111874
Run: 171 for target: signal Keeping : 1495 of 1956 = 0.7643149284253579
Run: 173 for target: signal Keeping : 1401 of 1820 = 0.7697802197802198
Run: 175 for target: signal Keeping : 1566 of 2015 = 0.7771712158808933
Run: 177 for target: signal Keeping : 1561 of 1955 = 0.7984654731457801
Run: 179 for target: signal Keeping : 1105 of 1671 = 0.6612806702573309
Run: 181 for target: signal Keeping : 2425 of 3426 = 0.7078225335668418
Run: 183 for target: signal Keeping : 2543 of 3550 = 0.716338028169014
Run: 185 for target: signal Keeping : 3033 of 3856 = 0.7865663900414938
Run: 187 for target: signal Keeping : 2712 of 3616 = 0.75
So definitely worse in that aspect.
Let's try cross entropy again, but with L1 or L2 regularization.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --ε 0.95 \ --modelOutpath ~/org/resources/nn_devel_mixing/19_03_23_l2_regularization/trained_model_incl_totalCharge_l2_regularization.pt
First attempt with:
SGDOptions.init(0.005).momentum(0.2).weight_decay(0.01)
does not really converge. I guess that is too large.. :) Trying again
with 0.001. This seems to work better.
Oh, it broke between epoch 10000 and 15000, but better again at 20000
(but worse than before). It stayed on a plateau above the previous
afterwards until the end. Also the distributions of the outputs are
quite different now.
./train_ingrid ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 --back ~/CastData/data/DataRuns2017_Reco.h5 --back ~/CastData/data/DataRuns2018_Reco.h5 --ε 0.95 --modelOutpath ~/org/resources/nn_devel_mixing/19_03_23_l2_regularization/trained_model_incl_totalCharge_l2_regularizationcheckpoint_epoch_100000_loss_0.0261_acc_0.9963.pt --predict --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/l2_regularization ./train_ingrid ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 --back ~/CastData/data/DataRuns2017_Reco.h5 --back ~/CastData/data/DataRuns2018_Reco.h5 --ε 0.95 --modelOutpath ~/org/resources/nn_devel_mixing/19_03_23_l2_regularization/trained_model_incl_totalCharge_l2_regularizationcheckpoint_epoch_10000_loss_0.0156_acc_0.9964.pt --predict --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/l2_regularization
./resources/nn_devel_mixing/19_03_23_l2_regularization/
./Figs/statusAndProgress/neuralNetworks/development/l2_regularization/
Looking at the prediction of the final checkpoint
(*_final_checkpoint.pdf) we see that we still have the same kind of
shift in the data. However, after epoch 10000
()
we see a much clearer overlap between the two (but likely also more
background?).
Still interesting, maybe L2 reg is useful if optimized to a good
parameter.
Let's look at the effective efficiencies of this particular checkpoint
and comparing with the very last one:
First the last:
./effective_eff_55fe \ -f ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/19_03_23_l2_regularization/trained_model_incl_totalCharge_l2_regularizationcheckpoint_epoch_100000_loss_0.0261_acc_0.9963.pt \ --ε 0.95
Run: 118 for target: signal Keeping : 1357 of 1651 = 0.8219261053906723
Run: 120 for target: signal Keeping : 2828 of 3413 = 0.8285965426311164
Run: 122 for target: signal Keeping : 4668 of 5640 = 0.8276595744680851
Run: 126 for target: signal Keeping : 2097 of 2596 = 0.8077812018489985
Run: 128 for target: signal Keeping : 6418 of 7899 = 0.8125079123939739
Run: 145 for target: signal Keeping : 2960 of 3646 = 0.811848601206802
Run: 147 for target: signal Keeping : 1731 of 2107 = 0.8215472235405791
Run: 149 for target: signal Keeping : 1588 of 1936 = 0.8202479338842975
Run: 151 for target: signal Keeping : 1482 of 1839 = 0.8058727569331158
Run: 153 for target: signal Keeping : 1565 of 1908 = 0.820230607966457
Run: 155 for target: signal Keeping : 1434 of 1777 = 0.806978052898143
Run: 157 for target: signal Keeping : 1457 of 1817 = 0.8018712162905889
Run: 159 for target: signal Keeping : 2914 of 3634 = 0.8018712162905889
Run: 161 for target: signal Keeping : 2929 of 3632 = 0.8064427312775331
Run: 163 for target: signal Keeping : 1474 of 1841 = 0.8006518196632265
Run: 165 for target: signal Keeping : 3134 of 3881 = 0.8075238340633857
Run: 167 for target: signal Keeping : 1609 of 2008 = 0.8012948207171314
Run: 169 for target: signal Keeping : 4738 of 5828 = 0.8129718599862732
Run: 171 for target: signal Keeping : 1591 of 1956 = 0.8133946830265849
Run: 173 for target: signal Keeping : 1465 of 1820 = 0.804945054945055
Run: 175 for target: signal Keeping : 1650 of 2015 = 0.8188585607940446
Run: 177 for target: signal Keeping : 1576 of 1955 = 0.8061381074168797
Run: 179 for target: signal Keeping : 1339 of 1671 = 0.8013165769000599
Run: 181 for target: signal Keeping : 2740 of 3426 = 0.7997664915353182
Run: 183 for target: signal Keeping : 2856 of 3550 = 0.8045070422535211
Run: 185 for target: signal Keeping : 3146 of 3856 = 0.8158713692946058
Run: 187 for target: signal Keeping : 2962 of 3616 = 0.8191371681415929
Once again in the ballpark of 80% while at 95% for CDL. And for epoch 10,000?
./effective_eff_55fe \ -f ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/19_03_23_l2_regularization/trained_model_incl_totalCharge_l2_regularizationcheckpoint_epoch_10000_loss_0.0156_acc_0.9964.pt \ --ε 0.95
Run: 118 for target: signal Keeping : 1357 of 1651 = 0.8219261053906723
Run: 120 for target: signal Keeping : 2790 of 3413 = 0.8174626428362145
Run: 122 for target: signal Keeping : 4626 of 5640 = 0.8202127659574469
Run: 126 for target: signal Keeping : 2092 of 2596 = 0.8058551617873652
Run: 128 for target: signal Keeping : 6377 of 7899 = 0.8073173819470819
Run: 145 for target: signal Keeping : 2974 of 3646 = 0.8156884256719693
Run: 147 for target: signal Keeping : 1735 of 2107 = 0.8234456573327005
Run: 149 for target: signal Keeping : 1606 of 1936 = 0.8295454545454546
Run: 151 for target: signal Keeping : 1485 of 1839 = 0.8075040783034257
Run: 153 for target: signal Keeping : 1575 of 1908 = 0.8254716981132075
Run: 155 for target: signal Keeping : 1444 of 1777 = 0.8126055149127743
Run: 157 for target: signal Keeping : 1478 of 1817 = 0.8134287286736379
Run: 159 for target: signal Keeping : 2932 of 3634 = 0.8068244358833242
Run: 161 for target: signal Keeping : 2942 of 3632 = 0.8100220264317181
Run: 163 for target: signal Keeping : 1484 of 1841 = 0.8060836501901141
Run: 165 for target: signal Keeping : 3134 of 3881 = 0.8075238340633857
Run: 167 for target: signal Keeping : 1612 of 2008 = 0.8027888446215139
Run: 169 for target: signal Keeping : 4700 of 5828 = 0.8064516129032258
Run: 171 for target: signal Keeping : 1582 of 1956 = 0.8087934560327198
Run: 173 for target: signal Keeping : 1469 of 1820 = 0.8071428571428572
Run: 175 for target: signal Keeping : 1630 of 2015 = 0.8089330024813896
Run: 177 for target: signal Keeping : 1571 of 1955 = 0.8035805626598466
Run: 179 for target: signal Keeping : 1366 of 1671 = 0.817474566128067
Run: 181 for target: signal Keeping : 2734 of 3426 = 0.7980151780502043
Run: 183 for target: signal Keeping : 2858 of 3550 = 0.8050704225352112
Run: 185 for target: signal Keeping : 3122 of 3856 = 0.8096473029045643
Run: 187 for target: signal Keeping : 2937 of 3616 = 0.8122234513274337
Interesting! Despite the much nicer overlap in the prediction at this checkpoint, the end result is not that different. Not quite sure what to make of that.
Next we try Adam, starting with this:
var optimizer = Adam.init( model.parameters(), AdamOptions.init(0.005) )
./resources/nn_devel_mixing/19_03_23_adam_optim/ ./Figs/statusAndProgress/neuralNetworks/development/adam_optim/
-> Outputs are very funny. Extremely wide, need --clampOutput
O(10000) or more. CDL and 55Fe are quite separated though!
Enough for today.
[X]TryL1and L2 regularization of the network (weight decay parameter)[ ]Try L1 regularization[ ]Try Adam optimizer[ ]Try L2 with a value slightly larger and slightly smaller than0.001
1.12.1. TODOs [/]
[ ]If we want to include the energy into the NN training at some point we'd have to make sure to use the correct real energy for the CDL data and not theenergyFromChargecase! -> But currently we don't use the energy at all anyway.[ ]Using the energy could be a useful studying tool I imagine. Would allow to investigate behavior if e.g. only energy is changed etc.
[ ]Understand why seemingly nice L2 reg example at checkpoint 10,000 still has such distinction between CDL and 55Fe despite distributions 'promising' difference? Maybe one bin is just too big?
1.12.2. DONE Bug in withLogLFilterCuts? [/]
I just noticed that in the withLogLFilterCuts the following line:
chargeCut = data[igTotalCharge][i].float > cuts.minCharge and data[igTotalCharge][i] < cuts.maxCharge
is still present even for the fitByRun case. The body of the
template is inserted after the data array is filled. This means
that the cuts are applied to the combined data. That combined data
then is further filtered by this charge cut. For the fitByRun case
however, the minCharge and maxCharge field of the cuts variable
will be set to the values seen in the last run!
Therefore the cut wrongly removes many clusters based on the wrong
charge cut in this case!
The effect of this needs to be investigated ASAP. Both what the CDL distributions look like before and after, as well as what this implies for the lnL cut method!
Which tool generated CDL distributions by run?
cdl_spectrum_creation.
But, cdl_spectrum_creation uses the readCutCDL procedure in
cdl_utils. The heart of it is:
let cutTab = getXrayCleaningCuts() let grp = h5f[(recoDataChipBase(runNumber) & $chip).grp_str] let cut = cutTab[$tfKind] result = cutOnProperties(h5f, grp, cut.cutTo, ("rmsTransverse", cut.minRms, cut.maxRms), ("length", 0.0, cut.maxLength), ("hits", cut.minPix, Inf), ("eccentricity", 0.0, cut.maxEccentricity))
from the h5f.getCdlCutIdxs(runNumber, chip, tfKind) call, i.e. it
manually only applies the X-ray cleaning cuts! So it only ever looks
at the distributions of those and never actually the full
LogLFilterCuts equivalent of the above!
So we might have never noticed cutting away too much for each spectrum, ugh.
[X]We'll do the following: Add a set of plots that show for each ingrid property:
- Raw data
- cut using
readCutCDL withXrayReferenceCutwithLogLFilterCut
and then compare what we see. -> Instead of trying to implement this into
cdl_spectrum_creationwe wrote a separate small plotting script here: ./../CastData/ExternCode/TimepixAnalysis/Plotting/plotCdl/plotCdlDifferentCuts.nim
./plotCdlDifferentCuts -f ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 -c ~/CastData/data/CDL_2019/calibration-cdl-2018.h5
generates the files found in ./Figs/statusAndProgress/cdlCuts/with_charge_cut_bug/ today (before fixing the charge cut bug).
NOTE: The plots have been updated and now include the cleaning cut case mentioned a paragraph down! Especially look at the following two plots
- Figs/statusAndProgress/cdlCuts/with_charge_cut_bug/Cu-Ni-15kV_totalCharge_histogram_by_different_cut_approaches.pdf
- Figs/statusAndProgress/cdlCuts/with_charge_cut_bug/Cu-Ni-15kV_rmsTransverse_histogram_by_different_cut_approaches.pdf
the total charge plot indicates how much is thrown away comparing LogLCuts & XrayCuts with CDL cuts and the rmsTransverse indicates how much in percentage of the signal is lost comparing the two.
The big question looking at this plot right now though is why the
X-ray reference cut behaves exactly the same way as the LogL cut does!
The 'last cuts' should only be applied to all data in the case of the
LogL cut usage!
-> The reason is that the X-ray reference cut case uses the I think
wrong set of two cuts. The idea should have been to reproduce the same
cuts as the CDL applies! But it's exactly only those cuts that contain
the charge cut and are intended to cut to the main peak of the
spectrum…
I mean I suppose it makes sense from the name, now that I think about
it. We'll add a withXrayCleaningCuts.
So, with the cleaning cut introduced, we get the behavior we would have expected. The LogL filter and XrayRef cuts lose precisely the peaks of the not last peak.
We'll fix it by not applying the charge cut in the case where we use
fitByRun.
The new plots are in: Figs/statusAndProgress/cdlCuts/charge_cut_bug_fixed/ and the same plots:
- Figs/statusAndProgress/cdlCuts/charge_cut_bug_fixed/Cu-Ni-15kV_totalCharge_histogram_by_different_cut_approaches.pdf
- Figs/statusAndProgress/cdlCuts/charge_cut_bug_fixed/Cu-Ni-15kV_rmsTransverse_histogram_by_different_cut_approaches.pdf
We can see we now keep the correct information!
This has implications for all the background rates and all the limits to an extent of course.
[X]Generate
likelihoodoutput with only lnL cut for Run-2 and Run-3 at 80% and compare with background rate from all likelihood combinations generated yesterday. That should give us an idea if it's necessary to regenerate all outputs and limits again. First need to regenerate the likelihood values in all the data files though.likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 --cdlYear 2018 --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 --computeLogL likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 --cdlYear 2018 --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 --computeLogL likelihood -f ~/CastData/data/CalibrationRuns2017_Reco.h5 --cdlYear 2018 --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 --computeLogL likelihood -f ~/CastData/data/CalibrationRuns2018_Reco.h5 --cdlYear 2018 --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 --computeLogL
and now for the likelihood calls:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crGold \ --signalEfficiency 0.8 \ --vetoSets "{fkLogL}" \ --out /t/playground \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --dryRun
and finally compare the background rates:
plotBackgroundRate \ ~/org/resources/lhood_limits_automation_with_nn_support/lhood/likelihood_cdl2018_Run2_crGold_lnL.h5 \ ~/org/resources/lhood_limits_automation_with_nn_support/lhood/likelihood_cdl2018_Run3_crGold_lnL.h5 \ /t/playground/likelihood_cdl2018_Run2_crGold_signalEff_0.8_lnL.h5 \ /t/playground/likelihood_cdl2018_Run3_crGold_signalEff_0.8_lnL.h5 \ --names "ChargeBug" --names "ChargeBug" \ --names "Fixed" --names "Fixed" \ --centerChip 3 \ --title "Background rate from CAST data, lnL@80, charge cut bug" \ --showNumClusters --showTotalTime \ --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_cast_lnL_80_charge_cut_bug.pdf \ --outpath /t/playground/ \ --quiet
The generated plot is:
As we can see we remove a few clusters, but the difference is absolutely minute. That fortunately means we don't need to rerun all the limits again!
Might still be beneficial for the NN training as the impact on other variables might be bigger.
1.13.
Continuing from yesterday, but before we do that, we need to generate
the new expected limits table using the script in
StatusAndProgress.org sec. [BROKEN LINK: sec:limit:expected_limits_different_setups_test].
[X]Generate limits table[ ]Regenerate all limits once more to have them with the correct eccentricity cut off value in the files -> Should be done, but not priority right now. Our band aid fix relying on the filename is fine for now.[ ]continue NN training / investigation[ ]Update systematics due todetermineEffectiveEfficiencyusing fixed code (correct energies & data frames) in thesis[ ]fix that same code forfitByRun
1.13.1. NN training
Let's try to reduce the number of neurons on the hidden layer of the network and see where that gets us in the output distribution.
(back using SGD without L2 reg):
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --ε 0.95 \ --modelOutpath ~/org/resources/nn_devel_mixing/20_03_23_hidden_layer_100neurons/trained_model_hidden_layer_100.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/hidden_layer_100neurons/
The CDL vs. 55Fe distribution is again slightly different (tested on checkpoint 35000). Btw: also good to know that we can easily run e.g. a prediction of a checkpoint while the training is ongoing. Not a problem whatsoever.
Next test a network that only uses the three variables used for the lnL cut! Back using 500 hidden neurons. Let's try that training while the other one is still running…
If this one shows the same distinction in 55Fe vs CDL data that is actually more damning for our current approach in some sense than anything else. If not however, than we can analyze which variable is the main contributor in giving us that separation in the predictions!
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --ε 0.95 \ --modelOutpath ~/org/resources/nn_devel_mixing/20_03_23_only_lnL_vars/trained_model_only_lnL_vars.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/only_lnL_vars
It seems like in this case the prediction is actually even in the opposite direction! Now the CDL data is more "background like" than the 55Fe data. ./Figs/statusAndProgress/neuralNetworks/development/only_lnL_vars/all_predictions.pdf What do the effective 55Fe numbers say in this case?
./effective_eff_55fe -f ~/CastData/data/CalibrationRuns2017_Reco.h5 --model ~/org/resources/nn_devel_mixing/20_03_23_only_lnL_vars/trained_model_only_lnL_varscheckpoint_epoch_100000_loss_0.1237_acc_0.9504.pt --ε 0.95
Error: unhandled cpp exception: Could not run 'aten::emptystrided' with arguments from the 'CUDA' backend. This could be because the operator doesn't exist for this backend, or was omitted during the selective/custom build process (if using custom build). If you are a Facebook employee using PyTorch on mobile, p lease visit https://fburl.com/ptmfixes for possible resolutions. 'aten::emptystrided' is only available for these backends: [CPU, Meta, BackendSelect, Pytho n, Named, Conjugate, Negative, ZeroTensor, ADInplaceOrView, AutogradOther, AutogradCPU, AutogradCUDA, AutogradXLA, AutogradLazy, AutogradXPU, AutogradMLC, Au togradHPU, AutogradNestedTensor, AutogradPrivateUse1, AutogradPrivateUse2, AutogradPrivateUse3, Tracer, AutocastCPU, Autocast, Batched, VmapMode, Functionali ze, PythonTLSSnapshot].
CPU: registered at aten/src/ATen/RegisterCPU.cpp:21249 [kernel] Meta: registered at aten/src/ATen/RegisterMeta.cpp:15264 [kernel] BackendSelect: registered at aten/src/ATen/RegisterBackendSelect.cpp:606 [kernel] Python: registered at ../aten/src/ATen/core/PythonFallbackKernel.cpp:77 [backend fallback] Named: registered at ../aten/src/ATen/core/NamedRegistrations.cpp:7 [backend fallback] Conjugate: fallthrough registered at ../aten/src/ATen/ConjugateFallback.cpp:22 [kernel] Negative: fallthrough registered at ../aten/src/ATen/native/NegateFallback.cpp:22 [kernel] ZeroTensor: fallthrough registered at ../aten/src/ATen/ZeroTensorFallback.cpp:90 [kernel] ADInplaceOrView: fallthrough registered at ../aten/src/ATen/core/VariableFallbackKernel.cpp:64 [backend fallback] AutogradOther: registered at ../torch/csrc/autograd/generated/VariableType2.cpp:12095 [autograd kernel] AutogradCPU: registered at ../torch/csrc/autograd/generated/VariableType2.cpp:12095 [autograd kernel] AutogradCUDA: registered at ../torch/csrc/autograd/generated/VariableType2.cpp:12095 [autograd kernel] AutogradXLA: registered at ../torch/csrc/autograd/generated/VariableType2.cpp:12095 [autograd kernel] AutogradLazy: registered at ../torch/csrc/autograd/generated/VariableType2.cpp:12095 [autograd kernel] AutogradXPU: registered at ../torch/csrc/autograd/generated/VariableType2.cpp:12095 [autograd kernel] AutogradMLC: registered at ../torch/csrc/autograd/generated/VariableType2.cpp:12095 [autograd kernel] AutogradHPU: registered at ../torch/csrc/autograd/generated/VariableType2.cpp:12095 [autograd kernel] AutogradNestedTensor: registered at ../torch/csrc/autograd/generated/VariableType2.cpp:12095 [autograd kernel] AutogradPrivateUse1: registered at ../torch/csrc/autograd/generated/VariableType2.cpp:12095 [autograd kernel] AutogradPrivateUse2: registered at ../torch/csrc/autograd/generated/VariableType2.cpp:12095 [autograd kernel] AutogradPrivateUse3: registered at ../torch/csrc/autograd/generated/VariableType2.cpp:12095 [autograd kernel] Tracer: registered at ../torch/csrc/autograd/generated/TraceType2.cpp:12541 [kernel] AutocastCPU: fallthrough registered at ../aten/src/ATen/autocastmode.cpp:462 [backend fallback] Autocast: fallthrough registered at ../aten/src/ATen/autocastmode.cpp:305 [backend fallback] Batched: registered at ../aten/src/ATen/BatchingRegistrations.cpp:1059 [backend fallback] VmapMode: fallthrough registered at ../aten/src/ATen/VmapModeRegistrations.cpp:33 [backend fallback] Functionalize: registered at ../aten/src/ATen/FunctionalizeFallbackKernel.cpp:52 [backend fallback] PythonTLSSnapshot: registered at../aten/src/ATen/core/PythonFallbackKernel.cpp:81 [backend fallback]
Uhh, this fails with a weird error… I love the "If you are a
Facebook employee" line!
Oh, never mind, I simply forgot the -d:cuda flag when compiling,
oops.
Run: 83 for target: signal Keeping : 823 of 916 = 0.898471615720524 Run: 88 for target: signal Keeping : 820 of 911 = 0.9001097694840834 Run: 93 for target: signal Keeping : 692 of 787 = 0.8792884371029225 Run: 96 for target: signal Keeping : 5079 of 5635 = 0.9013309671694765 Run: 102 for target: signal Keeping : 1409 of 1588 = 0.8872795969773299 Run: 108 for target: signal Keeping : 2714 of 3055 = 0.888379705400982 Run: 110 for target: signal Keeping : 1388 of 1554 = 0.8931788931788932 Run: 116 for target: signal Keeping : 1541 of 1717 = 0.8974956319161328 Run: 118 for target: signal Keeping : 1480 of 1651 = 0.8964264082374318 Run: 120 for target: signal Keeping : 3052 of 3413 = 0.8942279519484324 Run: 122 for target: signal Keeping : 4991 of 5640 = 0.8849290780141844 Run: 126 for target: signal Keeping : 2274 of 2596 = 0.8759630200308166 Run: 128 for target: signal Keeping : 6973 of 7899 = 0.8827699708823902 Run: 145 for target: signal Keeping : 3287 of 3646 = 0.9015359297860669 Run: 147 for target: signal Keeping : 1887 of 2107 = 0.8955861414333175 Run: 149 for target: signal Keeping : 1753 of 1936 = 0.9054752066115702 Run: 151 for target: signal Keeping : 1662 of 1839 = 0.9037520391517129 Run: 153 for target: signal Keeping : 1731 of 1908 = 0.9072327044025157
The numbers are hovering around 90% for the 95%
desired. Interesting. And not what we might have expected. I suppose
the different distributions in the CDL output then are related to
different CDL targets. Some are vastly more left than others? What
would the prediction look like if we restrict ourselves to the
MnCr12kV target?
Modified one line in predictAll, added this:
.filter(f{`Target` == "Mn-Cr-12kV"})
let's run that on the same model (last checkpoint) and see how it
compares in 55Fe vs CDL.
Indeed, the CDL data now is more compatible with the 55Fe data (and
likely slightly more to the right explaining the 90% for the target
95).
Be that as it may, the difference in the ROC curves of one of our "good" networks and this one is pretty stunning. Where the good ones are almost a right angled triangle, this one is pretty smooth: Figs/statusAndProgress/neuralNetworks/development/only_lnL_vars/roc_curve.pdf
1.13.2. DONE Expected limits table
cd $TPA/Tools/generateExpectedLimitsTable ./generateExpectedLimitsTable --path ~/org/resources/lhood_limits_automation_with_nn_support/limits
| εlnL | Scinti | FADC | εFADC | Septem | Line | eccLineCut | εSeptem | εLine | εSeptemLine | Total eff. | Limit no signal | Expected Limit |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.9 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.7587 | 3.7853e-21 | 7.9443e-23 |
| 0.9 | true | false | 0.98 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.7742 | 3.6886e-21 | 8.0335e-23 |
| 0.9 | true | true | 0.98 | false | true | 1.2 | 0.7841 | 0.8794 | 0.7415 | 0.7757 | 3.6079e-21 | 8.1694e-23 |
| 0.8 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.6744 | 4.0556e-21 | 8.1916e-23 |
| 0.8 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.6744 | 4.0556e-21 | 8.1916e-23 |
| 0.9 | true | true | 0.98 | false | true | 1.4 | 0.7841 | 0.8946 | 0.7482 | 0.7891 | 3.5829e-21 | 8.3198e-23 |
| 0.8 | true | true | 0.98 | false | true | 1.2 | 0.7841 | 0.8794 | 0.7415 | 0.6895 | 3.9764e-21 | 8.3545e-23 |
| 0.8 | true | true | 0.9 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.6193 | 4.4551e-21 | 8.4936e-23 |
| 0.9 | true | true | 0.98 | false | true | 1.6 | 0.7841 | 0.9076 | 0.754 | 0.8005 | 3.6208e-21 | 8.5169e-23 |
| 0.8 | true | true | 0.98 | false | true | 1.4 | 0.7841 | 0.8946 | 0.7482 | 0.7014 | 3.9491e-21 | 8.6022e-23 |
| 0.8 | true | true | 0.98 | false | true | 1.6 | 0.7841 | 0.9076 | 0.754 | 0.7115 | 3.9686e-21 | 8.6462e-23 |
| 0.9 | true | false | 0.98 | true | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.6593 | 4.2012e-21 | 8.6684e-23 |
| 0.7 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.5901 | 4.7365e-21 | 8.67e-23 |
| 0.9 | true | true | 0.98 | true | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.6461 | 4.3995e-21 | 8.6766e-23 |
| 0.7 | true | false | 0.98 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.6021 | 4.7491e-21 | 8.7482e-23 |
| 0.8 | true | true | 0.98 | true | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.5743 | 4.9249e-21 | 8.7699e-23 |
| 0.8 | true | true | 0.98 | false | false | 0 | 0.7841 | 0.8602 | 0.7325 | 0.784 | 3.6101e-21 | 8.8059e-23 |
| 0.8 | true | true | 0.8 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.5505 | 5.1433e-21 | 8.855e-23 |
| 0.7 | true | true | 0.98 | false | true | 1.2 | 0.7841 | 0.8794 | 0.7415 | 0.6033 | 4.4939e-21 | 8.8649e-23 |
| 0.8 | true | true | 0.98 | true | false | 0 | 0.7841 | 0.8602 | 0.7325 | 0.6147 | 4.5808e-21 | 8.8894e-23 |
| 0.9 | true | false | 0.98 | true | false | 0 | 0.7841 | 0.8602 | 0.7325 | 0.7057 | 3.9383e-21 | 8.9504e-23 |
| 0.7 | true | true | 0.98 | false | true | 1.4 | 0.7841 | 0.8946 | 0.7482 | 0.6137 | 4.5694e-21 | 8.9715e-23 |
| 0.8 | true | true | 0.9 | true | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.5274 | 5.3406e-21 | 8.9906e-23 |
| 0.9 | true | true | 0.9 | true | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.5933 | 4.854e-21 | 9e-23 |
| 0.8 | false | false | 0.98 | false | false | 0 | 0.7841 | 0.8602 | 0.7325 | 0.8 | 3.5128e-21 | 9.0456e-23 |
| 0.8 | true | false | 0.98 | false | false | 0 | 0.7841 | 0.8602 | 0.7325 | 0.8 | 3.5573e-21 | 9.0594e-23 |
| 0.7 | true | true | 0.98 | false | true | 1.6 | 0.7841 | 0.9076 | 0.754 | 0.6226 | 4.5968e-21 | 9.0843e-23 |
| 0.7 | true | true | 0.98 | true | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.5025 | 5.627e-21 | 9.1029e-23 |
| 0.8 | true | true | 0.9 | false | false | 0 | 0.7841 | 0.8602 | 0.7325 | 0.72 | 3.8694e-21 | 9.1117e-23 |
| 0.8 | true | true | 0.9 | true | false | 0 | 0.7841 | 0.8602 | 0.7325 | 0.5646 | 4.909e-21 | 9.2119e-23 |
| 0.7 | true | false | 0.98 | true | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.5128 | 5.5669e-21 | 9.3016e-23 |
| 0.7 | true | false | 0.98 | true | false | 0 | 0.7841 | 0.8602 | 0.7325 | 0.5489 | 5.3018e-21 | 9.3255e-23 |
| 0.7 | true | true | 0.9 | true | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.4615 | 6.1471e-21 | 9.4509e-23 |
| 0.8 | true | true | 0.8 | false | false | 0 | 0.7841 | 0.8602 | 0.7325 | 0.64 | 4.5472e-21 | 9.5113e-23 |
| 0.8 | true | true | 0.8 | true | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.4688 | 5.8579e-21 | 9.5468e-23 |
| 0.8 | true | true | 0.8 | true | false | 0 | 0.7841 | 0.8602 | 0.7325 | 0.5018 | 5.6441e-21 | 9.5653e-23 |
1.14.
From yesterday open TODOs:
[ ]Regenerate all limits once more to have them with the correct eccentricity cut off value in the files -> Should be done, but not priority right now. Our band aid fix relying on the filename is fine for now.[ ]continue NN training / investigation[ ]Update systematics due todetermineEffectiveEfficiencyusing fixed code (correct energies & data frames) in thesis
[ ]fix that same code forfitByRun
Additional:
[X]look at prediction of our best trained network (and maybe the lnL variable one?) for all the different CDL datasets separately. Maybe a ridgeline plot of the different "sets", i.e. background, 55Fe photo, 55Fe escape, CDL sets[ ]Do the same thing with the Run-2 and Run-3 calibration / background data split?[ ]Do the same thing, but using thelikelihooddistributions for each instead of the NN predictions!
[ ]Investigate whether effective efficiency (from tool) is correlated to mean gas gain of each calibration run. create a plot of the effective efficiency vs the mean gas gain of each run, per photo & escape type -> If this is strongly correlated it means we understand where the fluctuation comes from! If true, then can look at CDL data as well and check if this explains the variation.
1.14.1. Structured information about MLP layout
Instead of having to recompile the code each time to make changes to
the layout, I now made it all run time configurable using the
MLPDesc object. It stores the model and plot path, the number of
input neurons, hidden neurons and which datasets were used.
In order to make the 'old' models work a --writeMLPDesc option was
added.
For the with_total_charge:
./train_ingrid \ --modelOutpath ~/org/resources/nn_devel_mixing/19_03_23_with_total_charge/trained_model_incl_totalChargecheckpoint_epoch_100000_loss_0.0102_acc_0.9976.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/with_total_charge \ --numHidden 500 \ --writeMLPDesc
For the with_total_charge:
./train_ingrid \ --modelOutpath ~/org/resources/nn_devel_mixing/18_03_23/trained_mlp_mixed_data.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/mixing_data/ \ --numHidden 500 \ --datasets igEccentricity \ --datasets igSkewnessLongitudinal \ --datasets igSkewnessTransverse \ --datasets igKurtosisLongitudinal \ --datasets igKurtosisTransverse \ --datasets igLength \ --datasets igWidth \ --datasets igRmsLongitudinal \ --datasets igRmsTransverse \ --datasets igLengthDivRmsTrans \ --datasets igRotationAngle \ --datasets igFractionInTransverseRms \ --writeMLPDesc
For the charge_bug_fixed:
./train_ingrid \ --modelOutpath ~/org/resources/nn_devel_mixing/19_03_23_charge_bug_fixed/trained_model_charge_cut_bug_fixed.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/charge_cut_bug_fixed/ \ --numHidden 500 \ --writeMLPDesc
For the l1_loss:
./train_ingrid \ --modelOutpath ~/org/resources/nn_devel_mixing/19_03_23_l1_loss/trained_model_incl_totalCharge_l1_loss.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/l1_loss/ \ --numHidden 500 \ --writeMLPDesc
For the l2_regularization:
./train_ingrid \ --modelOutpath ~/org/resources/nn_devel_mixing/19_03_23_l2_regularization/trained_model_incl_totalCharge_l2_regularization.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/l2_regularization/ \ --numHidden 500 \ --writeMLPDesc
For the hidden_layer_100neurons:
./train_ingrid \ --modelOutpath ~/org/resources/nn_devel_mixing/20_03_23_hidden_layer_100neurons/trained_model_hidden_layer_100.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/hidden_layer_100neurons/ \ --numHidden 100 \ --writeMLPDesc
For the only_lnL_vars:
./train_ingrid \ --modelOutpath ~/org/resources/nn_devel_mixing/20_03_23_only_lnL_vars/trained_model_only_lnL_vars.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/only_lnL_vars/ \ --numHidden 500 \ --datasets igEccentricity \ --datasets igLengthDivRmsTrans \ --datasets igFractionInTransverseRms \ --writeMLPDesc
In the future this will likely also include the used optimizer, learning rate etc.
1.14.2. Prediction by target/filter
To do this I added an additional plot that also generates a ridgeline
in the predictAll case.
./train_ingrid ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --ε 0.95 \ --modelOutpath ~/org/resources/nn_devel_mixing/19_03_23_with_total_charge/trained_model_incl_totalChargecheckpoint_epoch_100000_loss_0.0102_acc_0.9976.pt --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/with_total_charge \ --predict
And for the network with 2500 hidden neurons:
./train_ingrid ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/21_03_23_hidden_2500neurons/trained_model_hidden_2500neurons.pt \ --predict
I suppose the best thing to do is to use a scaling transformation similar to what Cristina does. Transform CAST data into CDL data by a scaling factor and then transform other CDL energies back into CAST energies.
1.14.3. Train MLP with 2500 hidden neurons [/]
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/21_03_23_hidden_2500neurons/trained_model_hidden_2500neurons.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/hidden_500neurons/ \ --numHidden 2500
This one is damn good! Especially considering that test is better than train essentially the entire time up to 100,000 epochs!
[ ]Maybe try even larger?
1.15.
Continue with the jobs from yesterday:
Additional:
[X]look at prediction of our best trained network (and maybe the lnL variable one?) for all the different CDL datasets separately. Maybe a ridgeline plot of the different "sets", i.e. background, 55Fe photo, 55Fe escape, CDL sets
[ ]Do the same thing with the Run-2 and Run-3 calibration / background data split?[X]Do the same thing, but using thelikelihooddistributions for each instead of the NN predictions![X]Investigate whether effective efficiency (from tool) is correlated to mean gas gain of each calibration run. create a plot of the effective efficiency vs the mean gas gain of each run, per photo & escape type -> If this is strongly correlated it means we understand where the fluctuation comes from! If true, then can look at CDL data as well and check if this explains the variation.
That is: implement the lnL variant into the 'prediction' ridge line plots. And potentially look at the Run-2 vs Run-3 predictions.
[ ]Look at the background rate of the 90% lnL cut variant. How much background do we have in that case? How does it compare to 99% accuracy MLP prediction?[ ]maybe try even larger MLP?
As a bonus:
[ ]look at thehidden_2500neuronnetwork for the background rate[ ]try to use the neural network for a limit calculation it its "natural" prediction. i.e close to 99% accuracy! That should give us quite the amazing signal (but of course decent background!). Still, as alternative combined with line veto and/or FADC could be very competitive![X]Make notes about ROC curve plots[X]Next up: -> Look at effective efficiency again and how it varies -> Implement CAST ⇔ CDL transformation for cut values
1.15.1. Notes
- old ROC curves often filtered out the lnL = Inf cases for the lnL
method! (not everywhere, but in
likelihood.nimfor example!) - Apparently there are only 418 events < 0.4 keV in the whole background dataset. ROC curves for lnL at lowest target are very rough for that reason. Why weren't they rough before though?
1.15.2. Comparison of the MLP predictions & lnL distributions for each 'type'
This now also produces a plot
all_predictions_ridgeline_by_type_lnL.pdf which is the equivalent of
the MLP prediction ridgeline, but using the likelihood distributions:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/21_03_23_hidden_2500neurons/trained_model_hidden_2500neurons.pt \ --predict
Running the prediction now also produces the distributions for the likelihood data as well as ROC curves for both. Note that the ROC curves contain both CAST and CDL data for each target. As such they are a bit too 'good' for CAST and a bit too 'bad' for the CDL. In case of the LnL data they match better, because the likelihood distribution matches better between CAST and CDL.
See the likelihood distributions:
Note: All likelihood data at 50 and above has been cut off, as
otherwise the peak at 50 dominates the background data such that we
don't see the tail. Keep that in mind, the background contribution
that is in the range of the X-ray data is a very small fraction!
Compare that with the MLP output of this network (2500 hidden neurons): Figs/statusAndProgress/neuralNetworks/development/hidden_2500neurons/all_predictions_by_type_ridgeline_mlp.pdf
First of all we see that the MLP distributions are much better defined and not as wide (but keep in mind only about half the width in this plot than the other). Pay close attention to the CAST photo peak ('photo') and compare it with the Mn-Cr-12kV target. It should in theory be the same distribution, but it the CAST distribution is shifted slightly to the left! This is precisely the reason why the effective efficiency for the CAST data is always lower than expected (based on CDL that is).
Interestingly even in the lnL case these two distributions are not identical! Their mean is very similar, but the shape differs a bit.
Regarding the ROC curves: Old ROC curves often filtered out the lnL = Inf cases for the lnL method! Therefore, they appeared even worse than they actually are. If you include all data and only look at the mean (i.e. all data at the same time) it is not actually that bad! Which makes sense because by itself the lnL veto is already pretty powerful after all.
The ROC curve for all data MLP vs. LnL:
Look at the y scale! 0.96 is the minimum! So LnL really does a good
job. It's just that the MLP is even significantly better!
Now for the ROC curve split by different targets:
The first obvious thing is how discrete the low energy cases look! The
C-EPIC-0.6kV case in particular is super rugged. Why is that? And why
was that not the case in the past when we looked at the ROC curves for
different targets?
At the moment I'm not entirely sure, but my assumption is that we
(accidentally?) used all background data when computing the
efficiencies for each target, but only the X-rays corresponding to
each CDL dataset (note that in the past we never had any CAST 55Fe
data in there either).
As it turns out though at energies below 0.4 keV (lowest bin) in the
whole background dataset, there are only about ~400 clusters!
(checked using
verbose option in the targetSpecificRoc proc).
So this is all very interesting. And it reassures us that using such an MLP is definitely a very interesting avenue. But in order to use it we need to understand the differences in the output distributions for the 5.9 keV data in each of the datasets. One obvious difference between CDL and CAST data is, as we very well know, the temperature drifts that cause gas gain drifts. Therefore next we look at the behavior of the effective efficiency for the data in relation to the gas gain in each run.
1.15.3. Effective efficiency of MLP veto and gas gain dependence
We added reading of the gas gain information and plotting it against the effective efficiencies for each run into ./../CastData/ExternCode/TimepixAnalysis/Tools/NN_playground/effective_eff_55fe.nim
In order to look at all data we added the ability to hand multiple input files and also hand the CDL data file so that we can compare that too.
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/21_03_23_hidden_2500neurons/trained_model_hidden_2500neurons.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5
which now generates the plots in:
Where the first is just the effective efficiency of the CDL and CAST data (split by the target energy, 3.0 ~ escape peak and 5.9 ~ photo peak). It may look a bit confusing at first, but the green is simply the normalized gain and the purple is the effective efficiency if cut at a 95% value based on the local energy cuts using CDL data.
The green points hide a set of green crosses that are simply not visible because they overlap exactly the green dots (same run numbers for escape and photo peak, same for CDL data!). In the right part at higher run numbers is the CDL dataset (all datasets contain some events around 5.9 keV, most very few, same for 3.0 keV data). Everything is switched around, because the efficiency there is close to the target 95%, but in relative terms the gas gain is much lower.
Staring at this a bit longer indeed seems to indicate that there is a correlation between gas gain and effective efficiency!
This gets more extreme when considering the second plot, which maps the gas gain against the effective efficiency directly in a scatter plot. The left pane shows all data around the 5.9 keV data and the right around the 3.0 keV data. In both panes there is a collection of points in the 'bottom right' and one in the 'top left'. The bottom right contains high gain data at low effective efficiencies, this is the CAST data. The top left is the inverse, high effective efficiencies at low gain. The CDL data.
As we can see especially clearly in the 5.9 keV data, there is a very strong linear correlation between the effective efficiency and the gas gain! The two blobs visible in the CAST data at 5.9 keV correspond to the Run-2 data (the darker points) and Run-3 data (the brighter points). While they differ they seem to follow generally a very similar slope.
This motivates well to use a linear interpolation based on fits found for the CAST data in Run-2 and Run-3, which then is used together with the target efficiency and cut value at that efficiency in the CDL data to correct the cut value for each efficiency!
1.16.
From yesterday:
[ ]Look at the background rate of the 90% lnL cut variant. How much background do we have in that case? How does it compare to 99% accuracy MLP prediction?[ ]maybe try even larger MLP?As a bonus:
[ ]look at thehidden_2500neuronnetwork for the background rate[ ]try to use the neural network for a limit calculation it its "natural" prediction. i.e close to 99% accuracy! That should give us quite the amazing signal (but of course decent background!). Still, as alternative combined with line veto and/or FADC could be very competitive!
And in addition:
[ ]Implement a fit that takes the effective efficiency and gas gain correlation into account and try it to correct for the efficiencies at CAST![ ]Look at how the distributions change between different CDL runs with different gas gains.
1.17.
First look into the energyFromCharge for the CDL data and see if it
changes the cut values.
Important thought: ~3 keV escape events are not equivalent to 3 keV X-rays! escape events are effectively 5.9 keV photons that only deposit 3 keV! Real 3 keV X-rays have a much longer absorption length. That explains why 55Fe 3 keV data is shifted to a lower cut value, but CDL 3 keV data to a higher cut value when compared to the 5.9 keV data in each!
So our prediction of the cut value for the escape events via the slope of the 5.9 keV data is therefore too large, because the real events look "less" like X-rays to the network.
Generate two sets of fake events:
[ ]Events of the same energy, but at an effectively different diffusion length, by taking transverse diffusion and the distance and 'pulling' all electrons of an event towards the center of the cluster -> e.g. generate 'real' 3 keV events from the escape peak 3 keV events[ ]Events of an artificially lower Timepix threshold of same energy. Look at how many electrons calibration set 1 has compared to 2. Then throw away this many electrons, biased by those of the lowest charges (or inversely fix a threshold in electrons and remove all pixels below and see where we end up). Problem is that total number of recorded electrons (i.e. ToT value) itself also depends on the threshold.[ ](potential) a third set could just be looking at fake lower energy events generated in the way that we already do it in the lnL effective efficiency code!
These can then all be used to evaluate the MLP with.
[ ]Investigate fake events!
1.18.
Continuing from yesterday… Fake events and other stuff..
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/21_03_23_hidden_2500neurons/trained_model_hidden_2500neurons.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets
[ ]Why is skewness of all CAST data (incl. fake data!) slightly positive? It should be centered around 0, no?[ ]rmsTransverse, length and width are as expected: fake data and escape peak data is larger than real 3 keV data! Due to different absorption lengths
1.19.
And more continue, continue, continue…!
[X]First start with reordering the ridgeline plots according to energy[ ]Then implement two different other fake data generation
Generation of data at different diffusion: Change the effective diffusion of the drawn event.
- get the diffusion (transverse) coefficient
σT = getDiffusion() using the existing energy and target energies compute the required distance we 'move' the cluster from and to. That is: Assuming we have a diffusion equivalent to 3cm (conversion at cathode) and want to 'move' that to a diffusion of 2 cm (conversion 1cm away from cathode). We can compute the transverse diffusion by
σ_T * √x cm (x ∈ [2, 3])Each of the resulting numbers is the standard deviation of a normal distribution around the center position of the cluster![X]Verify how this relates to (see below) At 3cm the standard deviation is σ = √(6 D t) (3 dim)
With the distributions we expect we now have a few options to generate new events
- simplest and deterministic: push all electrons to the equivalent value of the PDF (longer distance: shallower PDF. Find P(xi) = P'(xi') and move each xi to xi'.
- draw from the P' distribution for each pixel. The resulting x' is the location in distance from existing cluster center to place the pixel at.
- We could maybe somehow generate a 'local' PDF for each pixel (based on how far away each already is) and drawing from that. So a mix of 1 and 2?
For now let's go with 2. Simpler to implement as we don't need to find an equivalent point on the PDF (which would be doable using
lowerBound)- define the a gaussian with mean of resulting diffusion around that distance (what sigma does it have?) -> Or: define gaussian of the transverse diffusion coefficient and simply multiply!
- for each pixel sample it and move each pixel the resulting distance towards the center / away from the center (depending)
1.19.1. About diffusion confusion
-> Normal distribution describes position! See also: file:///home/basti/org/Papers/gas_physics/randomwalkBerg_diffusion.pdf <x²> = 2 D t ( 1 dimension ) <x²> = 4 D t ( 2 dimensions ) <x²> = 6 D t ( 3 dimensions ) Also look into Sauli book again (p. 82 eq. (4.5) and eq. (4.6)). Also: file:///home/basti/org/Papers/Hilke-Riegler2020_Chapter_GaseousDetectors.pdf page 15 sec. 4.2.2.2 The latter mentions on page 15 that there is a distinction between: D = diffusion coefficient for which σ = √(2 D t) (1 dim) is valid and D* = diffusion constant for which: σ = D* √(z) is valid!
From PyBoltz source code in Boltz.pyx
self.TransverseDiffusion1 = sqrt(2.0 * self.TransverseDiffusion / self.VelocityZ) * 10000.0
which proves the distinction in the paper: √(2 D t) = D* √x ⇔ D* = √(2 D t) / √x = √(2 D t / x) = √(2 D / v) (with x = v t)
Check this with script:
import math let D = 4694.9611 * 1e-6 # cm² / s to cm² / μs let Dp = 644.22619 # μm / √cm let v = 22.6248 / 10.0 # mm/μs to cm/μs echo sqrt(4.0 * D / v) * 10_000.0 # cm to μm
Uhhhhh…. Doesn't make any sense :(
1.20.
And continue working on the fake event generation…!
[X]Adjusting the diffusion down to low values (e.g. 400) does not move thefractionInTransverseRmsto lower values![X]ImplementlogLandtrackingsupport intorunAnalysisChainto really make it do (almost) everything
1.20.1. DONE Figure out why skewness has a systematic bias
About the skewness being non zero: I just noticed that the transverse skewness is always slightly positive, but at the same time the longitudinal skewness is slightly negative by more or less the same amount! Why is that? It's surely some bias in our calculation that has this effect?
About the skewness discussion see: ./LLM_discussions/BingChat/skewness_of_clusters/ the images for the discussion and the code snippets for the generated code.
Based on that I think it is more or less safe that at least algorithmically our approach should not yield any skewed data. However, why does our fake data still show that? Let's try rotating each point by a random φ and see if it still is the case.
[X]I applied this, using a flat rotation angle for the data and the problem persisted.
After this I looked into the calculation of the geometry and wanted to play around with it. But immediately I found the issue.. I was being too overzealous in a recent bug fix. The following introduced the problem:
https://github.com/Vindaar/TimepixAnalysis/commit/8d4813d405bf3be6f2e98ef32fe0b1f178cdca01
Here we did not actually define any "axis" for the data, but instead simply took the larger value for each variable as the longitudinal one. That's rubbish of course!
Define the long axis (do it based on length & width!), but then stick to that.
Implemented and indeed has the 'desired' result. We now get 0 balanced skewness!
1.20.2. TODO Rerun the CAST reconstruction
Due to the skewness (and related) bug we need to regenerate all the data.
In addition we can already look at how the fake data is being handled by the MLP as a preview. -> Ok, it seems like our new fake events are considered "more" signal like (higher cut value) than the real data.
Before recalc, let's check if the other calibration file has the same skewness offset. Maybe it was generated before we introduced the bug? -> Yeah, also seems to have it already.
Some figures of the state as is right now (with skewness bug in real
CAST data) can be found here:
./runAnalysisChain -i ~/CastData/data \ --outpath ~/CastData/data \ --years 2017 --years 2018 \ --calib --back \ --reco --logL --tracking
Finished running, but the tracking wasn't added yet due to the wrong path to the log files!
1.20.3. Important realization about fraction in transverse RMS and normal distribution
I just realized that the fraction in transverse RMS is strongly connected to the probability density within a 1σ region around a bivariate normal distribution! https://en.wikipedia.org/wiki/Multivariate_normal_distribution#Geometric_interpretation
| Dimensionality | Probability |
|---|---|
| 1 | 0.6827 |
| 2 | 0.3935 |
| 3 | 0.1987 |
| 4 | 0.0902 |
| 5 | 0.0374 |
| 6 | 0.0144 |
| 7 | 0.0052 |
| 8 | 0.0018 |
| 9 | 0.0006 |
| 10 | 0.0002 |
Our fraction for the fake data is actually closer to the 40% than the real 5.9 keV data! I suppose a difference is visible in the first place, due to us always looking at the shorter axis. The actual standard deviation of our cluster is the average between the transverse and the longitudinal RMS after all! So we expect to capture less than the expected 39.35%!
1.21.
[ ]Rerun the tracking log info![ ]re evaluate the fake datasets & efficiencies in general using the non skewed data!
[X]Understand weird filtering leaving some unwanted events in there -> Ohhh, the reason is that we filter on event numbers only, and not the actual properties of the clusters! Because one event can have multiple clusters. One of them will be passing, but the other likely not.
Ok, finally done with all the hick hack of coming up with working fake event generation etc etc.
As it seems right now:
Fake 5.9 keV data using correct diffusion gives 'identical' results in terms of efficiency than real CAST data. I.e. using the gain fit is correct and useful.
For the 3.0 keV case it is not as simple. The fake data is considered 'more' X-ray like (larger cut values), but quite clearly they don't fit onto the same slope as the 5.9 keV data!
What therefore might be a reasonable option:
- Generate X-ray data for all lines below 5.9 keV
- Use the generated 'runs' to fit the gas gain curve for each dataset
- Use that gas gain curve for each energy range. Lower energy ranges are likely to have somewhat shallower gas gain dependencies? Or maybe it's events with shorter absorption length. We'll have to test.
[ ]Include one other energy, e.g. 930 eV, due to the very low absorption length. See how that behaves.
[ ]Generalize the effective efficiency code to also include other CDL lines
[ ]Rerun effective eff code and place efficiencies and plots somewhere[ ]Rerun old skewness model with--predictoption intrain_ingrid[X]Train a new MLP with same parameters as 2500 hidden neuron model, but using the corrected skewness data!
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/28_03_23_hidden_2500_fixed_skew/trained_model_hidden_2500_fixed_skew.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/hidden_2500neurons_fixed_skew/ \ --numHidden 2500
-> This was done, but we haven't looked at it yet!
[ ]In https://www.youtube.com/watch?v=kCc8FmEb1nY Andrej mentions 3e-4 is a good learning rate for AdamW. We've only tried Adam. Let's try AdamW as well.
1.21.1. DONE Fix the memory corruption bug
I think we managed to fix the memory corruption bug that plagued
us. The code that now includes also a 1 keV data line (fake and CDL)
crashed essentially every single time. In cppstl we put back the
original destructor code (i.e. that does nothing in Nim land) and
modified the NN code such that it compiles:
Instead of relying on emitTypes which, as we know, caused issues
with the shared_ptr file that is generated not knowing about
MLPImpl. In order to get anything to work we tried multiple
different things, but in the end the sanest solution seems to be to
write an actual C++ header file for the model definition and then
wrap that using the header pragma. So the code now looks as
follows:
type MLPImpl* {.pure, header: "mlp_impl.hpp", importcpp: "MLPImpl".} = object of Module hidden*: Linear classifier*: Linear MLP* = CppSharedPtr[MLPImpl] proc init*(T: type MLP): MLP = result = make_shared(MLPImpl) result.hidden = result.register_module("hidden_module", init(Linear, 13, 500)) result.classifier = result.register_module("classifier_module", init(Linear, 500, 2))
with the header file:
#include "/home/basti/CastData/ExternCode/flambeau/vendor/libtorch/include/torch/csrc/api/include/torch/torch.h" struct MLPImpl: public torch::nn::Module { torch::nn::Linear hidden{nullptr}; torch::nn::Linear classifier{nullptr}; }; typedef std::shared_ptr<MLPImpl> MLP;
(obviously the torch path should not be hardcoded). When compiling
this it again generates a .cpp file for the smartptrs Nim
module, but now it looks as follows:
#include "nimbase.h" #include <memory> #include "mlp_impl.hpp" #include "/home/basti/CastData/ExternCode/flambeau/vendor/libtorch/include/torch/csrc/api/include/torch/torch.h" #undef LANGUAGE_C #undef MIPSEB #undef MIPSEL #undef PPC #undef R3000 #undef R4000 #undef i386 #undef linux #undef mips #undef near #undef far #undef powerpc #undef unix #define nimfr_(x, y) #define nimln_(x, y) typedef std::shared_ptr<MLPImpl> TY__oV7GoY52IhMupsxgwx3HYQ; N_LIB_PRIVATE N_NIMCALL(void, eqdestroy___nn95predict_28445)(TY__oV7GoY52IhMupsxgwx3HYQ& dst__cnkLD5UfZbclV0XFs9bD47w) { }
so it contains the include required for the type definition, which
makes it all work without any memory corruption now I believe!
Note: it might be a good idea to change the current Flambeau code
to not use emitTypes but instead to write a header file in the
same form as above (having to include the Torch path!) and then use
the header pragma in the same way I do. This should be pretty simple
to do I believe and it would automate it.
1.22.
We'll continue from yesterday:
[X]Rerun the tracking log info![ ]re evaluate the fake datasets & efficiencies in general using the non skewed data![X]Include one other energy, e.g. 930 eV, due to the very low absorption length. See how that behaves.[ ]Generalize the effective efficiency code to also include other CDL lines[ ]Rerun effective eff code and place efficiencies and plots somewhere[ ]Rerun old skewness model with--predictoption intrain_ingrid[X]In https://www.youtube.com/watch?v=kCc8FmEb1nY Andrej mentions 3e-4 is a good learning rate for AdamW. We've only tried Adam. Let's try AdamW as well.
New TODOs for today:
[ ]Generate a plot of the cut values that contains all types of data in one with color being the data type
1.22.1. DONE Rerun tracking log info
We only need to rerun the tracking log info, so we can just do:
./runAnalysisChain \ -i ~/CastData/data \ --outpath ~/CastData/data \ --years 2017 --years 2018 \ --back --tracking
1.22.2. Re-evaluate fake & efficiency data using non skewed data
We re ran the code yesterday and today again
1.22.3. STARTED Train MLP with AdamW
Using the idea from Andrej, let's first train AdamW with a learning rate of 1e-3 and then with 3e-4.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/29_03_23_adamW_2500_1e-3/trained_model_adamW_2500_1e-3.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/adamW_2500_1e-3/ \ --numHidden 2500 \ --learningRate 1e-3
This one seems to achieve:
Train set: Average loss: 0.0002 | Accuracy: 1.000 Test set: Average loss: 0.0132 | Accuracy: 0.9983 Epoch is:15050
and from here nothing is changing anymore. I guess we're pretty much approaching the best possible separation. The training data must be overtrained already after all, with an accuracy at 1. :O
And the second model with 3e-4:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/29_03_23_adamW_2500_3e-4/trained_model_adamW_2500_3e-4.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/adamW_2500_3e-4/ \ --numHidden 2500 \ --learningRate 3e-4
1.22.4. What plots & numbers to generate
- Verification of the fake data generation
[ ]ridge line plots comparing real data (as we have it) with- fake data using pure remove pixels
- fake data using correct diffusion behavior
- Definition of cut values and gain dependence
[ ]plot showing 5.9 keV data linear dependence of gain & cut value[ ][ ]Numbers for effective efficiency comparing real data & fake diffusion data. 5.9 line matches essentially exactly![ ]For each CDL dataset:- plot of CDL data + fake data w/ diffusion
[ ]Some plot correlating NN cut value, fake data, gas gain behavior & absorption length. Something something.
- Difference in Run-2 and Run-3 behavior
1.22.5. Fake data generation
It seems like when generating very low energy events (1 keV) the
diffusion we simulate is significantly larger than what is seen in the
CDL data, see:
(consider length, width, RMS plots)
This is using
df.add ctx.handleFakeData(c, "8_Fake1.0", 0.93, FakeDesc(kind: fkDiffusion, λ: 1.0 / 7.0))
Of course it is quite possible that the 1/7 is not quite right (we don't compute it ourselves yet after all). But even if it was 1/6 or 1/5 it wouldn't change anything significantly. The CDL events are simply quite a bit smaller.
But of course, note that the CDL data has much fewer hits in it than the equivalent fake data. This will likely strongly impact what we would see. The question of course is whether the change is due to fewer electrons or also just less diffusion.
The CDL data was mostly taken with a hotter detector. With rising
temperatures the diffusion should increase though, no? At least
according to the PyBoltz simulation (see
sec. [BROKEN LINK: sec:simulation:diffusion_coefficients_cast] in
StatusAndProgress).
I'm not quite sure what to make of that.
Let's rerun the fake data gen code for 1 keV with a lower diffusion,
e.g. 540. We generate the plots in /tmp/
The result is slightly too small:
See the RMS and width / length plots.
Let's try 580.
So somewhere in the middle. Maybe 560 or 570.
According to Magboltz (see below) the value should indeed lie somewhere around 660 or so even in the case of 1052 mbar pressure as seen in the CDL data. PyBoltz gives smaller numbers, but still larger than 620.
One interesting thought:
[X]What does the average length look like for a cluster with even less energy than the 930 eV case? One similar to the number of hits of the CDL 930 eV data? -> It clearly seems like at least the RMS values are fully unaffected by having less hits. The width and length become slightly smaller, but not significantly as to this be the deciding factor between the CDL data and fake data.
1.22.6. DONE Testing Magboltz & computing diffusion based on pressure
IMPORTANT: The description on https://magboltz.web.cern.ch/magboltz/usage.html seems to be WRONG. In it it says the first 'input card' has 3 inputs. But nowadays apparently it has 5 inputs.
Compile:
gfortran -o magboltz -O3 magboltz-11.16.f
Argon isobutane test file at 25°C and 787.6 Torr = 1050 mbar and an electric field of 500 V/cm.
2 5 0 1 0.0 2 11 80 80 80 80 97.7 2.3 0.0 0.0 0.0 0.0 25.0 787.6 500.0 0.0 0.0 0
I ran it with different input files now and also ran PyBoltz in different cases.
All cases are the same gas and voltage, and at 25°C.
1050 mbar, 5e7 collisions (same as above):
2 5 0 1 0.0 2 11 80 80 80 80 97.7 2.3 0.0 0.0 0.0 0.0 25.0 787.6 500.0 0.0 0.0 0
./resources/magboltz_results/output_optimized_ar_iso_1050mbar_5e7.txt The main results section:
Z DRIFT VELOCITY = 0.2285E+02 MICRONS/NANOSECOND +- 0.06%
Y DRIFT VELOCITY = 0.0000E+00 MICRONS/NANOSECOND +- 0.00%
X DRIFT VELOCITY = 0.0000E+00 MICRONS/NANOSECOND +- 0.00%
DIFFUSION IN CM**2/SEC.
TRANSVERSE DIFFUSION = 0.4380D+04 +- 13.96%
= 0.95831 EV. +- 13.956%
= 619.132 MICRONS/CENTIMETER**0.5 +- 6.98%
LONGITUDINAL DIFFUSION = 0.7908D+03 +- 5.8%
= 0.1730 EV. +- 5.84%
= 263.090 MICRONS/CENTIMETER**0.5 +- 2.92%
1052 mbar, 5e7 collisions:
2 5 0 1 0.0 2 11 80 80 80 80 97.7 2.3 0.0 0.0 0.0 0.0 25.0 789.0 500.0 0.0 0.0 0
./resources/magboltz_results/output_optimized_ar_iso_1052mbar_5e7.txt Results:
Z DRIFT VELOCITY = 0.2287E+02 MICRONS/NANOSECOND +- 0.06%
Y DRIFT VELOCITY = 0.0000E+00 MICRONS/NANOSECOND +- 0.00%
X DRIFT VELOCITY = 0.0000E+00 MICRONS/NANOSECOND +- 0.00%
DIFFUSION IN CM**2/SEC.
TRANSVERSE DIFFUSION = 0.3984D+04 +- 9.35%
= 0.87119 EV. +- 9.350%
= 590.320 MICRONS/CENTIMETER**0.5 +- 4.67%
LONGITUDINAL DIFFUSION = 0.7004D+03 +- 7.8%
= 0.1531 EV. +- 7.76%
= 247.499 MICRONS/CENTIMETER**0.5 +- 3.88%
1050 mbar, 1e8 collisions:
2 10 0 1 0.0 2 11 80 80 80 80 97.7 2.3 0.0 0.0 0.0 0.0 25.0 787.6 500.0 0.0 0.0 0
./resources/magboltz_results/output_optimized_ar_iso_1050mbar_1e8.txt Results:
Z DRIFT VELOCITY = 0.2286E+02 MICRONS/NANOSECOND +- 0.05%
Y DRIFT VELOCITY = 0.0000E+00 MICRONS/NANOSECOND +- 0.00%
X DRIFT VELOCITY = 0.0000E+00 MICRONS/NANOSECOND +- 0.00%
DIFFUSION IN CM**2/SEC.
TRANSVERSE DIFFUSION = 0.5027D+04 +- 6.39%
= 1.09929 EV. +- 6.394%
= 663.109 MICRONS/CENTIMETER**0.5 +- 3.20%
LONGITUDINAL DIFFUSION = 0.8695D+03 +- 11.6%
= 0.1901 EV. +- 11.59%
= 275.781 MICRONS/CENTIMETER**0.5 +- 5.79%
1052 mbar, 1e8 collisions:
2 10 0 1 0.0 2 11 80 80 80 80 97.7 2.3 0.0 0.0 0.0 0.0 25.0 789.0 500.0 0.0 0.0 0
./resources/magboltz_results/output_optimized_ar_iso_1052mbar_1e8.txt Results:
Z DRIFT VELOCITY = 0.2288E+02 MICRONS/NANOSECOND +- 0.06%
Y DRIFT VELOCITY = 0.0000E+00 MICRONS/NANOSECOND +- 0.00%
X DRIFT VELOCITY = 0.0000E+00 MICRONS/NANOSECOND +- 0.00%
DIFFUSION IN CM**2/SEC.
TRANSVERSE DIFFUSION = 0.4960D+04 +- 6.39%
= 1.08401 EV. +- 6.386%
= 658.486 MICRONS/CENTIMETER**0.5 +- 3.19%
LONGITUDINAL DIFFUSION = 0.6940D+03 +- 10.3%
= 0.1517 EV. +- 10.26%
= 246.304 MICRONS/CENTIMETER**0.5 +- 5.13%
1050 mbar, 3e8 collisions:
2 30 0 1 0.0 2 11 80 80 80 80 97.7 2.3 0.0 0.0 0.0 0.0 25.0 787.6 500.0 0.0 0.0 0
./resources/magboltz_results/output_optimized_ar_iso_1050mbar_3e8.txt Results:
Z DRIFT VELOCITY = 0.2285E+02 MICRONS/NANOSECOND +- 0.02%
Y DRIFT VELOCITY = 0.0000E+00 MICRONS/NANOSECOND +- 0.00%
X DRIFT VELOCITY = 0.0000E+00 MICRONS/NANOSECOND +- 0.00%
DIFFUSION IN CM**2/SEC.
TRANSVERSE DIFFUSION = 0.5062D+04 +- 2.83%
= 1.10750 EV. +- 2.826%
= 665.582 MICRONS/CENTIMETER**0.5 +- 1.41%
LONGITUDINAL DIFFUSION = 0.6860D+03 +- 3.8%
= 0.1501 EV. +- 3.78%
= 245.029 MICRONS/CENTIMETER**0.5 +- 1.89%
1052 mbar, 3e8 collisions:
2 30 0 1 0.0 2 11 80 80 80 80 97.7 2.3 0.0 0.0 0.0 0.0 25.0 789.0 500.0 0.0 0.0 0
./resources/magboltz_results/output_optimized_ar_iso_1052mbar_3e8.txt
Z DRIFT VELOCITY = 0.2286E+02 MICRONS/NANOSECOND +- 0.02%
Y DRIFT VELOCITY = 0.0000E+00 MICRONS/NANOSECOND +- 0.00%
X DRIFT VELOCITY = 0.0000E+00 MICRONS/NANOSECOND +- 0.00%
DIFFUSION IN CM**2/SEC.
TRANSVERSE DIFFUSION = 0.5016D+04 +- 4.98%
= 1.09691 EV. +- 4.982%
= 662.394 MICRONS/CENTIMETER**0.5 +- 2.49%
LONGITUDINAL DIFFUSION = 0.7799D+03 +- 5.0%
= 0.1705 EV. +- 4.98%
= 261.183 MICRONS/CENTIMETER**0.5 +- 2.49%
Compare the transverse diffusion coefficients and their uncertainty. Magboltz is very bad at estimating uncertainties… The final numbers using 3e8 collisions seems to be the most likely correct number.
PyBoltz does not very any better, it actually is worse. When running with 5e7 it spits out numbers from 640 (1050) to 520 (1052)! At also 3e8 it says (./../src/python/PyBoltz/examples/test_argon_isobutane.py)
Running with Pressure: 787.6 Input Decor_Colls not set, using default 0 Input Decor_LookBacks not set, using default 0 Input Decor_Step not set, using default 0 Input NumSamples not set, using default 10 Trying 5.6569 Ev for final electron energy - Num analyzed collisions: 29900000 Calculated the final energy = 5.6568542494923815 Velocity Position Time Energy DIFXX DIFYY DIFZZ 22.9 2.0 86520417.1 1.1 2587.6 4848.9 0.0 22.9 4.0 172684674.2 1.1 3408.6 4620.1 0.0 22.9 5.9 259670177.0 1.1 4610.1 4246.2 565.9 22.9 7.9 346216335.9 1.1 4754.5 4376.4 530.8 22.9 9.9 432794455.8 1.1 4330.8 4637.1 589.1 22.9 11.9 519476567.6 1.1 4518.5 4490.4 681.0 22.9 13.9 606130691.2 1.1 4794.2 4499.9 661.0 22.9 15.9 692948117.3 1.1 5149.8 4469.2 687.0 22.9 17.8 779615117.3 1.1 5307.6 4350.5 650.8 22.9 19.8 866106715.1 1.1 5072.8 4376.6 644.3 Running with Pressure: 789.0 Trying 5.6569 Ev for final electron energy - Num analyzed collisions: 29900000 Calculated the final energy = 5.6568542494923815 Velocity Position Time Energy DIFXX DIFYY DIFZZ 22.9 2.0 86667248.5 1.1 3048.9 3483.7 0.0 22.9 4.0 173345020.6 1.1 4088.2 4967.2 0.0 22.9 5.9 260048909.2 1.1 3789.2 5445.7 495.5 22.9 7.9 346875330.4 1.1 4351.8 5540.9 631.9 22.9 9.9 433290608.8 1.1 3928.3 5060.4 979.6 22.9 11.9 519876593.3 1.1 4255.1 4938.9 910.2 22.9 13.9 606223061.1 1.1 4085.9 4610.8 862.2 22.9 15.9 693197771.7 1.1 4050.8 4661.8 818.2 22.9 17.9 780134417.2 1.1 4260.6 4753.5 803.4 22.9 19.9 866928793.5 1.1 4149.1 4876.4 808.5 time taken1544.6015286445618 α = 0.0 E = 500.0, P = 787.6, V = 22.88327213959691, DT = 4724.663347492957 DT1 = 642.6009590747105, DL = 644.3327626774859, DL1 = 237.30726957910778 α = 0.0 E = 500.0, P = 789.0, V = 22.912436886892305, DT = 4512.734382794393 DT1 = 627.6235672201768, DL = 808.4762345648321, DL1 = 265.65193649208607
642 vs 627. Still a rather massive difference here!
All this is very annoying, but we can be sure that higher pressures lead to lower diffusion. The extent that this is visible in the CDL data though seems to imply that there is something else going on at the same time.
1.22.7. TODO Think about rmsTransverse cuts
Christoph used the transverse RMS cuts at about 1.0 or 1.1 as the 'X-ray cleaning cuts'.
In the paper ./Papers/gridpix_energy_dependent_features_diffusion_krieger_1709.07631.pdf he reports a diffusion coefficient of ~470μm/√cm which is way lower than what we get from Magboltz.
With that number and the plots presented in that paper the 1.0 or 1.1 RMS transverse number is justifiable. However, in our data it really seems like we are cutting away some good data for the CDL data when cutting it (or similarly when we apply those cleaning cuts elsewhere).
I'm not sure how sensible that is.
However, one interesting idea would be to look at the 2014/15 data under the same light as done in the effective efficiency tool, i.e. plot a ridgeline of the distributions for escape and photo peaks. Do we reproduce the same RMS transverse numbers that Christoph gets?
One possibility is that our transverse RMS number is calculated differently?
If our code reproduces Christophs RMS values -> in the data, if not -> in the algorithm.
1.22.8. TODO Look at dependence of NN cut value depending on diffusion coefficient & absorption length
If we make the same plot as for the gas gain but using the diffusion coefficient & the absorption length, but leaving everything else the same, how does the cut value change?
[ ]NN cut value @ desired efficiency vs diffusion coefficient (different coefficient fake data!)[ ]NN cut value @ desired efficiency vs absorption length (different absorption length fake data!)[ ]NN cut value @ desired efficiency vs energy at real absorption lengths from fake data
1.23.
Can we make a fit to the rms transverse data of each 55Fe run, then use that upper limit of the transverse RMS to compute the diffusion and finally determine the cut value based on fake data with that diffusion and gas gain?
Goal: Determine NN cut value to use for a given cluster to achieve an efficiency of a desired value.
I have:
- CDL data that can be used to determine cut values for different energy. They have different gas gains and diffusion coefficients.
- Fake data for arbitrary energies, absorption lengths and diffusion coefficients
- real 5.9 keV data at different gas gains and diffusion coefficients.
What do I need: A relationship that maps a cut value from a CDL energy range to a cluster of different diffusion and gas gain.
How do I understand the dependence of diffusion and gas gain on NN cut value? Use upper percentile (e.g. 95) of rmsTransverse as an easy to use proxy for the diffusion in a dataset. Compute that value for each CDL run. Do same for every 55Fe run. Plot gas gain vs rmsTransverse for all this data.
Thoughts on diffusion & gas gain:
[ ]INSERT PLOT OF RMS T VS GAS GAIN & CUT VAL
For hidden 2500 with skewed fixed:
- ./Figs/statusAndProgress/neuralNetworks/development/hidden_2500neurons_fixed_skew/rmsTransverse_vs_NN_cutVal.pdf
- ./Figs/statusAndProgress/neuralNetworks/development/hidden_2500neurons_fixed_skew/rmsTransverse_vs_gas_gain.pdf
For AdamW@3e-4 lr:
- ./../../../org/Figs/statusAndProgress/neuralNetworks/development/adamW_2500_3e-4/rmsTransverse_vs_NN_cutVal.pdf
- ./../../../org/Figs/statusAndProgress/neuralNetworks/development/adamW_2500_3e-4/rmsTransverse_vs_gas_gain.pdf
Higher gas gains are associated with longer transverse RMS values and lower NN cut values. Higher gas gains are associated with lower temperatures. Lower temperatures mean higher densities at the same pressure. Higher densities imply shorter absorption lengths. Shorter absorption lengths imply longer drift distances. Longer drift distances imply larger diffusion. Larger diffusion implies larger rms transverse.
So it may not actually be that the gas diffusion changes significantly (or only?), but that the change in density implies a change in average diffusion value.
Keep in mind that the crosses for 3 keV are the escape photons and not real 3 keV data!
The two are certainly related though.
Maybe rms transverse of CDL data is different due to different pressure?
[ ]Make the same plot but instead of 3 keV escape photons generate 3 keV events at different diffusions
1.23.1. STARTED Train MLP with rmsTransverse cutoff closer to 1.2 - 1.3
This changed the rms transverse cut in the X-ray cleaning cuts and logL cuts to 1.2 to 1.3 (depending on energy range).
To see whether a larger rms transverse in the training data changes the way the model sees something as X-ray.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/30_03_23_adamW_2500_3e-4_largerRmsT/trained_model_adamW_2500_3e-4_largerRmsT.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/adamW_2500_3e-4_largerRmsT/ \ --numHidden 2500 \ --learningRate 3e-4
And with SGD once more:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/30_03_23_SGD_2500_3e-4_largerRmsT/trained_model_SGD_2500_3e-4_largerRmsT.pt \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/development/SGD_2500_3e-4_largerRmsT/ \ --numHidden 2500 \ --learningRate 3e-4
[ ]Think about introducing dropout layer? So that we might reduce overtraining especially in the AdamW case?
[ ]Plot all datasets against the cut value. For the AdamW model different rmsT values are almost independent of the cut value. For SGD it is extremely linear.[X]Plot raw NN prediction value against all datasets. -> This one is not so helpful (but we still generate it), but more useful is a version that only looks at the mean values of the lower, mid and upper 33% quantiles.
Interestingly the different models react quite differently in terms of what affects the cut efficiency!
[ ]Add plots
[ ]Try generating fake data and determining cut value from that, then use it on 55Fe
[ ]Why not just generate fake data at the energies used in CDL for all runs and use those as reference for cut?
1.24.
[ ]VerifyplotDatasetsdistributions of all fake data events! -> make this plot comparing CDL & fake data of same 'kind' by each set
1.24.1. Diffusion from data rmsTransverse
./../CastData/ExternCode/TimepixAnalysis/Tools/determineDiffusion/determineDiffusion.nim
-> Very useful! Using it now to fit to the rms transverse dataset to extract the diffusion from real data runs. Then generate fake data of a desired energy that matches this diffusion. It matches very well it seems!
We should make plots of the data distributions for the real vs fake data, but also on a single run by run basis.
1.25.
Interesting observation:
Today we were trying to debug the reason why our sampled data seems to have a mean in the centerX and in particular centerY position that is not centered at ~7mm (128). Turns out, our original real data from which we sample has the same bias in the centerY position. Why is that? Don't we apply the same cuts in fake gen data reading as in the effective efficiency code?
-> The only difference between the two sets of cuts in the data reading is that in the fake data reading we do not apply an energy cut to the data. We only apply the xray cleaning cuts!
1.26.
Finally implemented NN veto with run local cut values based on fake
data in likelihood
[ ]INSERT FIGURES OF EFFECTIVE EFFICIENCY USING FAKE CUT VALUES
likelihood \ -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out /tmp/testing/run2_only_mlp_local_0.95.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/nn_devel_mixing/30_03_23_SGD_2500_3e-4_largerRmsT/trained_model_SGD_2500_3e-4_largerRmsT.pt \ --nnSignalEff 0.95 \ --nnCutKind runLocal \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --readonly
Ah, but we cannot directly sample fake events in our current approach for the background datasets, because we still rely on real events as a starting point for the sampling.
Therefore: develop sampling of full events using gas gain and polya sampling.
- check what info is stored in H5 files regarding polya and gas gain -> nothing useful about threshold
- SCurve should in theory tell us something about threshold
[ ]investigate if all polyas in a single file (Run-2 or Run-3) have the same threshold if determined from data, e.g. using quantile 1
- sample from existing fixed parameter polya and check it looks reasonable
- open H5 file run by run, look at minima and quantile 1 data for each gas gain slice. -> The minima is fixed for each set of chip calibrations! Run-2: Run-2: minimum: 1027.450870326596, quantile 1: 1247.965130336289 or 1282.375318596229 Run-3: minimum: 893.4812944899318, quantile 1: 1014.494678806292 or 1031.681385601604 -> this is useful! Means we can just sample with fixed cutoffs for each dataset and don't need raw charge data for anything for a run, but only gas gain slice fit parameters! -> Raises question though: How does then this relate to # of hits in photo peak? Lower gain, closer to cutoff, thus less pixels, but opposite? How do we get to too many pixels?
- plot photo peak hit positions against gas gain
- sample from a poyla using parameters as read from gas gain slices, plot against polya dataset -> Looks reasonable. Now need to include the cutoff. -> Looks even better. Note that the real data looks ugly due to the equal bin widths that are not realistic.
Ahh! Idea: we can reuse the gas gain vs energy calibration factor fit! We get the gas gain of a run for which to generate fake data. That gas gain is fed into that fit. The result is a calibration factor that tells us the charge corresponding to a given energy (or its inverse). Place our target energy into the function to get the desired charge. Then sample from a normal distribution around the target charge as the target for each event. Generate pixels until total charge close to target. -> Until total charge close or alternatively: Given gas gain calculate number based on target charge: e.g. 600,000 target and gain of 3,000) then target hits is target / per hit = 200 in this case. In this case we can get less due to threshold effects, but not more. So: better to do the former? Draw from target charge distribution and then accumulate pixels until the total drawn is matched?
Question: In the real data, are the total charge and the number of hits strongly correlated? It's important to understand how to go from a deposited energy to a number of electrons. There are multiple reasons to lose electrons and charge from a fixed input energy:
- amplifications below the threshold of a pixel
- higher / lower than average ionization events than Wi = 26 eV
We can model the former, but the latter is tricky.
1.27.
Meeting with Klaus today:
Main take away: Train an MLP using purely generated data (for X-rays) with:
- an extra input neuron that corresponds to the transverse diffusion of the 'dataset' each.
- different MLPs, one for each input of the diffusion parameter
The former seems better to me. Just generate data with a uniform distribution in some range of diffusion parameters. Then each input actually has different values. When actually applying the network then we only have few distinct values (one for each run) of course. But that's fine.
Start a training run with the σT dataset!
With 100,000 generated events for each calibration file (why?). Still using the fixed cutoff of Run-3! This also uses a uniform distribution between G = 2400 .. 4500 σT = 550 .. 700 θ = 2.1 .. 2.4
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/12_04_23_sgd_sim_data_diffusion/trained_mlp_sgd_sim_data_diffusion.pt \ --plotPath ~/Sync/sgd_sim_data_diffusion/ \ --numHidden 2500 \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets σT \ --learningRate 3e-4 \ --simulatedData
At first glance it seems like the cut values determined from generating more fake data of the σT and gain of the real runs yields effective efficiencies that are all too high (95%, only some CDL runs approaching 80% target).
Now with normal distribution in G and σT:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/12_04_23_sgd_sim_gauss_diffusion/trained_mlp_sgd_sim_gauss_diffusion.pt \ --plotPath ~/Sync/sgd_sim_gauss_diffusion/ \ --numHidden 2500 \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets σT \ --learningRate 3e-4 \ --simulatedData
-> Continue training this if this is useful. Still a lot of downward trend in loss visible!
In addition it may be a good idea to also try it with the gain as another input?
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/12_04_23_sgd_sim_diffusion_gain/trained_mlp_sgd_sim_diffusion_gain.pt \ --plotPath ~/Sync/sgd_sim_diffusion_gain/ \ --numHidden 2500 \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets σT \ --datasets gasGain \ --learningRate 3e-4 \ --simulatedData
1.28.
[ ]Understand effective efficiency with gas gain parameter ofFakeDesc. E.g. rerun the gauss diffusion network again after changing code etc[ ]verify the number of neighboring pixels that are actually active anywhere! -> We'll write a short script that extracts that information from a given file.
[ ]Correlate with gas gain and extracted diffusion![ ]Compare with generated fake data without artificial neighbor activation and with![ ]How do neighbors relate in their charge?
In ./../CastData/ExternCode/TimepixAnalysis/Tools/countNeighborPixels:
./countNeighborPixels -f ~/CastData/data/CalibrationRuns2017_Reco.h5
Average neighbors in run 110 = 55.09144736842105 Average neighbors in run 175 = 57.15461200585651 Average neighbors in run 122 = 47.4565240584658 Average neighbors in run 126 = 52.52208235545125 Average neighbors in run 183 = 59.16529930112428 Average neighbors in run 161 = 66.33755847291384 Average neighbors in run 116 = 51.58539765319426 Average neighbors in run 155 = 68.88583638583638 Average neighbors in run 173 = 57.67820710973725 Average neighbors in run 151 = 66.58298001211386 Average neighbors in run 153 = 68.5710128055879 Average neighbors in run 108 = 55.27580484566877 Average neighbors in run 93 = 55.01024327784891 Average neighbors in run 147 = 61.76867469879518 Average neighbors in run 179 = 63.32529743268628 Average neighbors in run 159 = 66.94385479157053 Average neighbors in run 163 = 63.48872858431019 Average neighbors in run 118 = 52.85084521047398 Average neighbors in run 102 = 54.66866666666667 Average neighbors in run 177 = 56.74977000919963 Average neighbors in run 181 = 60.51956253850894 Average neighbors in run 165 = 59.47996965098634 Average neighbors in run 167 = 61.3946587537092 Average neighbors in run 185 = 57.19969558599696 Average neighbors in run 149 = 67.73385167464114 Average neighbors in run 157 = 69.89698937426211 Average neighbors in run 187 = 58.77159520807061 Average neighbors in run 171 = 59.13408330799635 Average neighbors in run 128 = 52.05005608524958 Average neighbors in run 169 = 58.22026431718061 Average neighbors in run 145 = 61.53792611101196 Average neighbors in run 83 = 51.74536148432502 Average neighbors in run 88 = 46.48597521200261 Average neighbors in run 120 = 49.65804645033767 Average neighbors in run 96 = 51.49257633765991
./countNeighborPixels -f ~/CastData/data/CalibrationRuns2018_Reco.h5
Average neighbors in run 304 = 83.96286764705883 Average neighbors in run 286 = 83.78564713256033 Average neighbors in run 294 = 84.31031159653068 Average neighbors in run 277 = 89.82936363636364 Average neighbors in run 241 = 81.1551888289432 Average neighbors in run 284 = 89.4854306756324 Average neighbors in run 260 = 85.9630966706779 Average neighbors in run 255 = 88.56640625 Average neighbors in run 292 = 85.56208945886769 Average neighbors in run 288 = 80.55608820709492 Average neighbors in run 247 = 77.27826358525921 Average neighbors in run 257 = 88.13119266055045 Average neighbors in run 239 = 73.20087064676616 Average neighbors in run 302 = 86.77864992150707 Average neighbors in run 249 = 79.53715365239294 Average neighbors in run 271 = 79.5505486808312 Average neighbors in run 300 = 90.83248730964468 Average neighbors in run 296 = 85.71424050632912 Average neighbors in run 243 = 80.90277344967279 Average neighbors in run 264 = 83.18354637823664 Average neighbors in run 280 = 88.5948709880428 Average neighbors in run 253 = 85.09293373659609 Average neighbors in run 251 = 77.57475909232204 Average neighbors in run 262 = 82.7622203811102 Average neighbors in run 290 = 79.6481004507405 Average neighbors in run 275 = 89.19542053956019 Average neighbors in run 269 = 82.60137931034483 Average neighbors in run 266 = 83.25234248788368 Average neighbors in run 273 = 81.36689741976086 Average neighbors in run 245 = 78.7516608668143 Average neighbors in run 259 = 86.49635416666666 Average neighbors in run 282 = 90.84639199809479
And using fake data, for the case of no simulated neighbors: Run-2
./countNeighborPixels -f ~/CastData/data/CalibrationRuns2017_Reco.h5 --fake
Average neighbors in run 161 = 38.16306522609044 Average neighbors in run 128 = 29.87515006002401 Average neighbors in run 183 = 34.27851140456183 Average neighbors in run 185 = 33.6796718687475 Average neighbors in run 88 = 28.7222 Average neighbors in run 179 = 34.7138 Average neighbors in run 187 = 32.83693477390956 Average neighbors in run 155 = 39.0502 Average neighbors in run 163 = 35.952 Average neighbors in run 118 = 30.9526 Average neighbors in run 171 = 32.7596 Average neighbors in run 126 = 29.8646 Average neighbors in run 151 = 37.3554 Average neighbors in run 169 = 32.7694 Average neighbors in run 120 = 28.37575030012005 Average neighbors in run 102 = 30.4712 Average neighbors in run 159 = 37.93177270908363 Average neighbors in run 153 = 38.0882 Average neighbors in run 157 = 40.6742 Average neighbors in run 167 = 34.4742 Average neighbors in run 96 = 29.2946 Average neighbors in run 175 = 33.7786 Average neighbors in run 177 = 33.1542 Average neighbors in run 93 = 31.7502 Average neighbors in run 116 = 29.2908 Average neighbors in run 83 = 29.9446 Average neighbors in run 145 = 34.6312 Average neighbors in run 147 = 34.9114 Average neighbors in run 108 = 31.13945578231293 Average neighbors in run 122 = 27.9868 Average neighbors in run 181 = 34.3359343737495 Average neighbors in run 165 = 32.90716286514606 Average neighbors in run 173 = 32.5428 Average neighbors in run 149 = 38.0322 Average neighbors in run 110 = 31.2116
Run-3
./countNeighborPixels -f ~/CastData/data/CalibrationRuns2018_Reco.h5 --fake
Average neighbors in run 284 = 41.75550220088035 Average neighbors in run 259 = 39.1072 Average neighbors in run 292 = 39.506 Average neighbors in run 286 = 39.2786 Average neighbors in run 239 = 37.4598 Average neighbors in run 288 = 37.3952 Average neighbors in run 251 = 36.9704 Average neighbors in run 255 = 40.5622 Average neighbors in run 262 = 39.1088 Average neighbors in run 260 = 40.5608 Average neighbors in run 294 = 38.376 Average neighbors in run 280 = 40.51660664265706 Average neighbors in run 271 = 37.5112 Average neighbors in run 296 = 39.0084 Average neighbors in run 275 = 40.3642 Average neighbors in run 269 = 38.63645458183273 Average neighbors in run 302 = 38.698 Average neighbors in run 304 = 38.2386 Average neighbors in run 266 = 37.8624 Average neighbors in run 243 = 37.6958 Average neighbors in run 264 = 39.4124 Average neighbors in run 257 = 40.6936 Average neighbors in run 282 = 41.2402 Average neighbors in run 290 = 37.7166 Average neighbors in run 277 = 40.49369369369369 Average neighbors in run 253 = 38.238 Average neighbors in run 249 = 38.3926 Average neighbors in run 273 = 38.073 Average neighbors in run 247 = 37.3596 Average neighbors in run 245 = 36.2114 Average neighbors in run 241 = 37.1948 Average neighbors in run 300 = 39.7052
So the number of neighbors:
| Period | Type | ~Neighbors per event |
|---|---|---|
| Run-2 | Real | 50-60 |
| Run-3 | Real | 80-90 |
| Run-2 | Fake | 30-35 |
| Run-3 | Fake | 35-40 |
Activating neighbor sharing we can get those numbers up, but that's for later.
Next: Plot charge of pixels with neighbors against something.
Note: For run 288 the following:
if charge > 3500.0: # whatever # possibly activate a neighbor pixel! let activateNeighbor = rnd.rand(1.0) < 0.5 if activateNeighbor: let neighbor = rand(3) # [up, down, right, left] let totNeighbor = rnd.sample(psampler) / 2.0 # reduce amount case neighbor
yields very good agreement already, with the exception of the hits
data if the gain.G * 0.85 is not used (but instead G itself).
1.29.
In ./../CastData/ExternCode/TimepixAnalysis/Tools/countNeighborPixels we can now produce also histograms of the ToT values / charges recorded by the center chip (split by number of neighbors of a pixel). Three different versions, density of ToT, raw charge and density of charge.
The three are important because of the non-linearity of the ToT to charge conversion. The question is: which distribution should we really sample from to get the correct distribution of charges seen by the detector?
Running:
./countNeighborPixels -f ~/CastData/data/CalibrationRuns2017_Reco.h5 --chargePlots
yields the plots in:
We can see that the density based version of the charge histograms looks very much not like a Polya. The histogram of ToT looks somewhat like it and the raw charge one looks closest I would say.
The other thing we see here is that the real data shows a shift to larger charges for the data with more neighbors. The effect is not extreme, but visible. Much more so in the Run-3 data than in the Run-2 data though, which matches our expectations (see table from yesterday).
For the fake data:
./countNeighborPixels -f ~/CastData/data/CalibrationRuns2018_Reco.h5 --chargePlots --fake
yields: Figs/statusAndProgress/gasGainAndNeighbors/charges_neighbors_fake.pdf
At the very least the distributions currently generated do not match. Multiple reasons:
- our scaling of gas gain using G * 0.85 is bad (yes)
- currently we're sampling from a polya of the ToT values
We will now study the gas gains on a specific run, say 241. We'll try to imitate the look of the real 241 data in the fake data. Using:
./countNeighborPixels -f ~/CastData/data/CalibrationRuns2018_Reco.h5 --chargePlots --fake --run 241
we'll make changes and try to get it to look better.
Starting point:
#let gInv = invert(gain.G * 0.85, calibInfo) let gInv = invert(gain.G, calibInfo) # not using scaling # ... let ToT = rnd.sample(psampler) # sampling from ToT # and no neighbor activation logic
pdfunite *neighbor_run_charges_241* ~/org/Figs/statusAndProgress/gasGainAndNeighbors/run_241_comparison_start.pdf
First step, what does our 0.85 scaling actually do?
let gInv = invert(gain.G * 0.85, calibInfo) # ... let ToT = rnd.sample(psampler) # sampling from ToT # and no neighbor activation logic
./countNeighborPixels -f ~/CastData/data/CalibrationRuns2018_Reco.h5 --chargePlots --fake --run 241
pdfunite *neighbor_run_charges_241* ~/org/Figs/statusAndProgress/gasGainAndNeighbors/run_241_comparison_0.85_scaling.pdf
As expected, it makes the gains a bit smaller.
Notice how the ToT histogram of the real data has a much shorter tail than the fake data. At ToT = 150 it is effectively 0, but fake data still has good contribution there! Try scaling further down to see what that does.
First step, what does our 0.85 scaling actually do?
let gInv = invert(gain.G * 0.6, calibInfo) # ... let ToT = rnd.sample(psampler) # sampling from ToT # and no neighbor activation logic
./countNeighborPixels -f ~/CastData/data/CalibrationRuns2018_Reco.h5 --chargePlots --fake --run 241
pdfunite *neighbor_run_charges_241* ~/org/Figs/statusAndProgress/gasGainAndNeighbors/run_241_comparison_0.6_scaling.pdf
The tail now looks more correct (almost zero at 150 ToT in fake), but:
- the peak is way too far left compared to real data
[X]why does the real data have ToT values up to 0, but the generated data has a sharp cutoff at ~10 or so?
charge 3516.613062845426 from ToT 64.00852228399629 for 1 is 893.4812944899318 charge 1405.389895032497 from ToT 26.41913859339533 for 1 is 893.4812944899318 charge 6083.063184722204 from ToT 88.29237432771937 for 1 is 893.4812944899318
This matches my expectation, but not the data. Ah! It's because of our 1.15 scaling, no?
charge 2295.67900197231 from ToT 47.40724170682395 for 1 is 893.4812944899318 for cutoff: 1027.503488663421 inverted cutoff: 9.0 charge 4940.614026607494 from ToT 78.30781008751393 for 1 is 893.4812944899318 for cutoff: 1027.503488663421 inverted cutoff: 9.0 charge 1318.261093845941 from ToT 23.29152025807923 for 1 is 893.4812944899318 for cutoff: 1027.503488663421 inverted cutoff: 9.0 charge 2643.805171369868 from ToT 52.90409160403075 for 1 is 893.4812944899318 for cutoff: 1027.503488663421 inverted cutoff: 9.0 charge 2102.98283203606 from ToT 43.92681742421028 for 1 is 893.4812944899318 for cutoff: 1027.503488663421 inverted cutoff: 9.0
Exactly!
The ToT behavior makes me worried in one sense: I really feel like the issue is that certain pixels have different thresholds, which makes the distribution so ugly.
Next, go back to sampling from actual polya and see what that looks like (without any scaling of gas gain):
let params = @[gain.N, gain.G, gain.theta] let psampler = initPolyaSampler(params, frm = 0.0, to = 20000.0) #invert(20000.0, calibInfo)) # ... let charge = rnd.sample(psampler) # sampling from ToT let ToT = invert(charge, calibInfo)
./countNeighborPixels -f ~/CastData/data/CalibrationRuns2018_Reco.h5 --chargePlots --fake --run 241
pdfunite *neighbor_run_charges_241* ~/org/Figs/statusAndProgress/gasGainAndNeighbors/run_241_comparison_from_polya.pdf
This does look much more realistic! So sampling from the real polya seems more sensible after all I think. Maybe it's a bit too wide though?
Let's look at scaling the theta parameter down by 30% (50%)
let params = @[gain.N, gain.G, gain.theta * 0.7] # 0.5 let psampler = initPolyaSampler(params, frm = 0.0, to = 20000.0) # ... let charge = rnd.sample(psampler) # sampling from ToT let ToT = invert(charge, calibInfo)
./countNeighborPixels -f ~/CastData/data/CalibrationRuns2018_Reco.h5 --chargePlots --fake --run 241
pdfunite *neighbor_run_charges_241* ~/org/Figs/statusAndProgress/gasGainAndNeighbors/run_241_comparison_polya_theta_smaller30.pdf
In the 50% versin of the density charge plot a difference becomes visible. The behavior towards the peak on the left is different. The peak moves a bit to smaller values approaching the cutoff more. But the width towards the tail does not really change.
In case the individual pixel thresholds are very different we could maybe approximate that using an exponential distribution from which we draw, which implies 100% not activating a pixel at cutoff and exponentially more likely to activate approaching the peak.
We implemented this by using the function:
proc expShift(x: float, u, l, p: float): float = ## Shifted exponential distribution that satisfies: ## f(l) = p, f(u) = 1.0 let λ = (u - l) / ln(p) result = exp(- (x - u) / λ) # ... ## XXX: make dependent on Run-2 or Run-3 data! const cutoffs = @[1027.450870326596, # Run-2 893.4812944899318] # Run-3 let cutoff = cutoffs[1] * 1.15 let actSampler = (proc(rnd: var Rand, x: float): bool = let activateThreshold = expShift(x, gain.G, cutoff, 1e-1) result = x > cutoff and rnd.rand(1.0) < activateThreshold echo "Charge ", x, " threshold: ", activateThreshold, " is ", result, " and cutoff ", cutoff ) let activatePixel = actSampler(rnd, charge) if not activatePixel: #charge < cutoff: continue
This is way too extreme:
pdfunite *neighbor_run_charges_241* ~/org/Figs/statusAndProgress/gasGainAndNeighbors/run_241_comparison_polya_exp_activation.pdf
the latter with p = 0.3 instead of 0.1.
I think this is the wrong approach. It pushes us even stronger to lager values. In the real charge density plot we need to be lower rather than higher.
Not using an exponential cutoff, but modifying both the gas gain and
theta parameters yields the best result. But it's still
ugly. Fortunately our latest network does not rely on the total charge
anymore.
Let's run the effective efficiency for the dataset plots of run 241 comparison:
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/12_04_23_sgd_sim_diffusion_gain/trained_mlp_sgd_sim_diffusion_gain.pt \ --ε 0.8 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/sgd_sim_diffusion_gain/effective_eff \ --run 241
Fake:
- too many hits
- energy mismatch
- eccentricity way too small!
- charge a bit too large
-> very bad match. Some of it is due to not using neighbors. But realistically at this gas gain we cannot justify more hits! So we need a higher gain to get less hits again.
Using
let params = @[gain.N, gain.G * 0.75, gain.theta / 3.0]
yields a good agreement in the histograms.
with
showing similar issues as above.
But first let's try to look at neighbors again:
if charge > 3500.0: # whatever # possibly activate a neighbor pixel! let activateNeighbor = rnd.rand(1.0) < 0.5 if activateNeighbor: let neighbor = rand(3) # [right, left, up, down] let totNeighbor = rnd.sample(psampler) / 2.0 # reduce amount case neighbor of 0: insert(xp + 1, yp, totNeighbor) of 1: insert(xp - 1, yp, totNeighbor) of 2: insert(xp, yp + 1, totNeighbor) of 3: insert(xp, yp - 1, totNeighbor) else: doAssert false totalCharge += calib(totNeighbor, calibInfo) insert(xp, yp, ToT)
same settings of gain etc as last above yields
The issues are obvious. The neighbor histogram shows stark jumps
(expected now that I think about it) and the eccentricity is still too
low while energy a bit too high already.
Next: let's implement a strategy for smoother activation of neighbors. Let's go with a linear activation from 1000 to 10000 electrons, 0 to 1 chance and see what it looks like.
let neighSampler = (proc(rnd: var Rand, x: float): bool = let m = 1.0 / 9000.0 let activateThreshold = m * x - 1000 * m result = rnd.rand(1.0) < activateThreshold ) # ... if neighSampler(rnd, charge): # charge > 3500.0: # whatever # possibly activate a neighbor pixel! #let activateNeighbor = rnd.rand(1.0) < 0.5 let chargeNeighbor = rnd.sample(psampler) / 2.0 # reduce amount let activateNeighbor = actSampler(rnd, chargeNeighbor) if true: # activateNeighbor: let neighbor = rand(3) # [right, left, up, down] #let chargeNeighbor = rnd.sample(psampler) / 2.0 # reduce amount let totNeighbor = invert(chargeNeighbor, calibInfo) case neighbor of 0: insert(xp + 1, yp, totNeighbor) of 1: insert(xp - 1, yp, totNeighbor) of 2: insert(xp, yp + 1, totNeighbor) of 3: insert(xp, yp - 1, totNeighbor) else: doAssert false totalCharge += chargeNeighbor
This yields:
./countNeighborPixels -f ~/CastData/data/CalibrationRuns2018_Reco.h5 --chargePlots --fake --run 241 ./effective_eff_55fe ~/CastData/data/CalibrationRuns2018_Reco.h5 --model ~/org/resources/nn_devel_mixing/12_04_23_sgd_sim_diffusion_gain/trained_mlp_sgd_sim_diffusion_gain.pt --ε 0.8 --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 --evaluateFit --plotDatasets --plotPath ~/Sync/sgd_sim_diffusion_gain/effective_eff --run 241
Looking better. The neighbor distribution is more sensible, in particular in the ToT plot we can see a similar drift for larger neighboring cases as in the real data. However, the energy is now even bigger and the number of hits way too large (peak 300 compared to 260).
The total charge and energy being too large is probably simply an issue with our gas gain. Given that we don't use the charge or energy (but the gain in one network!) it's not that big of an issue. But the number of hits better be believable.
Given that we still have probably less higher neighbor cases, too low eccentricity and too many hits, likely implies:
- add higher neighbor cases with lower chance
- scale down the target charge by our modification of the gain?
The former first implementation:
let neighSampler = (proc(rnd: var Rand, x: float): int = ## Returns the number of neighbors to activate! let m = 1.0 / 9000.0 let activateThreshold = m * x - 1000 * m let val = rnd.rand(1.0) if val * 4.0 < activateThreshold: result = 4 elif val * 3.0 < activateThreshold: result = 3 elif val * 2.0 < activateThreshold: result = 2 elif val < activateThreshold: result = 1 else: result = 0 #result = rnd.rand(1.0) < activateThreshold ) # ... let numNeighbors = neighSampler(rnd, charge) if numNeighbors > 0: # charge > 3500.0: # whatever # possibly activate a neighbor pixel! #let activateNeighbor = rnd.rand(1.0) < 0.5 var count = 0 type Neighbor = enum Right, Left, Up, Down var seen: array[Neighbor, bool] while count < numNeighbors: let chargeNeighbor = rnd.sample(psampler) / 2.0 # reduce amount #let activateNeighbor = actSampler(rnd, chargeNeighbor) let neighbor = block: var num = Neighbor(rnd.rand(3)) while seen[num]: num = Neighbor(rnd.rand(3)) # [right, left, up, down] num seen[neighbor] = true #let chargeNeighbor = rnd.sample(psampler) / 2.0 # reduce amount let totNeighbor = invert(chargeNeighbor, calibInfo) case neighbor of Right: insert(xp + 1, yp, totNeighbor) of Left: insert(xp - 1, yp, totNeighbor) of Up: insert(xp, yp + 1, totNeighbor) of Down: insert(xp, yp - 1, totNeighbor) totalCharge += chargeNeighbor insert(xp, yp, ToT) totalCharge += charge inc count
Yeah, a look at the count histogram shows this is clearly rubbish. At
the very least we managed to produce something significantly more
eccentric than the real data, yet has still significantly less hits!
Sigh, bug in the insertion code…! The insert(xp, yp, ToT)
shouldn't be in that part!
This is looking half way reasonable for the neighbor histograms!
Wow, this looks pretty convincing in the property histograms! Aside
from having too many hits still, it looks almost perfect.
Let's be so crazy and run the effective efficiencies on all runs:
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/12_04_23_sgd_sim_gauss_diffusion/trained_mlp_sgd_sim_gauss_diffusion.pt \ --ε 0.8 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/sgd_sim_gauss_diffusion/effective_eff ./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/12_04_23_sgd_sim_diffusion_gain/trained_mlp_sgd_sim_diffusion_gain.pt --ε 0.8 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/sgd_sim_diffusion_gain/effective_eff
For the gauss diffusion network
The results are not as good as I hoped. But there is a high chance the
numbers are also bad because the training data was not the way it
looks now? Didn't run with the network knowing the gain due to the
problems of having the gain be equivalent between real and fake data.
Let's look at the worst performing run, 282 from this plot:
In this case the eccentricity is still too small compared to the real
case.
Let's first fix the number of hits we see.
let gainToUse = gain.G * 0.75 let calibFactor = linearFunc(@[calibInfo.bL, calibInfo.mL], gainToUse) * 1e-6
introduced this variable that's used everywhere instead of
gain.G. There were still some places we only used gain.G!
(Note: oh: we achieved an efficiency of 81% on this run with the gain
diffusion network)
Takeaway:
- energy way too low
- hits way too low (190 compared to 260)
- eccentricity slightly too large
(Note: weird: changing the gain the reconstruct fake event call to use
the gainInfo.G * 0.75 we currently use, makes the effective efficiency
be 18% only, but the energy histogram does not change?? Ahh, it's
because of the gain being an input neuron! That of course means it
breaks the expectation of the trained network. Implies same for
network without gain should yield good result. -> Checked, it
does. Bigger question: why does the energy reconstruction still yield
such a low energy? Shouldn't that change if we change the apparent
gain?
-> OHH, it's because we use the hardcoded gain from the calibInfo
argument in the computeEnergy function! Updating… Yup, energy is
looking perfect now:
but of course the effective efficiency is still completely off in this
network!)
So, next target is getting the hits in order. The reason must be the fact that our neighbor tot/charge histograms have a longer tail than the real data. Hmm, but the tail looks almost identical now…
I tried different gain values after all again and played around with counting the added neighboring pixels in the total charge or not counting them. It seems to me like going back to the original gain is the better solution. We still produce too few hits (to be thought about more), but at least we natively get the correct energy & charge. The histograms of the gain curves also don't look so horrible with our theta/3 hack.
and the properties:
Fraction in transverse RMS is slightly too low though. But the
effective efficiencies look quite good:
So next points to do:
[ ]Maybe number of neighbors should include diagonal elements after all? If UV photons are the cause the distance can be larger maybe. Currently our code says > 110 neighbors for the fake data but O(90) for the real data in Run-3![ ]investigate more into the number of hits we see. Can we fix it?[ ]Look into how properties look now for CDL data. Maybe our slope etc of the neighbor logic needs to be adjusted for lower gains?[ ]train a new network that uses the correct neighboring charges as training data
1.30.
First we changed the amount of charge for neighbors from div 2 to div 3:
let chargeNeighbor = rnd.sample(psampler) / 3.0 # reduce amount
This already improved the numbers a bit, also lowering the number of neighbors to a mean of 100.
Then we lowered the gas gain from 1.0 to 0.9 but not for the target charge.
let gainToUse = gain.G * 0.9 let calibFactor = linearFunc(@[calibInfo.bL, calibInfo.mL], gain.G) * 1e-6
i.e. leaving the calibration factor unchanged. Why? I have no idea, but it gets the job done.
Running the effective efficiencies of all runs now:
-> This makes the efficiencies worse! But that could be an effect of
having a network trained on the wrong data.
So now let's try to train a new network using our now better fake data and trying again. This now uses all our new changes and:
- gain 0.9 for everything but target charge
- possibly up to 4 neighbors
- neighbors receive 1/3 of sampled charge
- linear chance to activate neighbors
Also: we increase the range of theta parameters in the generation of
fake events from 2.1 .. 2.4 to 0.8 to 2.4.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/16_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusion.pt \ --plotPath ~/Sync/16_04_23_sgd_sim_diffusion_gain/ \ --numHidden 2500 \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets σT \ --learningRate 3e-4 \ --simulatedData
(Note: we had a short run by accident with only 10000 photons and
theta 2.1 to 2.4 that is stored in a directory with a 10000photons
part in the name)
There's some weirdness going on with the training. The outputs look
fine and the accuracy is good, but the training data loss function is
O(>100,000). Uhh.
In the meantime:
looking at run 340 for energy 0.525 keV that has effective efficiency
of ~90%.
The biggest difference is in the length dataset! Our fake events are
too long at the moment. Is the diffusion still bad for these datasets?
YES. Hmmm.
[ ]Why energy so wide in CDL data?[ ]why length too long?
SO to understand this:
[ ]check if CDL data we use in plotting of all properties is representative. Shouldn't it have cuts to CDL charges?[ ]check rmsTransverse fit plots for this run. Is the fit good? Does it have still too much rubbish in there? Explanation of too much data for energy could be.
shows the energy seen in the CuEPIC0.9kV dataset (0.525 keV data)
split by run based on CDL energy and charge energy. Run 340 has "all"
energies, and is not correctly filtered?? What the heck. The code
should be filtering both though. Is it our calibration of the energy
being broken? I don't understand.
I think it's the following:
if not fileIsCdl: ## We don't need to write the calibrated dataset to the `calibration-cdl` file, because ## the data is copied over from the `CDL_Reco` file! let df = df.filter(f{idx("Cut?") == "Raw"}) .clone() ## Clone to make sure we don't access full data underlying the DF! let dset = h5f.create_dataset(grpName / igEnergyFromCdlFit.toDset(), df.len, dtype = float, overwrite = true, filter = filter) dset.unsafeWrite(df["Energy", float].toUnsafeView(), df.len)
in cdl_spectrum_creation. Combining the toUnsafeView with the
filter call is unsafe. Hence I added the clone line there now!
See all plots with the bug present here:
UPDATE: Upon further inspection after fixing the code in other ways
again, it rather seems to be a different issue.
The updated plots are here:
(Note that the missing of the energy in Mn-Cr and Cu-Nu-15 in the
former plots is a cut on < 3 keV data in the plotting code).
With one exception (run 347) all runs that show behavior of very
varying energies in the charge energy dataset are those from runs
without the FADC! Why is that?
UPDATE I finally figured out the real
problem. It was related to the above snippet, but not in the way I
initially thought! The unsafe view was not the real culprit. The
actual issue is that the calculated energies were not ordered
correctly! We were just writing the data in the order in which it was
in the data frame, which at this point in the code sometimes does not
align correctly with the other datasets (due to a groups call over
run numbers it did line up correctly for exactly one run I think).
This is now fixed and the histograms of the energy
(And yes, some of the runs really have that little statistics after
the cuts! Check out:
 for an overview of the fits etc)
for an overview of the fits etc)
Let's look at the plot of the rmsTransverse from run 340 again:
It has changed a bit. Can't say it's very obvious, but the shape is
clearly less gaussian than before and the width to the right is a bit
larger?
What do the properties look like comparing run 340 now?
IMPORTANT The size difference has actually become worse now!
However, the effective efficiency still improves despite that. We
probably should look at the determination of the diffusion again now
that we actually look at the correct data for the CDL! Maybe the gauss
fit is now actually viable.
Now that we actually look at the correct data for all the CDL runs,
let's look at the effective efficiencies again using the gauss
diffusion network without gain from :
Aside from the 270 eV datasets all effective efficiencies have come
much closer to the target efficiency! This is pretty great. See below
for again looking at datasets comparing properties for the worst.
Let's also look at this using the new network that was trained on the new fake data!
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/16_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusion.pt \ --ε 0.8 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/16_04_23_sgd_sim_diffusion_gain/effective_eff
It looks surprisingly similar to the other network. It seems like
regardless they learned similar things in spite of the other network
having seen quite different data.
[X]Look atrmsTransversefit of run from earlier today again![X]compare number of counts in the histograms before and after fix!
We'll continue the training of the model we created above
[ ]LOOK At diffusion determination again after energy bug is fixed! Maybe the gauss fit method is now viable. In particular think about run 340 and how its length in the generated data is still too large! Also try to understand why runs for 250 eV are still so bad.^– These are the properties of the worst run in terms of effective efficiency. By far the biggest issues are related to the size of the generated events and their eccentricity. We apparently generate way too many neighbors in this case? Investigate maybe.
[ ]STILL have to adjust the gas gain and diffusion ranges for the fake data in training to be more sensible towards the real numbers!
1.31.
Let's start by fixing the code to continue the training process.
We've added the accuracy and loss fields to the MLPDesc, updated
serialize.nim to deal with missing data better (just left empty) and
will no rerun the training of the network from yesterday. We'll remove
the full network and start from 0 to 100,000 epochs and then start it
again to test (but this time containing the loss and accuracy values
in MLPDesc H5 file).
So again (see MLP section below) While MLP is running back to the diffusion from the bad runs, 340 and also 342. 342 the sizes and diffusion is actually quite acceptable, but the eccentricity is pretty wrong, likely due to too many neighbors? Maybe need a lower threshold after all? What is the gas gain of 342 compared to other CDL runs? 2350 or so, compared to 2390 for 340. So comparable.
First 340:
using gaussian fit with scale 1.65 again instead of limiting to 10% of
height of peak.
Better length & diffusion, but still too large. Let's check run 241
though before we lower further:
-> This is already too small! So the fit itself is not going to be a
good solution.
Comparing the RMS plots:
Given that drop off on RHS is MUCH stronger in 55Fe run, let's try a
fixed value as
peak + fixed. The run 241 data indicates something
like 0.15 to the peak position (0.95 + 0.15 = 1.1).
Run 241:
And 340:
This matches well for run 241, but in case of 340 it is way too short a cutoff.
What defines the hardness of the cutoff? Number of pixels? Not quite, run 342 is narrower than 340. But 342 used FADC and 340 did not! Fixed cutoff is also a fail for 342 though! Could we determine it by simulation? Do a simple optimization that uses fit or something as start, rmsTransverse of events using diffusion and energy, then if small enough, cutoff? Then maybe use Kolmogorov-Smirnov to determine if agreement? Seems like maybe the most sane approach?
[ ]Should we also store the training data of the net inMLPDesc?
1.31.1. MLP training
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/17_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusion.pt \ --plotPath ~/Sync/17_04_23_sgd_sim_diffusion_gain/ \ --numHidden 2500 \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets σT \ --learningRate 3e-4 \ --simulatedData
Now let's try to continue the training:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/17_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusion.pt \ --plotPath ~/Sync/17_04_23_sgd_sim_diffusion_gain/ \ --learningRate 3e-4 \ --simulatedData \ --continueAfterEpoch 100000
Given that the loss is still monotonically decreasing, I'll start another 100,000 epochs. And another 100,000 now!
1.32.
We've implemented the ideas from yesterday to determine the diffusion based on simulating events itself using some optimization strategy (currently Newton combined with Cramér-von Mises).
The annoying part is we need to look at not only one axis, but actually both and then determine the long and short axes. Otherwise our estimate of the rms transverse ends up too large, because we look at the mean RMS instead of transverse RMS.
Having implemented it (with probably improvable parameters) and
running it yields:
which is actually quite good.
Let's look at run 333 (1.49 keV), one of the lowest ones in the
efficiency. Was our value for the diffusion bad in that one?
Hmm, not really. It looks quite reasonable. But, as expected: this run
did not use the FADC! Probably NN filters out double photon events.
Let's check if all "bad" ones are no FADC runs (y = FADC, n = no FADC):
0.93 keV runs:
- 335 (y), 336 (n), 337 (n) -> 335 is the best one, 336 and 337 are indeed the ones with low efficiency!
0.525 keV runs:
- 339 (y), 340 (n) -> 339 is the good one, 340 is the bad one!
So yes, this really seems to be the cause!
Let's check our new network from yesterday on the same:
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/17_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_400000_loss_0.0115_acc_0.9975.pt \ --ε 0.8 --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/17_04_23_sgd_sim_diffusion_gain/effective_eff/
As one could have hoped, the results look a bit better still.
Let's also look at the Run-2 data:
./effective_eff_55fe \
~/CastData/data/CalibrationRuns2017_Reco.h5 \
--model ~/org/resources/nn_devel_mixing/17_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_400000_loss_0.0115_acc_0.9975.pt \
--ε 0.8 --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \
--evaluateFit \
--plotDatasets \
--plotPath ~/Sync/17_04_23_sgd_sim_diffusion_gain/effective_eff/
shows a plot of both run periods together.
We switched from Newton's method to gradient descent. It is much more
stable in the approach to the minimum:
n
The outliers to very low values are again all CDL runs without the
FADC!
The 5.9 CAST data does indeed still have values quite a bit below
target. But the spread is quite nice.
The CDL data generally comes closer to the target, but overall has a
larger spread (but notice not within a target! with the exception of
the no FADC runs).
From here: look at some more distributions, but generally this is good to go in my opinion. Need to implement logic into application of the MLP method now that takes care of not only calculating the correct cut for each run & energy, but also writes the actual efficiency that the real data saw for the target to the output files, as well as the variance of all runs maybe.
Idea: Can we somehow find out which events are thrown out by the network that should be kept according to the simulated data cut value? More or less the 'difference' between the 80% cut based on the real data and the 80% cut based on the simulated data. The events in that region are thrown out. Look at some and generate distributions of those events to understand what they are? Maybe they are indeed "rubbish"? -> Events in the two 80% cuts of real and simulated. UPDATE: Hmm, at a first glance there does not seem to be much to see there. To a "human eye" they look like X-rays I would say. It's likely the correlation between different variables that is making them more background like than X-ray like. I implemented a short function that filters the data between the cut region of real and simulated data at target efficiency:
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/17_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_400000_loss_0.0115_acc_0.9975.pt \ --ε 0.8 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/analyze_5.9/ \ --run 241
which gives the following property plots:
Next: Check how many elements we really need to generate for our Newton optimization. And can we:
- make the difference smaller again? -> Let's try with diffusion difference of 1 in numerical derivative (at 10k samples)
- use
Dualafter all? -> No. Tried again, but Cramér-von Mises also destroys all derivative information. - only use about 1000 elements instead of 10000?
-> at 1000 the spread becomes quite a bit bigger than at 10k!
[ ]Put 3.0 escape photon data for CAST back into plot!
1.33.
So, from yesterday to summarize: Using gradient descent makes the determination of the diffusion from real data fast enough and stable.
First continue a short look at the 'intermediate' data between the cut
values of simulated and real data. We will plot the intermediate
against the data that passes both.
Running the same command as yesterday for run 241 yields:
In direct comparison we can see that the events that fail to pass the
simulated cut are generally a bit more eccentric and longer / wider /
slightly bigger RMS. Potentially events including a captured escape
photon?
Let's extract the event numbers of those intermediate events:
Event numbers that are intermediate: Column of type: int with length: 107
contained Tensor: Tensor[system.int] of shape "[107]" on backend "Cpu"
104 109 123 410 537 553 558 1042 1272 1346 1390 1447 1527 1583 1585 1594 1610 1720 1922 1965 2082 2155 2176 2198 2419 2512 2732 2800 2801 3038 3072 3095 3296 3310 3473 3621 3723 3820 4088 4145 4184 4220 4250 4308 4347 4353 4466 4558 4590 4725 4843 4988 5204 5234 5288 5497 5637 5648 5661 5792 5814 5848 5857 6090 6175 6187 6312 6328 6359 6622 6657 6698 6843 6944 6951 7121 7137 7162 7192 7350 7436 7472 7545 7633 7634 7730 7749 7788 7936 8003 8014 8075 8214 8364 8441 8538 8549 8618 8827 8969 8991 9008 9026 9082 9102 9260 9292
Now we'll use plotData to generate event displays for them:
plotData \ --h5file ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --runType rtCalibration \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --runs 241 \ --eventDisplay \ --septemboard \ --events 104 --events 109 --events 123 --events 410 --events 537 --events 553 --events 558 --events 1042 --events 1272 --events 1346 --events 1390 --events 1447 --events 1527 --events 1583 --events 1585 --events 1594 --events 1610 --events 1720 --events 1922 --events 1965 --events 2082 --events 2155 --events 2176 --events 2198 --events 2419 --events 2512 --events 2732 --events 2800 --events 2801 --events 3038 --events 3072 --events 3095 --events 3296 --events 3310 --events 3473 --events 3621 --events 3723 --events 3820 --events 4088 --events 4145 --events 4184 --events 4220 --events 4250 --events 4308 --events 4347 --events 4353 --events 4466 --events 4558 --events 4590 --events 4725 --events 4843 --events 4988 --events 5204 --events 5234 --events 5288 --events 5497 --events 5637 --events 5648 --events 5661 --events 5792 --events 5814 --events 5848 --events 5857 --events 6090 --events 6175 --events 6187 --events 6312 --events 6328 --events 6359 --events 6622 --events 6657 --events 6698 --events 6843 --events 6944 --events 6951 --events 7121 --events 7137 --events 7162 --events 7192 --events 7350 --events 7436 --events 7472 --events 7545 --events 7633 --events 7634 --events 7730 --events 7749 --events 7788 --events 7936 --events 8003 --events 8014 --events 8075 --events 8214 --events 8364 --events 8441 --events 8538 --events 8549 --events 8618 --events 8827 --events 8969 --events 8991 --events 9008 --events 9026 --events 9082 --events 9102 --events 9260 --events 9292

It really seems like the main reason for them being a bit more eccentric is the presence of potentially higher number of single pixels that are outliers? I suppose it makes sense that these would be rejected with a larger likelihood. However, why would such events appear more likely in the 5.9 keV CAST data than in simulated data?
For now I'm not sure what to make of this. Of course one could attempt to do something like use a clustering algorithm that ignores outliers etc. But all that doesn't seem that interesting, at least not if we don't understand why we have this distinction from simulated to real data in the first place.
So for now let's just continue.
First: add 3.0 keV data with 5.9 keV absorption length into effective efficiency plot and see how the efficiency fares:
Run-2 @ 99%:
likelihood \ -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out ~/Sync/run2_17_04_23_mlp_local_0.99.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/nn_devel_mixing/17_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_400000_loss_0.0115_acc_0.9975.pt \ --nnSignalEff 0.99 \ --nnCutKind runLocal \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --readonly
Run-3 @ 99%:
likelihood \ -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out ~/Sync/run3_17_04_23_mlp_local_0.99.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/nn_devel_mixing/17_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_400000_loss_0.0115_acc_0.9975.pt \ --nnSignalEff 0.99 \ --nnCutKind runLocal \ --calibFile ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --readonly
Need to run effective efficiency at 99% as well to know the real efficiencies.
NOTE: The gradient descent code unfortunately is still rather slow. Should we aim to cache the results for each run? Generally I'd prefer no caching and just making it faster though. Maybe we can pick an example that is pretty hard to converge and use that as a reference to develop? Ah, and we can cache stuff within a single program run, so that we only need to compute the stuff once for each run?
1.34.
- we know cache the results of the diffusion calculation, but still have to verify it works as intended
- for the background datasets the diffusion coefficient is now also
~correctly determined by sampling both the energy and position of
the data, i.e.
- uniform in drift distance (muons enter anywhere between cathode and anode)
- energy exponential distribution that has ~20-30% of flux at 10 keV of the flux at ~0 eV
[ ]ADD PLOT
- verify the diffusion coefficients are correctly determined by calculating all of them for background and calibration and creating a plot showing the determined numbers -> maybe a good option to show the plots of the RMS data used and the generated data for each?
[ ]implement calculation of variance and mean value for effective efficiencies for each run period (write to H5 out inlikelihood)[X]TAKE NOTES of the diffusion vs run plot[ ]ADD EFFECTICE EFFICIENCY PLOT WITH 3 keV escape data
1.35.
Yesterday we turned determineDiffusion into a usable standalone
tool to compute the diffusion parameters of all runs. It generates a
plot showing what the parameters are, colored by the 'loss' of the
best estimate (via Cramér-von Mises).
./determineDiffusion \ ~/CastData/data/DataRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/DataRuns2018_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5
yields the following plot of the diffusion values deduced using
gradient descent:
We can clearly see two important aspects:
- the background diffusion parameters are typically a good 20-30 μm/√cm larger than the 5.9 Fe points
- there are some cases where:
- the loss is very large (background, runs close to run 80)
- the diffusion is exactly 660, our starting value
The latter implies that the loss is already smaller than 2 (our current stopping criterion). The former implies the distributions look very different than the simulated distribution. I've already seen one reason: in some runs there is an excessive contribution at rmsTransvers < 0.2 or so which has a huge peak. Some kind of noise signal? We'll check that.
For now here are all the plots of the rmsTransverse dataset for sim and real data that were generated from the best estimates:

The worst run is run 86 (see page 158 in the PDF). Let's investigate what kind of events contribute such data to the transverse RMS. My assumption would be noisy events at the top of the chip or something like that? First the ingrid distributions:
plotData \ --h5file ~/CastData/data/DataRuns2017_Reco.h5 \ --runType rtBackground \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --runs 86 \ --ingrid \ --cuts '("rmsTransverse", 0.0, 0.2)' \ --applyAllCuts \ --chips 3
So clearly defined in centerX and centerY. The sparky point on the grid?

Yeah, that seems to be it. They are all pretty much below 20 hits anyway. So we'll just introduce an additional cut for non CDL data that clusters must be larger 20 hits. They match the pixels we consider noisy for the background cluster plot and in limit!
Added such a filter (hits > 20) and running again.
The diffusion values are now these. The largest is on the order of 3
and they look more reasonable. Of course the distinction between
background and calibration is still presetn.
Next we've removed the limit of ks > 2.0 as a stopping criterion and
added logic to reset to best estimate if half number of bad steps
already done.
Let's rerun again:
Now it actually looks pretty nice!
The difference between 5.9 and background is now pretty much
exactly 40. So we'll use that in getDiffusion as a constant for now.
Finally, let's try to run likelihood on one dataset (Run-2) at a
fixed efficiency to see what is what (it should read all diffusion values
from the cache).
The generated files are:
/Sync/run2_17_04_23_mlp_local_0.80.h5/Sync/run2_17_04_23_mlp_local_0.90.h5/Sync/run3_17_04_23_mlp_local_0.80.h5/Sync/run3_17_04_23_mlp_local_0.90.h5
Time to plot a background rate from both, we compare it to the LnL only method at 80%:
plotBackgroundRate \ ~/Sync/run2_17_04_23_mlp_local_0.90.h5 \ ~/Sync/run3_17_04_23_mlp_local_0.90.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ --names "MLP@90" \ --names "MLP@90" \ --names "LnL@80" \ --names "LnL@80" \ --centerChip 3 \ --title "Background rate from CAST data, LnL@80%, MLP@90%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_lnL_0.8_mlp_0.9.pdf \ --outpath ~/Sync/ \ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@90 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.7298e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 2.2748e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@90 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 3.4122e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 1.7061e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@90 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 9.6914e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.1537e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@90 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 1.0413e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 4.1650e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@90 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 3.3067e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 8.2667e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@90 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.9383e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.4229e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@90 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 9.3572e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.5595e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8mlp0.9.pdf
As we can see the rates are quite comparable. What is nice to see is that the Argon peak is actually more visible in the MLP data than in the LnL cut method. Despite higher efficiency the network performs essentially the same though. That's very nice! The question is what is the effective efficiency based on 5.9 keV data using 90%?
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/17_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_400000_loss_0.0115_acc_0.9975.pt \ --ε 0.9 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/run2_run3_gradient_descent_eff_0.9/
So we're actually looking at about 85% realistically.
And background rate with MLP@85%:
plotBackgroundRate \ ~/Sync/run2_17_04_23_mlp_local_0.85.h5 \ ~/Sync/run3_17_04_23_mlp_local_0.85.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ --names "MLP@85" \ --names "MLP@85" \ --names "LnL@80" \ --names "LnL@80" \ --centerChip 3 \ --title "Background rate from CAST data, LnL@80%, MLP@85%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_lnL_0.8_mlp_0.85.pdf \ --outpath ~/Sync/ \ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.4501e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 2.0418e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 1.3192e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 8.4250e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 1.8722e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9879e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5952e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.9022e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 7.2554e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.7114e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.1392e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.5130e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.4188e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8mlp0.85.pdf [WARNING]: Printing total background time currently only supported for single datasets.
So at this point the effective efficiency is pretty much 80%. This means the network is significantly better at lower energies (aside from first bin), but essentially the same everywhere else. Pretty interesting. Question is how it fares at higher signal efficiencies!
1.35.1. Running likelihood on SGD trained network after 500k epochs
Using 95% signal efficiency: Run-2:
likelihood -f \ ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out ~/Sync/run2_17_04_23_mlp_local_0.95_500k.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/nn_devel_mixing/17_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_500000_loss_0.0106_acc_0.9977.pt \ --nnSignalEff 0.95 \ --nnCutKind runLocal \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --readonly
likelihood -f \ ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out ~/Sync/run3_17_04_23_mlp_local_0.95_500k.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/nn_devel_mixing/17_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_500000_loss_0.0106_acc_0.9977.pt \ --nnSignalEff 0.95 \ --nnCutKind runLocal \ --calibFile ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --readonly
Yields the background:
plotBackgroundRate \ ~/Sync/run2_17_04_23_mlp_local_0.95_500k.h5 \ ~/Sync/run3_17_04_23_mlp_local_0.95_500k.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ --names "MLP@95" \ --names "MLP@95" \ --names "LnL@80" \ --names "LnL@80" \ --centerChip 3 \ --title "Background rate from CAST data, LnL@80%, MLP@95%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_lnL_0.8_mlp_0.95.pdf \ --outpath ~/Sync/ \ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 3.0605e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 2.5504e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 4.3972e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 2.1986e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1222e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.4937e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 1.2224e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 4.8897e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 3.9927e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 9.9816e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 2.2267e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.7834e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 1.0465e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.7442e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8mlp0.95.pdf [WARNING]: Printing total background time currently only supported for single datasets.
Comparing the now 85% at 500k:
plotBackgroundRate \ ~/Sync/run2_17_04_23_mlp_local_0.85_500k.h5 \ ~/Sync/run3_17_04_23_mlp_local_0.85_500k.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ --names "MLP@85" \ --names "MLP@85" \ --names "LnL@80" \ --names "LnL@80" \ --centerChip 3 \ --title "Background rate from CAST data, LnL@80%, MLP@85% 500k" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_lnL_0.8_mlp_0.85_500k.pdf \ --outpath ~/Sync/ \ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.4730e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 2.0608e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 2.7087e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 1.3543e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 8.4954e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 1.8879e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 9.3221e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.7288e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.8670e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 7.1674e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.7413e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.1766e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.4778e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.4130e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8mlp0.85500k.pdf [WARNING]: Printing total background time currently only supported for single datasets.
Phew, can it be any more similar?
Now we're also running 99% for the 500k epoch version.
plotBackgroundRate \ ~/Sync/run2_17_04_23_mlp_local_0.99_500k.h5 \ ~/Sync/run3_17_04_23_mlp_local_0.99_500k.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ --names "MLP@99" \ --names "MLP@99" \ --names "LnL@80" \ --names "LnL@80" \ --centerChip 3 \ --title "Background rate from CAST data, LnL@80%, MLP@99% 500k" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_lnL_0.8_mlp_0.99_500k.pdf \ --outpath ~/Sync/ \ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 3.8150e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 3.1792e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 7.7918e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.8959e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.5707e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 3.4904e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 1.7219e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 6.8878e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 4.9249e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 1.2312e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 2.8951e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 3.6189e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 1.2418e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 2.0696e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8mlp0.99500k.pdf [WARNING]: Printing total background time currently only supported for single datasets.
Effective efficiency at 99% with 500k:
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/17_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_500000_loss_0.0106_acc_0.9977.pt \ --ε 0.99 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/run2_run3_gradient_descent_eff_0.99/
yields:
Wow, so it actually works better than at 80%!
1.35.2. Training again with AdamW
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/21_04_23_adamW_gauss_diffusion/trained_mlp_adamW_gauss_diffusion.pt \ --plotPath ~/Sync/21_04_23_adamW_sim_diffusion_gain/ \ --numHidden 2500 \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets σT \ --learningRate 3e-4 \ --simulatedData
After the training I started another 100k epochs!
Running the likelihood with the trained network (at 85%):
likelihood -f ~/CastData/data/DataRuns2017_Reco.h5 --h5out ~/Sync/run2_21_04_23_adamW_local_0.85.h5 --region crGold --cdlYear 2018 --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 --mlp ~/org/resources/nn_devel_mixing/21_04_23_adamW_gauss_diffusion/trained_mlp_adamW_gauss_diffusion.pt --nnSignalEff 0.85 --nnCutKind runLocal --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5 --readonly likelihood -f ~/CastData/data/DataRuns2018_Reco.h5 --h5out ~/Sync/run3_21_04_23_adamW_local_0.85.h5 --region crGold --cdlYear 2018 --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 --mlp ~/org/resources/nn_devel_mixing/21_04_23_adamW_gauss_diffusion/trained_mlp_adamW_gauss_diffusion.pt --nnSignalEff 0.85 --nnCutKind runLocal --calibFile ~/CastData/data/CalibrationRuns2018_Reco.h5 --readonly
and plotting:
plotBackgroundRate \ ~/Sync/run2_21_04_23_adamW_local_0.85.h5 \ ~/Sync/run3_21_04_23_adamW_local_0.85.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ --names "MLP@85" \ --names "MLP@85" \ --names "LnL@80" \ --names "LnL@80" \ --centerChip 3 \ --title "Background rate from CAST data, LnL@80%, AdamW MLP@85%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_lnL_0.8_adamW_mlp_0.85.pdf \ --outpath ~/Sync/ \ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 1.9365e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.6138e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 3.6409e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 1.8204e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 8.5130e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 1.8918e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.0029e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.2012e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.8142e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 7.0355e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.5074e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 1.8842e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 7.5984e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.2664e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8adamWmlp0.85.pdf [WARNING]: Printing total background time currently only supported for single datasets.
This network now needs an effective efficiency:
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/21_04_23_adamW_gauss_diffusion/trained_mlp_adamW_gauss_diffusion.pt \ --ε 0.85 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/run2_run3_adamW_gradient_descent_eff_0.85/
Ouch, while the mean is good, the variance is horrific!
Maybe this is related to the horribly bad loss? The accuracy is still good, but the loss (that we optimize for after all) is crap.
And for 95%:
plotBackgroundRate \ ~/Sync/run2_21_04_23_adamW_local_0.95.h5 \ ~/Sync/run3_21_04_23_adamW_local_0.95.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ --names "MLP@95" \ --names "MLP@95" \ --names "LnL@80" \ --names "LnL@80" \ --centerChip 3 \ --title "Background rate from CAST data, LnL@80%, AdamW MLP@95%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_lnL_0.8_adamW_mlp_0.95.pdf \ --outpath ~/Sync/ \ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.4660e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 2.0550e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 4.7842e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 2.3921e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.0694e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.3764e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 1.0501e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 4.2002e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 3.0956e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 7.7391e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.8820e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.3525e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.8823e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.4804e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8adamWmlp0.95.pdf [WARNING]: Printing total background time currently only supported for single datasets.
1.35.3. Training an SGD network including total charge
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/21_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusion.pt \ --plotPath ~/Sync/21_04_23_sgd_sim_diffusion_gain/ \ --numHidden 2500 \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --learningRate 7e-4 \ --simulatedData
Note the larger learning rate, due to the very slow convergence before!
Trained for 400k epochs using learning rate 7e-4, then lowered to 3e-4 for another 100k.
Effective efficiency at 80% for this network:
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/21_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_500000_loss_0.0084_acc_0.9981.pt \ --ε 0.80 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/run2_run3_21_04_23_sgd_gradient_descent_eff_0.8/
So generally a good 7.5% too low. ~72.5% real efficiency compared to 80% desired. This does make some sense as the difference between charges is still quite different from what we target, I suppose. Maybe we should look into that again to see if we can get better alignment there?
Despite the low efficiency, let's look at the background rate of this case:
plotBackgroundRate \ ~/Sync/run2_21_04_23_mlp_local_0.80_500k.h5 \ ~/Sync/run3_21_04_23_mlp_local_0.80_500k.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ --names "MLP@80" \ --names "MLP@80" \ --names "LnL@80" \ --names "LnL@80" \ --centerChip 3 \ --title "Background rate from CAST data, LnL@80%, SGD w/ charge MLP@80%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_lnL_0.8_sgd_21_04_23_mlp_0.8.pdf \ --outpath ~/Sync/ \ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 1.9664e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.6387e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 1.9524e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 9.7618e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 7.3521e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 1.6338e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 6.2440e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 2.4976e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.2514e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 5.6284e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.3438e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 1.6797e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 7.4401e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.2400e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8sgd210423mlp0.8.pdf [WARNING]: Printing total background time currently only supported for single datasets.
But let's check 95% and see where we end up thered
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/21_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_500000_loss_0.0084_acc_0.9981.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/run2_run3_21_04_23_sgd_gradient_descent_eff_0.95/
Interesting! At this efficiency the match is much better than at
80%.
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/21_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_500000_loss_0.0084_acc_0.9981.pt \ --ε 0.99 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/run2_run3_21_04_23_sgd_gradient_descent_eff_0.99/
And at 98% it's even better! Nice.
What does the background look like at 95% and 99%?
plotBackgroundRate \ ~/Sync/run2_21_04_23_mlp_local_0.95_500k.h5 \ ~/Sync/run3_21_04_23_mlp_local_0.95_500k.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ --names "MLP@95" \ --names "MLP@95" \ --names "LnL@80" \ --names "LnL@80" \ --centerChip 3 \ --title "Background rate from CAST data, LnL@80%, SGD w/ charge MLP@95%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_lnL_0.8_sgd_21_04_23_mlp_0.95.pdf \ --outpath ~/Sync/ \ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.9180e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 2.4317e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 3.9399e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 1.9699e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.0465e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.3256e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 1.1521e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 4.6083e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 3.5705e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 8.9263e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 2.0948e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.6185e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 9.8146e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.6358e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8sgd210423mlp0.95.pdf [WARNING]: Printing total background time currently only supported for single datasets.
plotBackgroundRate \ ~/Sync/run2_21_04_23_mlp_local_0.99_500k.h5 \ ~/Sync/run3_21_04_23_mlp_local_0.99_500k.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ --names "MLP@99" \ --names "MLP@99" \ --names "LnL@80" \ --names "LnL@80" \ --centerChip 3 \ --title "Background rate from CAST data, LnL@80%, SGD w/ charge MLP@99%" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_only_lnL_0.8_sgd_21_04_23_mlp_0.99.pdf \ --outpath ~/Sync/ \ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 3.5705e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 2.9754e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.7541e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.3771e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.4352e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 3.1894e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 1.5707e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 6.2827e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 4.4851e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 1.1213e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 2.6823e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 3.3529e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 1.1679e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.9465e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8sgd210423mlp0.99.pdf [WARNING]: Printing total background time currently only supported for single datasets.
1.35.4. Investigate huge loss values for AdamW and sometimes SGD
Let's see what happens there. Fortunately have a network that reproduces it!
Ah, but the loss is directly the output of the sigmoid_cross_entropy
function…
Chatting with BingChat helped understand some things, but didn't
really answer why it might lead to a big loss.
Instead of our current forward definition:
var x = net.hidden.forward(x).relu() return net.classifier.forward(x).squeeze(1)
we can use:
var x = net.hidden.forward(x).relu() return net.classifier.forward(x).tanh().squeeze(1)
which then should make using something like MSEloss or L1loss more stable I think. Could be worth a try.
1.35.5. TODO Train a network with hits as an input?
Maybe that helps a lot?
-> We're currently training another net with total charge as input.
1.35.6. TODO Can we train a network that focuses on…
separating background and calibration data as far away as possible?
But then again this is what the loss effectively does anyway, no?
The question is how does BCE loss actually work. Need to understand that. Does it penalize "more or less wrong" things? Or does it focus on "correct" vs "wrong"?
1.35.7. TODO Network with 2 hidden layers
Two very small ones:
- 100 neurons with tanh
- 100 neurons with gelu activation
I also tried ELU and Leaky_ReLU, but they gave only NaNs in the
loss. No idea why.
Still maybe try something with tanh/sigmoid on output layer and MSE loss.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/22_04_23_sgd_gauss_diffusion_2hidden/trained_mlp_sgd_gauss_diffusion_2hidden.pt \ --plotPath ~/Sync/22_04_23_sgd_sim_diffusion_gain_2hidden/ \ --numHidden 100 \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --learningRate 7e-4 \ --simulatedData
We continue training this network for 100k epochs more. Using 100k simulated events now instead of 30k.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/22_04_23_sgd_gauss_diffusion_2hidden/trained_mlp_sgd_gauss_diffusion_2hidden.pt \ --plotPath ~/Sync/22_04_23_sgd_sim_diffusion_gain_2hidden/ \ --numHidden 100 \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --learningRate 7e-4 \ --simulatedData
Now repeat tanh network, but with all training data & 1000 hidden neurons each:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/22_04_23_sgd_gauss_diffusion_2hidden_tanh1000/trained_mlp_sgd_gauss_diffusion_2hidden_tanh1000.pt \ --plotPath ~/Sync/22_04_23_sgd_sim_diffusion_gain_2hidden_tanh1000/ \ --numHidden 1000 \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --learningRate 7e-4 \ --simulatedData
1.35.8. Effective efficiency with escape photon data
Back to the effective efficiency:
is the plot including the effective efficiency based on the escape
peak data in the CAST data. It used:
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/17_04_23_sgd_gauss_diffusion/trained_mlp_sgd_gauss_diffusioncheckpoint_epoch_400000_loss_0.0115_acc_0.9975.pt \ --ε 0.8 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/run2_run3_incl_escape_photons
So unfortunately the values for the escape data are actually even a bit lower. This is working by generated 3.0 keV data with the absorption length of the 5.9 keV data. Surely the equivalence is not perfect.
1.36.
Wrote a small helper studyMLP that runs:
- effective efficiency
- likelihood for 2017 and 2018 data
for a list of target efficiencies for a given MLP model.
./studyMLP 0.85 0.95 0.99 \ --model ~/org/resources/nn_devel_mixing/22_04_23_sgd_gauss_diffusion_2hidden/trained_mlp_sgd_gauss_diffusion_2hiddencheckpoint_epoch_400000_loss_0.0067_acc_0.9984.pt \ --plotPath ~/Sync/22_04_23_sgd_sim_diffusion_gain_2hidden/ \ --h5out ~/Sync/22_04_23_tanh_hidden2_100.h5
Let's see!
Hmm, we have some issue that the likelihood output files are on the
order of 1.5 - 2.5 GB!
Investigating…
Ah: I didn't recompile likelihood, so it still used the single layer
MLP instead of the two layer. Therefore the weights obviously didn't
properly match.
The above ran properly now and produced the following effective efficiency plots:
Background rate tanh 2 layer 100 neurons 85%:
plotBackgroundRate ~/Sync/22_04_23_tanh_hidden2_100eff_0.85_run2.h5 ~/Sync/22_04_23_tanh_hidden2_100eff_0.85_run3.h5 ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 --names "MLP@85" --names "MLP@85" --names "LnL@80" --names "LnL@80" --centerChip 3 --title "Background rate from CAST data, LnL@80%, SGD tanh100 MLP@85%" --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge --outfile background_rate_only_lnL_0.8_sgd_tanh100_22_04_23_mlp_0.85.pdf --outpath ~/Sync/ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 1.9594e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.6328e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 2.0579e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 1.0289e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 7.4577e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 1.6573e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 5.5405e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 2.2162e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.5328e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.3320e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.3033e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 1.6292e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@85 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 7.7918e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.2986e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8sgdtanh100220423mlp0.85.pdf [WARNING]: Printing total background time currently only supported for single datasets.
plotBackgroundRate ~/Sync/22_04_23_tanh_hidden2_100eff_0.95_run2.h5 ~/Sync/22_04_23_tanh_hidden2_100eff_0.95_run3.h5 ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 --names "MLP@95" --names "MLP@95" --names "LnL@80" --names "LnL@80" --centerChip 3 --title "Background rate from CAST data, LnL@80%, SGD tanh100 MLP@95%" --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge --outfile background_rate_only_lnL_0.8_sgd_tanh100_22_04_23_mlp_0.95.pdf --outpath ~/Sync/ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.6278e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 2.1898e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 3.3771e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 1.6885e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 9.8673e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.1927e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 9.1462e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.6585e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 3.2715e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 8.1788e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.8275e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.2843e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@95 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 9.5155e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.5859e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8sgdtanh100220423mlp0.95.pdf [WARNING]: Printing total background time currently only supported for single datasets.
plotBackgroundRate ~/Sync/22_04_23_tanh_hidden2_100eff_0.99_run2.h5 ~/Sync/22_04_23_tanh_hidden2_100eff_0.99_run3.h5 ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 --names "MLP@99" --names "MLP@99" --names "LnL@80" --names "LnL@80" --centerChip 3 --title "Background rate from CAST data, LnL@80%, SGD tanh100 MLP@99%" --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge --outfile background_rate_only_lnL_0.8_sgd_tanh100_22_04_23_mlp_0.99.pdf --outpath ~/Sync/ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.3288e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.9406e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 3.3911e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 2.8259e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 5.9450e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 2.9725e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.3192e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.9315e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.9703e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.5881e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 1.4599e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 5.8395e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 4.0630e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 1.0158e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6551e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0689e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 2.5082e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 3.1352e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@99 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 1.0923e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.8204e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:DataFrame with 7 columns and 116 rows: Idx Energy Rate totalTime RateErr Dataset yMin yMax dtype: float float constant float string float float 0 0.4 5.98 3159 1.026 LnL@80 4.955 7.006 1 0.6 6.332 3159 1.055 LnL@80 5.277 7.387 2 0.8 5.98 3159 1.026 LnL@80 4.955 7.006 3 1 5.98 3159 1.026 LnL@80 4.955 7.006 4 1.2 3.166 3159 0.7462 LnL@80 2.42 3.912 5 1.4 2.99 3159 0.7252 LnL@80 2.265 3.715 6 1.6 2.814 3159 0.7036 LnL@80 2.111 3.518 7 1.8 3.166 3159 0.7462 LnL@80 2.42 3.912 8 2 1.583 3159 0.5277 LnL@80 1.055 2.111 9 2.2 1.759 3159 0.5562 LnL@80 1.203 2.315 10 2.4 0.8794 3159 0.3933 LnL@80 0.4861 1.273 11 2.6 1.583 3159 0.5277 LnL@80 1.055 2.111 12 2.8 2.287 3159 0.6342 LnL@80 1.652 2.921 13 3 5.101 3159 0.9472 LnL@80 4.154 6.048 14 3.2 5.277 3159 0.9634 LnL@80 4.313 6.24 15 3.4 4.221 3159 0.8617 LnL@80 3.36 5.083 16 3.6 3.342 3159 0.7667 LnL@80 2.575 4.109 17 3.8 2.111 3159 0.6093 LnL@80 1.501 2.72 18 4 0.7036 3159 0.3518 LnL@80 0.3518 1.055 19 4.2 1.055 3159 0.4308 LnL@80 0.6245 1.486
[INFO]:INFO: storing plot in /home/basti/Sync/backgroundrateonlylnL0.8sgdtanh100220423mlp0.99.pdf [WARNING]: Printing total background time currently only supported for single datasets.
All together:
So clearly we also mainly gain at low energies here. Even at 99% we still are effectively not seeing any degradation in most regions!
1.36.1. Train MLP with sigmoid on last layer and MLE loss!
Changed the MLP forward to:
proc forward*(net: MLP, x: RawTensor): RawTensor = var x = net.hidden.forward(x).tanh() x = net.hidden2.forward(x).tanh() return net.classifier.forward(x).sigmoid()
i.e. a sigmoid on the output.
Then as loss we use mse_loss instead of the sigmoid_cross_entropy.
Let's see!
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss.pt \ --plotPath ~/Sync/23_04_23_sgd_sim_diffusion_gain_tanh300_mse_loss/ \ --numHidden 300 \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --learningRate 7e-4 \ --simulatedData
Trained it up to 500k epochs.
First note: as expected due to the Sigmoid on the last layer, the output is indeed between -1 and 1 with very strong separation between the two.
I also then ran studyMLP for it:
./studyMLP 0.85 0.95 0.99 --model ~/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/trained_mlp_sgd_gauss_diffusion_tanh300_mse_losscheckpoint_epoch_500000_loss_0.0017_acc_0.9982.pt --plotPath ~/Sync/23_04_23_sgd_sim_diffusion_gain_tanh300_mse_loss --h5out ~/Sync/23_04_23_tanh300_mle_loss.h5
but now that I think about it I don't know if I actually recompiled
both the effective_eff_55fe and likelihood programs for the new
activation function. Oops.
Of course, if one uses this network without the sigmoid layer it will still produce output similar to the previous tanh (trained via sigmoid cross entropy loss) network. Rerunning at the moment.
(really need things like optimizer, number of layers, activation functions etc as part of MLPDesc)
1.36.2. DONE Things still todo from today!
[X]Need to implement number of layers into MLPDesc and handle loading correct network!
1.37. TODO
IMPORTANT:
- See if adjusting the rms transverse values down to the old ones (~1.0 to 1.1 instead of 1.2 - 1.3) gets us closer to the 80% in case of the e.g. 80% network efficiency!!!
1.38.
Finished the implementation of mapping all parameters of interest for
the model layout and training to the MLPDesc and loading / applying
the correct thing at runtime.
In order to update any of the existing mlp_desc.h5 files, we need to
provide the settings used for each network! At least the new settings
that is.
For example let's update the last tanh network we trained with a
sigmoid output layer. We do it by also continuing training on it for
another 100k epochs:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss.pt \ --plotPath ~/Sync/23_04_23_sgd_sim_diffusion_gain_tanh300_mse_loss/ \ --numHidden 300 \ --numHidden 300 \ --activationFunction tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --continueAfterEpoch 500000
As we have the existing H5 file for the MLPDesc we don't need to supply the datasets. But the additional new fields are required to update them in the file.
We implemented that the code raises an exception if only an old
MLPDesc file is found in effective_eff_fe55. Also the file name now
contains the version number itself for convenience. Also the version
is another field now that is serialized.
Let's check the following TODO from yesterday:
IMPORTANT:
- See if adjusting the rms transverse values down to the old ones (~1.0 to 1.1 instead of 1.2 - 1.3) gets us closer to the 80% in case of the e.g. 80% network efficiency!!!
-> We'll reset the cuts to their old values and then run the above
network on effective_eff_fe55
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss.pt \ --ε 0.8 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/run2_run3_23_04_23_tanh300_rmsT_old/
-> The plot
generally doesn't look too bad, but it's hard to read because there
are 2 escape peak values with negative efficiency! Need to fix that.
We'll look at only run 88, which should show that behavior.
Fake data for 3.0 at run 88 and energy 2.98 keV target Ag-Ag-6kV has a cut value 0.9976992011070251 and effective eff -0.01075268817204301
That's fishy. Computed from (kept vs total in that dataset):
Number kept: -1 vs 93
Only 93 escape events? And -1 kept? Ohhh! It's because of this line:
if pred.len < 100: return (-1, @[])
in predictCut!
Removed that line as it's not very useful nowadays. We still care about the number.
Rerunning and storing in:
This looks pretty much like the other case with wider rmsT, no?
Let's make sure and change again and run again.
-> Ok, it does improve the situation. The mean is now rather at 75-76 percent compared to 73% or so.
What to make of this? The more rms transverse we allow, the more ugly non X-ray events we will have in our data? Or just that we reproduce the ugly events with our fake data?
1.39.
Next steps:
implement computing the effective efficiencies for:
- the given network
- the desired efficiency
These will also be stored in some form of a cache (given that the calculation takes O(15 min) -> Implemented including caching. Just needs to be called from
likelihoodnow.VetoSettingshas the fields required. Should we compute it first or last? ->Compute in-> Not in the init, due to problems with circular imports. The effective eff fields are now filled ininitLikelihoodContext.likelihooditself. They are mainly needed for that anyway, so that should be fine.- fix the application of the septem veto etc when using an MLP. Use that to decide if a cluster passes instead of logL -> up next. -> Implemented now!
- automate MLP for
likelihoodincreateAllLikelihoodCombinations- this should be done via a
--mlpoption which takesseq[string]which are paths to the model file. For each we add afkMLPflag that will perform a call tolikelihoodas a replacementfkLogL
- this should be done via a
[X]IMPORTANT: We were still using the oldgetDiffusionworking with the fit for any code usingreadValidDsetsandreadCdlDset! Means both the diffusion values used in training for the background data as well as for anything with prediction based on real data likely had a wrong diffusion value! FIX and check the impact.
1.40.
Fixed the usage of the old
getDiffusioninio_helpers. Now using the correct value from the cache table. Let's see the effect of that on the effective efficiency!./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss.pt \ --ε 0.8 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/run2_run3_23_04_23_tanh300_correct_diffusion
The result is pretty much unchanged, if not possibly actually a bit worse than before.
Yeah, comparing with the old version it's a bit worse even. :/
We've implemented MLP support into createAllLikelihoodCombinations
now. The result is not super pretty, but it should work. The filename
of the MLP is added to the output file name, leading to very long
filenames. Ideally we'd have a better solution there.
Let's give it a test run, but before we do that, let's run a
likelihood combining the MLP with FADC, septem and line veto.
Run2:
likelihood \ -f ~/CastData/data/DataRuns2017_Reco.h5 \ --h5out /t/run_2_mlp_all_vetoes.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/23_04_23_tanh300_mse.pt \ --nnSignalEff 0.99 \ --vetoPercentile 0.99 \ --lineVeto \ --septemVeto \ --calibFile ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --readonly
likelihood \ -f ~/CastData/data/DataRuns2018_Reco.h5 \ --h5out /t/run_3_mlp_all_vetoes.h5 \ --region crGold \ --cdlYear 2018 \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --mlp ~/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/23_04_23_tanh300_mse.pt \ --nnSignalEff 0.99 \ --vetoPercentile 0.99 \ --lineVeto \ --septemVeto \ --calibFile ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --readonly
[ ]Well, issue:- need to find a way to better deal with the cache table for effective efficiencies! It uses strings in the keys, which we currently don't correctly support. Either change them to fixed length or fix the code for variable length strings as part compound types.
- when running likelihood our center event index is not assigned
correctly for the septem veto! Something is up there with the
predicted values or the cut values, such that we never enter the
branch the sets
centerEvIdx.
1.41.
[X]fixed thecenterEvIdxbug: the problem was our comparison of the NN prediction value to the cut value interpolator. I missed that the LnL and NN values are inverted!
Running the program to create all likelihood combinations:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll --regions crGold \ --signalEfficiency 0.8 --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 \ --fadcVetoPercentile 0.99 \ --vetoSets "{fkMLP, fkFadc, fkScinti, fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss.pt \ --out ~/org/resources/lhood_limits_23_04_23_mlp/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing
So we output the files to: ~/org/resources/lhoodlimits230423mlp
1.42.
Running the code last night more or less worked, but in some cases there was a segfault running the code.
I spent most of today debugging this issue. gdb while useful was
still pretty confusing. valgrind is still running…
It turns out that
- confusion and bad stack traces are due to injected destructor calls. those break the line information
- the issue is a crash in the destructor of a H5Group
- only triggered with our
try/exceptcode. We've changed that toif/elsenow. Saner anyway.
With this fixed, we can rerun the likelihood combinations. Both using the sigmoid output layer MLP from and if possible one of the linear output ones. For now start with one of them.
[ ]Note: when running with another MLP still need to regenerate the MLPDesc H5 file!
We'll rerun the command from yesterday. Still, can we speed up the process somehow?
I think we should introduce caching also for the
CutValueInterpolator.
1.43. &
Finally fixed all the HDF5 data writing stuff.. Will write more about that tomorrow.
Will test likelihood producing a sensible file now:
First a run to generate the file. Will take a short look, then read it
in a separate script to see what it contains. Then finally rerun the
same command to see if the fake event generation is then skipped.
likelihood -f /home/basti/CastData/data/DataRuns2018_Reco.h5 --h5out /t/blabla_broken.h5 --region=crGold --cdlYear=2018 --scintiveto --fadcveto --septemveto --lineveto --mlp /home/basti/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss.pt --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 --readOnly --calibFile=/home/basti/CastData/data/CalibrationRuns2018_Reco.h5 --vetoPercentile=0.99 --nnSignalEff=0.95
[X]segfault due to string[ ]HDF5-DIAG: Error detected in HDF5 (1.10.5) thread 0: #000: H5Tcompound.c line 354 in H5Tinsert(): unable to insert member major: Datatype minor: Unable to insert object #001: H5Tcompound.c line 446 in H5T__insert(): member extends past end of compound type major: Datatype minor: Unable to insert object-> partially fixed it but:
-> We have alignment issues. The H5 library also seems to align data in some cases when necessary. Our code currently assumes there is no such thing.
So I guess we need to go back to an approach that does actually generate some helper type or what?
-> I think I got it all working now. Merged the two approaches of
copying data to a Buffer and
Ok, the caching seems to work I think. And the generated file looks good.
Let's read the data and see what it contains:
import nimhdf5, tables const CacheTabFile = "/dev/shm/cacheTab_runLocalCutVals.h5" type TabKey = (int, string, float) # ^-- run number # ^-- sha1 hash of the NN model `.pt` file # ^-- target signal efficiency TabVal = seq[(string, float)] # ^-- CDL target # ^-- MLP cut value CacheTabTyp = Table[TabKey, TabVal] var tab = deserializeH5[CacheTabTyp](CacheTabFile) for k, v in tab: echo "Key: ", k, " = ", v
Key: (295, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.974539938569069), ("Al-Al-4kV", 0.9415906369686127), ("C-EPIC-0.6kV", 0.7813522785902023), ("Cu-EPIC-0.9kV", 0.8288368076086045), ("Cu-EPIC-2kV", 0.8751996099948883), ("Cu-Ni-15kV", 0.9722824782133103), ("Mn-Cr-12kV", 0.9686738938093186), ("Ti-Ti-9kV", 0.9641254603862762)] Key: (270, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9771067887544632), ("Al-Al-4kV", 0.9460293263196945), ("C-EPIC-0.6kV", 0.7850535094738007), ("Cu-EPIC-0.9kV", 0.8130916118621826), ("Cu-EPIC-2kV", 0.8937846541404724), ("Cu-Ni-15kV", 0.9715777307748794), ("Mn-Cr-12kV", 0.9703928083181381), ("Ti-Ti-9kV", 0.9622162997722625)] Key: (285, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9717331320047379), ("Al-Al-4kV", 0.9252435654401779), ("C-EPIC-0.6kV", 0.7428321331739426), ("Cu-EPIC-0.9kV", 0.7788086831569672), ("Cu-EPIC-2kV", 0.8636017471551896), ("Cu-Ni-15kV", 0.9687924206256866), ("Mn-Cr-12kV", 0.9663917392492294), ("Ti-Ti-9kV", 0.9504937410354615)] Key: (267, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.975317457318306), ("Al-Al-4kV", 0.9462290376424789), ("C-EPIC-0.6kV", 0.776226544380188), ("Cu-EPIC-0.9kV", 0.834472405910492), ("Cu-EPIC-2kV", 0.8766408234834671), ("Cu-Ni-15kV", 0.9714102745056152), ("Mn-Cr-12kV", 0.9717013716697693), ("Ti-Ti-9kV", 0.9653892040252685)] Key: (240, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9765782624483108), ("Al-Al-4kV", 0.9469905078411103), ("C-EPIC-0.6kV", 0.8053286731243133), ("Cu-EPIC-0.9kV", 0.8526969790458679), ("Cu-EPIC-2kV", 0.8962102770805359), ("Cu-Ni-15kV", 0.9759411454200745), ("Mn-Cr-12kV", 0.9700609385967255), ("Ti-Ti-9kV", 0.9632380992174149)] Key: (244, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9737197607755661), ("Al-Al-4kV", 0.936311411857605), ("C-EPIC-0.6kV", 0.7668724238872529), ("Cu-EPIC-0.9kV", 0.8007214874029159), ("Cu-EPIC-2kV", 0.8769152045249939), ("Cu-Ni-15kV", 0.9682861983776092), ("Mn-Cr-12kV", 0.9659816890954971), ("Ti-Ti-9kV", 0.9604099780321121)] Key: (287, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9702932089567184), ("Al-Al-4kV", 0.9345487594604492), ("C-EPIC-0.6kV", 0.7561751186847686), ("Cu-EPIC-0.9kV", 0.7987294286489487), ("Cu-EPIC-2kV", 0.8735345602035522), ("Cu-Ni-15kV", 0.9701228439807892), ("Mn-Cr-12kV", 0.9675930917263031), ("Ti-Ti-9kV", 0.9575580269098282)] Key: (278, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9720843970775604), ("Al-Al-4kV", 0.9305856913328171), ("C-EPIC-0.6kV", 0.741540789604187), ("Cu-EPIC-0.9kV", 0.7682088732719422), ("Cu-EPIC-2kV", 0.864213228225708), ("Cu-Ni-15kV", 0.9648653626441955), ("Mn-Cr-12kV", 0.9665578484535218), ("Ti-Ti-9kV", 0.9552424371242523)] Key: (250, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9766805320978165), ("Al-Al-4kV", 0.9418972045183182), ("C-EPIC-0.6kV", 0.76584292948246), ("Cu-EPIC-0.9kV", 0.8238443195819855), ("Cu-EPIC-2kV", 0.8775055557489395), ("Cu-Ni-15kV", 0.9738869041204452), ("Mn-Cr-12kV", 0.9712955445051193), ("Ti-Ti-9kV", 0.9667880535125732)] Key: (283, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9675714403390885), ("Al-Al-4kV", 0.9285876244306565), ("C-EPIC-0.6kV", 0.71473089158535), ("Cu-EPIC-0.9kV", 0.7696082562208175), ("Cu-EPIC-2kV", 0.8377785980701447), ("Cu-Ni-15kV", 0.9635965615510941), ("Mn-Cr-12kV", 0.9641305923461914), ("Ti-Ti-9kV", 0.9575453609228134)] Key: (301, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9705011487007141), ("Al-Al-4kV", 0.9279218137264251), ("C-EPIC-0.6kV", 0.7229891419410706), ("Cu-EPIC-0.9kV", 0.793598598241806), ("Cu-EPIC-2kV", 0.8582320868968963), ("Cu-Ni-15kV", 0.9688272416591645), ("Mn-Cr-12kV", 0.9668716788291931), ("Ti-Ti-9kV", 0.9546888172626495)] Key: (274, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.974557614326477), ("Al-Al-4kV", 0.9371200978755951), ("C-EPIC-0.6kV", 0.7444291532039642), ("Cu-EPIC-0.9kV", 0.7895265400409699), ("Cu-EPIC-2kV", 0.8598116517066956), ("Cu-Ni-15kV", 0.9712087035179138), ("Mn-Cr-12kV", 0.9688791006803512), ("Ti-Ti-9kV", 0.9589674681425094)] Key: (242, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9761280864477158), ("Al-Al-4kV", 0.9446888864040375), ("C-EPIC-0.6kV", 0.766591414809227), ("Cu-EPIC-0.9kV", 0.8117899149656296), ("Cu-EPIC-2kV", 0.8900630325078964), ("Cu-Ni-15kV", 0.971353754401207), ("Mn-Cr-12kV", 0.9718088060617447), ("Ti-Ti-9kV", 0.9645727574825287)] Key: (306, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9756848812103271), ("Al-Al-4kV", 0.9455605119466781), ("C-EPIC-0.6kV", 0.7938183635473252), ("Cu-EPIC-0.9kV", 0.8287457168102265), ("Cu-EPIC-2kV", 0.8792453199625015), ("Cu-Ni-15kV", 0.9696165889501571), ("Mn-Cr-12kV", 0.972235518693924), ("Ti-Ti-9kV", 0.9627663731575012)] Key: (303, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9724529892206192), ("Al-Al-4kV", 0.9305596590042114), ("C-EPIC-0.6kV", 0.7230549484491349), ("Cu-EPIC-0.9kV", 0.7953008472919464), ("Cu-EPIC-2kV", 0.8613291561603547), ("Cu-Ni-15kV", 0.9646958172321319), ("Mn-Cr-12kV", 0.9663623839616775), ("Ti-Ti-9kV", 0.9565762877464294)] Key: (291, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9746044754981995), ("Al-Al-4kV", 0.945956015586853), ("C-EPIC-0.6kV", 0.7661843031644822), ("Cu-EPIC-0.9kV", 0.8199316382408142), ("Cu-EPIC-2kV", 0.8820369154214859), ("Cu-Ni-15kV", 0.972326734662056), ("Mn-Cr-12kV", 0.9720319092273713), ("Ti-Ti-9kV", 0.9641989678144455)] Key: (281, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9678589403629303), ("Al-Al-4kV", 0.9328784346580505), ("C-EPIC-0.6kV", 0.7547005414962769), ("Cu-EPIC-0.9kV", 0.7789339125156403), ("Cu-EPIC-2kV", 0.8745017945766449), ("Cu-Ni-15kV", 0.9656407535076141), ("Mn-Cr-12kV", 0.9641033113002777), ("Ti-Ti-9kV", 0.9552679359912872)] Key: (276, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9702300161123276), ("Al-Al-4kV", 0.9325366169214249), ("C-EPIC-0.6kV", 0.7419947564601898), ("Cu-EPIC-0.9kV", 0.7747547417879105), ("Cu-EPIC-2kV", 0.8381759762763977), ("Cu-Ni-15kV", 0.9623401463031769), ("Mn-Cr-12kV", 0.9613375902175904), ("Ti-Ti-9kV", 0.9547658741474152)] Key: (279, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9683325439691544), ("Al-Al-4kV", 0.9248780459165573), ("C-EPIC-0.6kV", 0.7393383264541626), ("Cu-EPIC-0.9kV", 0.7804951041936874), ("Cu-EPIC-2kV", 0.8707629531621933), ("Cu-Ni-15kV", 0.9666939914226532), ("Mn-Cr-12kV", 0.9646541595458984), ("Ti-Ti-9kV", 0.9589365422725677)] Key: (268, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9719461470842361), ("Al-Al-4kV", 0.9368112564086915), ("C-EPIC-0.6kV", 0.7573535948991775), ("Cu-EPIC-0.9kV", 0.8275747984647751), ("Cu-EPIC-2kV", 0.8748720288276672), ("Cu-Ni-15kV", 0.9710170537233352), ("Mn-Cr-12kV", 0.9695031344890594), ("Ti-Ti-9kV", 0.9605758100748062)] Key: (258, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9745412111282349), ("Al-Al-4kV", 0.9339351028203964), ("C-EPIC-0.6kV", 0.7520094603300095), ("Cu-EPIC-0.9kV", 0.8004794657230377), ("Cu-EPIC-2kV", 0.8735634952783584), ("Cu-Ni-15kV", 0.9667491674423218), ("Mn-Cr-12kV", 0.9659819602966309), ("Ti-Ti-9kV", 0.9595000624656678)] Key: (248, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.977547001838684), ("Al-Al-4kV", 0.943942391872406), ("C-EPIC-0.6kV", 0.7833251833915711), ("Cu-EPIC-0.9kV", 0.8251269578933715), ("Cu-EPIC-2kV", 0.8876170873641968), ("Cu-Ni-15kV", 0.9738454699516297), ("Mn-Cr-12kV", 0.9718320488929748), ("Ti-Ti-9kV", 0.9669605851173401)] Key: (246, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9743694633245468), ("Al-Al-4kV", 0.9456877648830414), ("C-EPIC-0.6kV", 0.7678399056196212), ("Cu-EPIC-0.9kV", 0.8119639933109284), ("Cu-EPIC-2kV", 0.8899865686893463), ("Cu-Ni-15kV", 0.9739743441343307), ("Mn-Cr-12kV", 0.9730724036693573), ("Ti-Ti-9kV", 0.966443908214569)] Key: (254, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9705976217985153), ("Al-Al-4kV", 0.9223994761705399), ("C-EPIC-0.6kV", 0.7517837852239608), ("Cu-EPIC-0.9kV", 0.7894137501716614), ("Cu-EPIC-2kV", 0.8740812391042709), ("Cu-Ni-15kV", 0.9649749875068665), ("Mn-Cr-12kV", 0.9622029483318328), ("Ti-Ti-9kV", 0.9560768663883209)] Key: (299, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9735205113887787), ("Al-Al-4kV", 0.9309952080249786), ("C-EPIC-0.6kV", 0.7400766223669052), ("Cu-EPIC-0.9kV", 0.7843391090631485), ("Cu-EPIC-2kV", 0.8648605406284332), ("Cu-Ni-15kV", 0.967012819647789), ("Mn-Cr-12kV", 0.9662521809339524), ("Ti-Ti-9kV", 0.9518016576766968)] Key: (256, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9687688469886779), ("Al-Al-4kV", 0.9329577207565307), ("C-EPIC-0.6kV", 0.7124274164438248), ("Cu-EPIC-0.9kV", 0.7790258109569549), ("Cu-EPIC-2kV", 0.8657959043979645), ("Cu-Ni-15kV", 0.9670290321111679), ("Mn-Cr-12kV", 0.9633038669824601), ("Ti-Ti-9kV", 0.9567578852176666)] Key: (293, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9724262475967407), ("Al-Al-4kV", 0.9334424823522568), ("C-EPIC-0.6kV", 0.7237139195203781), ("Cu-EPIC-0.9kV", 0.8003135979175567), ("Cu-EPIC-2kV", 0.8604774057865143), ("Cu-Ni-15kV", 0.9647800117731095), ("Mn-Cr-12kV", 0.9671240687370301), ("Ti-Ti-9kV", 0.9593446969985961)] Key: (261, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.971569812297821), ("Al-Al-4kV", 0.9378552496433258), ("C-EPIC-0.6kV", 0.7479894399642945), ("Cu-EPIC-0.9kV", 0.8020979523658752), ("Cu-EPIC-2kV", 0.8605112731456757), ("Cu-Ni-15kV", 0.9698976039886474), ("Mn-Cr-12kV", 0.9648890495300293), ("Ti-Ti-9kV", 0.9587083220481872)] Key: (289, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9720789223909378), ("Al-Al-4kV", 0.9372004061937332), ("C-EPIC-0.6kV", 0.7466429948806763), ("Cu-EPIC-0.9kV", 0.7778348356485367), ("Cu-EPIC-2kV", 0.8603413850069046), ("Cu-Ni-15kV", 0.967164334654808), ("Mn-Cr-12kV", 0.9689937591552734), ("Ti-Ti-9kV", 0.9565973937511444)] Key: (298, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9661869823932647), ("Al-Al-4kV", 0.9258407413959503), ("C-EPIC-0.6kV", 0.7028941214084625), ("Cu-EPIC-0.9kV", 0.7626788705587387), ("Cu-EPIC-2kV", 0.8486940711736679), ("Cu-Ni-15kV", 0.9655997604131699), ("Mn-Cr-12kV", 0.9632629603147507), ("Ti-Ti-9kV", 0.9513149082660675)] Key: (265, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9717926532030106), ("Al-Al-4kV", 0.9275272369384766), ("C-EPIC-0.6kV", 0.7334349215030671), ("Cu-EPIC-0.9kV", 0.7848327666521072), ("Cu-EPIC-2kV", 0.879998528957367), ("Cu-Ni-15kV", 0.9686383992433548), ("Mn-Cr-12kV", 0.9665170550346375), ("Ti-Ti-9kV", 0.9604606479406357)] Key: (272, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9744620800018311), ("Al-Al-4kV", 0.9406907320022583), ("C-EPIC-0.6kV", 0.7583581209182739), ("Cu-EPIC-0.9kV", 0.803549861907959), ("Cu-EPIC-2kV", 0.8741284489631653), ("Cu-Ni-15kV", 0.9701647937297821), ("Mn-Cr-12kV", 0.9710357129573822), ("Ti-Ti-9kV", 0.9592038929462433)] Key: (297, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.970538979768753), ("Al-Al-4kV", 0.9283351331949234), ("C-EPIC-0.6kV", 0.7336910545825959), ("Cu-EPIC-0.9kV", 0.7939026236534119), ("Cu-EPIC-2kV", 0.8525126844644546), ("Cu-Ni-15kV", 0.9671412736177445), ("Mn-Cr-12kV", 0.9655984342098236), ("Ti-Ti-9kV", 0.9555086642503738)] Key: (263, "D7DBC196401F3CAC564FBE899306343BEB5022BA", 0.95) = @[("Ag-Ag-6kV", 0.9716018617153168), ("Al-Al-4kV", 0.9364291220903397), ("C-EPIC-0.6kV", 0.7137390047311782), ("Cu-EPIC-0.9kV", 0.7985885977745056), ("Cu-EPIC-2kV", 0.8676451265811921), ("Cu-Ni-15kV", 0.9684803396463394), ("Mn-Cr-12kV", 0.9639465093612671), ("Ti-Ti-9kV", 0.9574855715036392)]
And finally rerun all likelihood combinations:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll --regions crGold \ --signalEfficiency 0.8 --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 \ --fadcVetoPercentile 0.99 \ --vetoSets "{fkMLP, fkFadc, fkScinti, fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss.pt \ --out ~/org/resources/lhood_limits_23_04_23_mlp/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing
Running the command was somewhat of a failure:
- the default of 8 jobs per process is not enough. We use way more memory than expected. Even with only 4 jobs, there is a risk of one or more being killed!
sometimes (at least in
crAllcombinations) we still get the "broken event" quit error. In this case however we can spy the following:Cluster: 1 of chip 3 has val : 0.9999758005142212 copmare: 0.9467523097991943 Cluster: 0 of chip 5 has val : 0.02295641601085663 copmare: 0.9783756822347641 Cluster: 1 of chip 3 has val : 0.9999998807907104 copmare: 0.9467523097991943 Cluster: 0 of chip 4 has val : 5.220065759203862e-06 copmare: 0.99133480489254 Cluster: 1 of chip 3 has val : 0.9999876022338867 copmare: 0.9467523097991943 Cluster: 0 of chip 3 has val : nan copmare: 0.9545457273721695 Broken event! DataFrame with 3 columns and 1 rows: Idx eventIndex eventNumber chipNumber dtype: int int float 0 10750 35811 3-> Note the
nanvalue for cluster 0 in the last line before the DF print! This means the MLP predicted a value ofnanfor a cluster!
-> We need to debug the likelihood call for one of the cases and isolate the events for that.
[X]get a combination that causes this:
lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss_vQ_0.99.log
[X]extract which run is at fault -> Run 92 in this case[X]Doesnanappear more often? -> Yes! O(5) times in the same log file[ ]Rerun the equivalent command for testing and try to debug the cause. -> Reconstruct the command:
likelihood \ -f /home/basti/CastData/data/DataRuns2017_Reco.h5 \ --h5out /t/debug_broken_event.h5 \ --region=crAll \ --cdlYear=2018 \ --scintiveto \ --fadcveto \ --septemveto \ --lineveto \ --mlp /home/basti/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss.pt \ --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --readOnly \ --calibFile=/home/basti/CastData/data/CalibrationRuns2017_Reco.h5 \ --vetoPercentile=0.99 \ --nnSignalEff=0.85
[X]ModifycreateAllLikelihoodCombinationssuch that each job does not stop on a failure, but retries failed jobs -> Done[ ]I'll restart the create likelihood combinations command now that we've implemented restart on failure & disabled failing on NaN septem events (i.e. "no cluster found") -> FIX ME
[X]We've also implemented a version oftoH5anddeserializeH5that
retries the writing / reading if the file is locked. This should make it safer to run multiple instances in parallel which might try to access the same file.
1.44.
[X]Look into the origin of events with a NaN value! -> Likely they just pass by accident due to having NaN? That would be "good"[X]look at what such clusters look like
Having inserted printing the event number for run 92 with the NaN event, the output is:
Broken event! DataFrame with 3 columns and 1 rows:
Idx eventIndex eventNumber chipNumber
dtype: int int float
0 10750 35811 3
The event is: @[("eventNumber", (kind: VInt, num: 35811))]
Let's plot that event:
plotData \ --h5file ~/CastData/data/DataRuns2017_Reco.h5 \ --runType rtBackground \ --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml \ --runs 92 \ --eventDisplay \ --septemboard \ --events 35811
The event is:
Well, even some of the properties are NaN! No wonder it yields a NaN result for the NN prediction. We have to be careful not to accidentally consider them "passing" though! (Which seems to be the case currently)
We've changed the code to always veto NaN events. And as a result the broken event should never happen again, which is why we reintroduced the quit condition. Let's see if the command from yesterday now runs correctly.
Well, great. The process was killed due to memory usage of running 5 jobs concurrently. Guess I'll just wait for all the other jobs to finish now. We're ~half way done or so.
Small problem: In the limit calculation given that our effective efficiency is Run-2/3 specific we need to think about how to handle that. Can we use different efficiencies for different parts of the data? Certainly, but makes everything more complicated. -> Better to compute an average of the two different runs.
Sigh, it seems like even 2 jobs at the same time can use too much memory!
(oh, what a dummy: I didn't even add the 99% case to the createLogL call!)
Therefore:
For now let's try to see what we can get with exactly one setup:
- 99% MLP
- all vetoes except septem veto
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll --regions crGold \ --signalEfficiency 0.99 \ --fadcVetoPercentile 0.99 \ --vetoSets "{fkMLP, +fkFadc, +fkScinti, fkLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss.pt \ --out ~/org/resources/lhood_limits_23_04_23_mlp_sEff_0.99/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ #--multiprocessing
We're running without multiprocessing now. :( At least for the crAll cases. We'll do crGold with 2 jobs first.
Gold is finished and now running crAll with a single job. :/
So: Things we want for the meeting:
- background rate in gold region
- background clusters of MLP@99
- background clusters of MLP@99 + vetoes
- expected limits for MLP@99 & MLP@99+vetoes
1.44.1. Background rate in gold region
We will compare:
- lnL@80
- lnL@80 + vetoes
- MLP@99
- MLP@99 + vetoes
plotBackgroundRate \ ~/org/resources/lhood_limits_23_04_23_mlp_sEff_0.99/lhood_c18_R2_crGold_sEff_0.99_mlp_trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss.h5 \ ~/org/resources/lhood_limits_23_04_23_mlp_sEff_0.99/lhood_c18_R3_crGold_sEff_0.99_mlp_trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss.h5 \ ~/org/resources/lhood_limits_23_04_23_mlp_sEff_0.99/lhood_c18_R2_crGold_sEff_0.99_scinti_fadc_line_mlp_trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss_vQ_0.99.h5 \ ~/org/resources/lhood_limits_23_04_23_mlp_sEff_0.99/lhood_c18_R3_crGold_sEff_0.99_scinti_fadc_line_mlp_trained_mlp_sgd_gauss_diffusion_tanh300_mse_loss_vQ_0.99.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold_scinti_fadc_line_vetoPercentile_0.99.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold_scinti_fadc_line_vetoPercentile_0.99.h5 \ --names "MLP@99" --names "MLP@99" --names "MLP@99+V" --names "MLP@99+V" --names "LnL@80" --names "LnL@80" --names "LnL@80+V" --names "LnL@80+V" \ --centerChip 3 \ --title "Background rate CAST, LnL@80%, SGD tanh300 MLE MLP@99% + vetoes" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_run2_3_mlp_0.99_plus_vetoes.pdf \ --outpath ~/Sync/ \ --quiet
1.45.
UPDATE:
We changed the train_ingrid code slightly today, so that our
previous hardcoded change to replace subsetPerRun by 6 *
subsetPerRun for background data was taken out. That's why I modified
the command below to accommodate for that by adding the
--subsetPerRun 6000 argument!
- the fact that
mcmc_limitgets stuck ontoH5is because of theKDtree, which is massive with O(350 k) entries! [X]add filter option (energyMinandenergyMax) -> shows that in Run-2 alone without any energy filtering & without vetoes there are almost 200k cluster! -> cutting 0.2 < E < 12 and using vetoes cuts it in half O(40k) compared to O(75k) when 0 < E < 12.[X]Check data selection for background training sample -> OUCH: We still filter to THE GOLD REGION in the background sample. This likely explains why the background is so good there, but horrible towards the edges![X]write a custom serializer forKDTreeto avoid extremely nested H5 data structure -> We now only serialize the actual data of the tree. The tree can be rebuilt from that after all.[X]implement filtering on energy < 200 eV formcmc_limit[X]also check the impact of that on the limit! -> running right now -> very slight improvement to gae² = 6.28628480082639e-21 from gae² = 6.333984435685045e-21
[ ]analyze memory usage oflikelihoodwhen using NN veto[X]Train a new MLP of same architecture as currently, but using background data over the whole chip! -> [X] CenterX or Y is not part of inputs, right? -> nope, it's not
Here we go:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/10_05_23_sgd_gauss_diffusion_tanh300_mse_loss/mlp_tanh300_mse.pt \ --plotPath ~/Sync/10_05_23_sgd_tanh300_mse/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --subsetPerRun 6000
-> Trained up to 500k on .
1.46.
Continue from yesterday:
[ ]analyze memory usage oflikelihoodwhen using NN veto -> try withnimprof-> useless
Regenerating the cache tables:
./determineDiffusion \ ~/CastData/data/DataRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/DataRuns2018_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll --regions crGold \ --signalEfficiency 0.99 \ --fadcVetoPercentile 0.99 \ --vetoSets "{fkMLP, +fkFadc, +fkScinti, fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/10_05_23_sgd_gauss_diffusion_tanh300_mse_loss/mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.pt \ --out ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5
With these plots generated and the corresponding sections written, we are almost done with that part of the thesis.
[ ]Mini section about background rate with MLP[ ]Extend MLP w/ all vetoes to include MLP
1.47.
[X]Let's see if running
likelihoodwithlnLinstead of the MLP also eats more and more memory!likelihood -f /home/basti/CastData/data/DataRuns2017_Reco.h5 --h5out /t/dm_lnl_slowit.h5 --region=crAll --cdlYear=2018 --scintiveto --fadcveto --septemveto --lineveto --lnL --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 --readOnly --calibFile=/home/basti/CastData/data/CalibrationRuns2017_Reco.h5 --vetoPercentile=0.99 --signalEff=0.8
-> Not really. It stays nice and low.
Running the following script:
import nimhdf5, os, seqmath, sequtils, datamancer import ingrid / ingrid_types proc readAllChipData(h5f: H5File, group: H5Group, numChips: int): AllChipData = ## Read all data for all chips of this run that we need for the septem veto let vlenXY = special_type(uint8) let vlenCh = special_type(float64) result = AllChipData(x: newSeq[seq[seq[uint8]]](numChips), y: newSeq[seq[seq[uint8]]](numChips), ToT: newSeq[seq[seq[uint16]]](numChips), charge: newSeq[seq[seq[float]]](numChips)) for i in 0 ..< numChips: result.x[i] = h5f[group.name / "chip_" & $i / "x", vlenXY, uint8] result.y[i] = h5f[group.name / "chip_" & $i / "y", vlenXY, uint8] result.ToT[i] = h5f[group.name / "chip_" & $i / "ToT", vlenCh, uint16] result.charge[i] = h5f[group.name / "chip_" & $i / "charge", vlenCh, float] var h5f = H5open("~/CastData/data/DataRuns2017_Reco.h5", "r") let grp = h5f["/reconstruction/run_186/".grp_str] echo grp var df = newDataFrame() for i in 0 ..< 10: let data = readAllChipData(h5f, grp, 7) df = toDf({"x" : data.x.flatten.mapIt(it.float)}) echo "Read: ", i, " = ", getOccupiedMem().float / 1e6, " MB", " df len: ", df.len discard h5f.close()
under valgrind right now.
-> In this setup valgrind does not see any leaks.
Let's also run it under heaptrack to see where the 8GB of memory
come from!
-> Running without -d:useMalloc yields pretty much nothing (as it
seems to intercept malloc calls)
-> With -d:useMalloc it doesn't look unusual at all
-> the 8GB seen when running without -d:useMalloc seem to be the
standard Nim allocator doing its thing
-> So this code snippet seems fine.
[ ]let's try to trim downlikelihoodto still reproduce the memleak issuesee the memory usage of a cpp MLP run without vetoes -> so far seems to run without growing endlessely. -> it grows slightly, approaching 9GB Maybe we can run this under
heaptrack? -> Yeah, no problems without septem&line veto -> Now trying with only septem veto -> This already crashesheaptrack! -> Let's trylikelihoodwith lnLheaptrack likelihood -f /home/basti/CastData/data/DataRuns2017_Reco.h5 --h5out /t/more_lnl_slowit.h5 --region=crAll --cdlYear=2018 --scintiveto --fadcveto --septemveto --lnL --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 --readOnly --calibFile=/home/basti/CastData/data/CalibrationRuns2017_Reco.h5 --vetoPercentile=0.99 --signalEff=0.80
[X]Even withlnLand septem vetoheaptrackcrashes! Let's see if we understand where.
[ ]Debug the crashing of
heaptrack-> cause is in septem veto -> in geometry -> in DBScan reconstruction -> in nearestNeighbor call[X]serialized a bunch of pixels that cause the crash in/tmp/serial_pixels.h5
-> let's only run DBSCAN on these pixels!
import arraymancer proc callDb(data: Tensor[float]) = echo dbscan(data, 65.0, 3) import nimhdf5 type Foo = object shape: seq[int] data: seq[float] let foo = deserializeH5[Foo]("/tmp/serial_pixels.h5") var pT = foo.data.toTensor.reshape(foo.shape) for _ in 0 ..< 1000: callDb(pT)
-> cannot reproduce the problem this way
[X]Ok, given thatheaptrackalways seems to crash when using DBSCAN in that context, I just ran it using the default cluster algo for the septem veto logic. This ran correctly and showed no real memory leak. And a peak heap mem size of ~6 GB.[X]Running again now using default clusterer but using MLP! -> Reproduces the problem andheaptracktracks the issue. We're leaking in the H5 library due to identifiers not being closed. I've changed the code now to automatically close identifiers by attaching them to=destroycalls. -> Also changed the logic to only read the MLP H5 file a single time for the septem veto, because this was the main origin (reading the file for each single cluster!)
UPDATE:
-> We've replaced the distinct hid_t logic in nimhdf5 by an approach
that wraps the identifiers in a ref object to make sure we destroy
every single identifiers when it goes out of scope. This fixed the
memory leak that could finally be tracked properly with the following
snippet:
import nimhdf5 type MLPDesc* = object version*: int # Version of this MLPDesc object path*: string # model path to the checkpoint files including the default model name! modelDir*: string # the parent directory of `path` plotPath*: string # path in which plots are placed calibFiles*: seq[string] ## Path to the calibration files backFiles*: seq[string] ## Path to the background data files simulatedData*: bool numInputs*: int numHidden*: seq[int] numLayers*: int learningRate*: float datasets*: seq[string] # Not `InGridDsetKind` to support arbitrary new columns subsetPerRun*: int rngSeed*: int backgroundRegion*: string nFake*: int # number of fake events per run period to generate # activationFunction*: string outputActivation*: string lossFunction*: string optimizer*: string # fields that store training information epochs*: seq[int] ## epochs at which plots and checkpoints are generated accuracies*: seq[float] testAccuracies*: seq[float] losses*: seq[float] testLosses*: seq[float] proc getNumber(file: string): int = let desc = deserializeH5[MLPDesc](file) result = desc.numInputs proc main(fname: string) = for i in 0 ..< 50_000: echo "Number: ", i, " gives ", getNumber(fname) when isMainModule: import cligen dispatch main
running via:
heaptrack ./testmem3 -f ~/org/resources/nn_devel_mixing/23_04_23_sgd_gauss_diffusion_tanh300_mse_loss/mlp_desc.h5
The issue really was the many deserializeH5 calls, which suddenly
highlighted the memory leak!
In the meantime let's look at background clusters of the likelihood
outputs from last night:
1.48.
[ ]Try again to rerun
heaptrackon the DBSCAN code:heaptrack likelihood -f /home/basti/CastData/data/DataRuns2017_Reco.h5 --h5out /t/mlp_isit_faster_noleak_dbscan.h5 --region=crAll --cdlYear=2018 --scintiveto --fadcveto --septemveto --mlp ~/org/resources/nn_devel_mixing/10_05_23_sgd_gauss_diffusion_tanh300_mse_loss/mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.pt --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 --readOnly --calibFile=/home/basti/CastData/data/CalibrationRuns2017_Reco.h5 --vetoPercentile=0.99 --nnSignalEff=0.99
-> Still the same problem! So either a bug in
heaptrackor a bug in our code still. :/[ ]Run
perfon the code usingdefaultclustering algo. Then usehotspotto check the report:perf record -o /t/log_noleaks_slow.data --call-graph dwarf -- likelihood -f /home/basti/CastData/data/DataRuns2017_Reco.h5 --h5out /t/log_noleaks_debug_slowit. h5 --region=crAll --cdlYear=2018 --scintiveto --fadcveto --septemveto --lineveto --mlp ~/org/resources/nn_devel_mixing/10_05_23_sgd_gauss_diffusion_tanh300_mse_loss/mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.pt --cdlFile =/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 --readOnly --calibFile=/home/basti/CastData/data/CalibrationRuns2017_Reco.h5 --vetoPercentile=0.99 --nnSignalEff=0.99
Killed it after about 20 runs processed as the file already grew to over 15GB. Should be enough statistics… :) -> Ok, the perf data shows that time is spent in a variety of different places.
mergeChainof the cluster algo is a bigger one, so isforwardof the MLP and generally a whole lot of copying data. Nothing in particular jumps out though.I guess performance for this is acceptable for now.
Let's also look at
DBSCAN(same command as above): -> Yeah, as expected. ThequeryImplcall is by far the dominating part of the performance report when using DBSCAN. Theheapqueuesused make up>50% of the time with the distance and ~index_selectandtoTensorTuplethe rest. IntoTensorTuplethe dominating factor ispop.-> We've replaced the
HeapQueueby aSortedSeqnow for testing. With it thepopprocedure is much faster (but insert is a bit slower). We've finally added a--runoption tolikelihoodto now time the performance of the heapqueue vs the sorted seq. We'll run on run 168 as it is one of the longest runs. Sorted seq:likelihood -f /home/basti/CastData/data/DataRuns2017_Reco.h5 --h5out 314.52s user 3.16s system 100%
Heap queue:
likelihood -f /home/basti/CastData/data/DataRuns2017_Reco.h5 --h5out 324.78s user 3.12s system 100%
-> Well. Guess it is a bit faster than the heap queue approach, but it's a small improvement. I guess we mostly traded building the sorted seq by popping from it. NOTE: There was a segfault after the run was done?
[X]just ran heaptrack again on the default clustering case for the full case, because I didn't remember if we did that with the ref object IDs. All looking good now. Peak memory at9.5GB. High, but explained by the 3.7GB overhead of the CUDA (that is "leaked"). Oh, I just googled for the CUDA "leak": https://discuss.pytorch.org/t/memory-leak-in-libtorch-extremely-simple-code/38149/3 -> It's because we don't use "no grad" mode! Hmm, but we *are* using our ~no_grad_modetemplate innn_typesand effectively innn_predict. -> I introduced the "NoGradGuard" into Flamebau and used it in places in addition to theno_grad_modeand runheaptrackagain. Let's see. -> Didn't change anything!
So I guess that means we continue with our actual work. Performance is deemed acceptable now.
Let's go with a test run of 5 different jobs in
createAllLikelihoodCombinations:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll --regions crGold \ --signalEfficiency 0.95 --signalEfficiency 0.90 \ --fadcVetoPercentile 0.99 \ --vetoSets "{fkMLP, +fkFadc, +fkScinti, fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/10_05_23_sgd_gauss_diffusion_tanh300_mse_loss/mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.pt \ --out ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 5
[X]We get errors for dataspaces. Apparently ournimhdf5code is problematic in some cases. Let's try to fix that up, run all its tests. ->tread_writereproduces it -> fixed all the issues by another rewrite of the ID logic
1.49.
NOTE: I'm stopping the following limit calculation at :
shellCmd: mcmc_limit_calculation limit -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.99_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 -f /home/basti/org/resources/lhood_li mits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.99_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 --years 2017 --years 2018 --σ_p 0.05 --limitKind lkMCMC --nmc 1000 --suffix=_sEff_0.99_mlp_mlp_tanh300_msecheckpo int_epoch_485000_loss_0.0055_acc_0.9933 --path "" --outpath /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/limits/ shell 5127> files @["/home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.99_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5", "/home/basti/org/resources/lhood_limits_10_05_23_mlp_sEf f_0.99/lhood_c18_R3_crAll_sEff_0.99_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5"] shell 5127> shell 5127> @["/home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.99_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5", "/home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99 /lhood_c18_R3_crAll_sEff_0.99_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5"] shell 5127> [INFO]: Read a total of 340911 input clusters.
because each MCMC takes too long to build:
shell 5127> MC index 12 shell 5127> Building chain of 150000 elements took 126.9490270614624 s shell 5127> Acceptance rate: 0.2949866666666667 with last two states of chain: @[@[1.250804325635904e-21, 0.02908566421900792, -0.004614450435544698, 0.03164360408307887, 0.03619049049344585], @[1.250804325635904e-21, 0.02908566421900792, -0.004614450435544698, 0.03164360408307887, 0.03619049049344585]] shell 5127> Limit at 7.666033305250432e-21 shell 5127> Number of candidates: 16133 shell 5127> INFO: The integer column `Hist` has been automatically determined to be continuous. To overwrite this behavior add a `+ scale_x/y_discrete()` call to the plotting chain. Choose `x` or `y` depending on which axis this column refers to. Or apply a `factor` to the column name in the `aes` call, i.e. `aes(..., factor("Hist"), ...)`. shell 5127> MC index 12 shell 5127> Building chain of 150000 elements took 133.913836479187 s shell 5127> Acceptance rate: 0.3029 with last two states of chain: @[@[5.394302558724233e-21, 0.005117702932866319, -0.003854151251413666, 0.1140589851066757, -0.01650063836525805], @[5.394302558724233e-21, 0.005117702932866319, -0.003854151251413666, 0.1140589851066757, -0.01650063836525805]] shell 5127> Limit at 8.815294304890919e-21 shell 5127> Number of candidates: 16518 shell 5127> INFO: The integer column `Hist` has been automatically determined to be continuous. To overwrite this behavior add a `+ scale_x/y_discrete()` call to the plotting chain. Choose `x` or `y` depending on which axis this column refers to. Or apply a `factor` to the column name in the `aes` call, i.e. `aes(..., factor("Hist"), ...)`. shell 5127> MC index 12
(due to the ~16k candidates)
I'll add the file to processed.txt and start the rest now.
shell 5127> Initial chain state: @[3.325031213438127e-21, -0.005975150812670194, 0.2566543411242529, 0.1308918272537833, 0.3838098582402962] ^C Interrupted while running processing of file: /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.99_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 Check the `processed.txt` file in the output path to see which files were processed successfully!
Added that file manually to the processed.txt file now. Restarting:
./runLimits --path ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/ --outpath ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/limits/ --prefix lhood_c18_R2_crAll --nmc 1000
Doing the same with 90% MLP:
^C Interrupted while running processing of file: /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 Check the `processed.txt` file in the output path to see which files were processed successfully!
Restarted with the veto based ones for 90% left.
Running the expected limits table generator:
[X]I updated it to include the MLP efficiencies. Still have to change it so that it prints whether MLP or LnL was in use!
./generateExpectedLimitsTable \ -p ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/limits/ \ --prefix mc_limit_lkMCMC_skInterpBackground_nmc_1000
yields:
| εlnL | MLP | MLPeff | Scinti | FADC | εFADC | Septem | Line | eccLineCut | εSeptem | εLine | εSeptemLine | Total eff. | Limit no signal | Expected Limit |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.8 | 0.95 | 0.9107 | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7677 | 3.7078e-21 | 7.7409e-23 |
| 0.8 | 0.99 | 0.9718 | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.8192 | 3.509e-21 | 7.871e-23 |
| 0.8 | 0.9 | 0.8474 | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7143 | 3.8986e-21 | 7.9114e-23 |
| 0.8 | 0.95 | 0.9107 | false | false | 0.98 | false | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.9107 | 3.1115e-21 | 8.1099e-23 |
| 0.8 | 0.85 | 0.7926 | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6681 | 4.2397e-21 | 8.1234e-23 |
| 0.8 | 0.8 | 0.7398 | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6237 | 4.5115e-21 | 8.2423e-23 |
| 0.8 | 0.85 | 0.7926 | false | false | 0.98 | false | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7926 | 3.6449e-21 | 8.3336e-23 |
| 0.8 | 0.99 | 0.9718 | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6976 | 4.0701e-21 | 8.3474e-23 |
| 0.8 | 0.99 | 0.9718 | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7468 | 3.8991e-21 | 8.3492e-23 |
| 0.8 | 0.8 | 0.7398 | false | false | 0.98 | false | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7398 | 3.9209e-21 | 8.4438e-23 |
| 0.8 | 0.95 | 0.9107 | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6538 | 4.2749e-21 | 8.4451e-23 |
| 0.8 | 0.9 | 0.8474 | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6083 | 4.6237e-21 | 8.4821e-23 |
| 0.8 | 0.95 | 0.9107 | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6998 | 4.049e-21 | 8.5324e-23 |
| 0.8 | 0.9 | 0.8474 | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6511 | 4.2498e-21 | 8.5486e-23 |
| 0.8 | 0.85 | 0.7926 | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.569 | 4.9101e-21 | 8.7655e-23 |
| 0.8 | 0.85 | 0.7926 | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.609 | 4.6382e-21 | 8.7954e-23 |
| 0.8 | 0.8 | 0.7398 | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.5311 | 5.241e-21 | 8.8823e-23 |
| 0.8 | 0.8 | 0.7398 | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.5685 | 4.7938e-21 | 8.8924e-23 |
1.50.
Continue on from meeting with Klaus on :
[ ]Start by fixing the systematics -> start by extracting the systematic values used to compute the current number. The default values are:
σ_sig = 0.04692492913207222, σ_back = 0.002821014576353691, σ_p = 0.05,
computed from section
sec:systematics:combined_uncertaintiesin StatusAndProgress via the following code (stripped down to signal):import math, sequtils let ss = [3.3456, 0.5807, 1.0, 0.2159, 2.32558, 0.18521] #2.0] #1.727] ## ^-- This is the effective efficiency for 55Fe apparently. proc total(vals: openArray[float]): float = for x in vals: result += x * x result = sqrt(result) let ss0 = total(ss) let ss17 = total(concat(@ss, @[1.727])) let ss2 = total(concat(@ss, @[2.0])) echo "Combined uncertainty signal (Δ software eff = 0%): ", ss0 / 100.0 echo "Combined uncertainty signal (Δ software eff = 1.727%): ", ss17 / 100.0 echo "Combined uncertainty signal (Δ software eff = 2%): ", ss2 / 100.0
[X]There is one mystery here: The value that comes out of that
calculation is
0.458 instead of the ~0.469 used in the code. I don't know why that is exactly. -> *SOLVED*: The reason is the ~0.469 is from assuming 2%, but the ~0.458 is from using the value ~1.727!So now all we need to do is to combine the value without the software efficiency numbers with the effective efficiency from the MLP! (or using the 2% for LnL).
We do this by:
import math let ss = [3.3456, 0.5807, 1.0, 0.2159, 2.32558, 0.18521] proc total(vals: openArray[float]): float = for xf in vals: let x = xf / 100.0 result += x * x result = sqrt(result) let sStart = total(ss) # / 100.0 doAssert abs(sStart - 0.04244936953654317) < 1e-5 # add new value: let seff = 1.727 / 100.0 let s17 = sqrt( (sStart)^2 + seff^2 ) echo "Uncertainty including Δseff = 1.727% after the fact: ", s17
[X]Implemented
1.50.1. look into the cluster centers
…that are affected by the more noisy behavior of the LnL
[X]create background cluster plot for MLP@95% for Run-2 and Run-3 separately
plotBackgroundClusters \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ --zMax 30 --title "X-ray like clusters CAST MLP@95+no vetoes" \ --outpath ~/Sync/mlp_10_05_23_eff_95_find_noisy_pixels/ \ --suffix "10_05_23_mlp_0.95_only_mlp" \ --energyMax 12.0 --energyMin 0.2
yields file:///home/basti/Sync/mlp_10_05_23_eff_95_find_noisy_pixels/background_cluster_centers10_05_23_mlp_0.95_only_mlp.pdf]] and running with the existing noise filter:
plotBackgroundClusters \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ --zMax 30 --title "X-ray like clusters CAST MLP@95+no vetoes" \ --outpath ~/Sync/mlp_10_05_23_eff_95_find_noisy_pixels/ \ --suffix "10_05_23_mlp_0.95_only_mlp_filter_noisy" \ --energyMax 12.0 --energyMin 0.2 \ --filterNoisyPixels
~/Sync/mlp100523eff95findnoisypixels/backgroundclustercenters100523mlp0.95onlymlpfilternoisy.pdf]]
[X]Need to add pixel at bottom -> added(66, 107)[ ]add noisy thing in Run-2 smaller above[ ]add all "Deiche"
[X]look at plot with vetoes (including septem)
plotBackgroundClusters \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters CAST MLP@95+allv" \ --outpath ~/Sync/mlp_10_05_23_eff_95_find_noisy_pixels/ \ --suffix "10_05_23_mlp_0.95_mlp_with_vetoes" \ --energyMax 12.0 --energyMin 0.2 plotBackgroundClusters \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters CAST MLP@95+allv" \ --outpath ~/Sync/mlp_10_05_23_eff_95_find_noisy_pixels/ \ --suffix "10_05_23_mlp_0.95_mlp_with_vetoes_filter_noisy" \ --energyMax 12.0 --energyMin 0.2 \ --filterNoisyPixels
-> removes most things!
[X]without septem but with line:
plotBackgroundClusters \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters CAST MLP@95+noseptem" \ --outpath ~/Sync/mlp_10_05_23_eff_95_find_noisy_pixels/ \ --suffix "10_05_23_mlp_0.95_mlp_vetoes_noseptem" \ --energyMax 12.0 --energyMin 0.2 plotBackgroundClusters \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters CAST MLP@95+noseptem" \ --outpath ~/Sync/mlp_10_05_23_eff_95_find_noisy_pixels/ \ --suffix "10_05_23_mlp_0.95_mlp_vetoes_noseptem_filter_noisy" \ --energyMax 12.0 --energyMin 0.2 \ --filterNoisyPixels
[X]same plots for Run-3 (without filter noisy pixels, as there's no difference)
plotBackgroundClusters \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ --zMax 30 --title "X-ray like clusters CAST MLP@95+no vetoes" \ --outpath ~/Sync/mlp_10_05_23_eff_95_find_noisy_pixels/ \ --suffix "10_05_23_mlp_R3_0.95_only_mlp" \ --energyMax 12.0 --energyMin 0.2 plotBackgroundClusters \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters CAST MLP@95+allv" \ --outpath ~/Sync/mlp_10_05_23_eff_95_find_noisy_pixels/ \ --suffix "10_05_23_mlp_R3_0.95_mlp_with_vetoes" \ --energyMax 12.0 --energyMin 0.2 plotBackgroundClusters \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters CAST MLP@95+noseptem" \ --outpath ~/Sync/mlp_10_05_23_eff_95_find_noisy_pixels/ \ --suffix "10_05_23_mlp_R3_0.95_mlp_vetoes_noseptem" \ --energyMax 12.0 --energyMin 0.2
-> Looks SO MUCH better! Why the heck is that?
We can gather:
- No need for filtering of noisy pixels in the Run-3 dataset!
- just add "Deiche" and the individual noisy thing in the Run-2 dataset still visible above the "known" point.
For the secondary cluster we'll redo the Run-2 noisy pixel filter plot without vetoes:
plotBackgroundClusters \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ --zMax 30 --title "X-ray like clusters CAST MLP@95+no vetoes" \ --outpath ~/Sync/mlp_10_05_23_eff_95_find_noisy_pixels/ \ --suffix "10_05_23_mlp_0.95_only_mlp_filter_noisy_find_clusters" \ --energyMax 12.0 --energyMin 0.2 \ --filterNoisyPixels
Ok, I think I've eliminated all important pixels. We could think
about taking out "one more radius" essentially. If plotting without
--zMax there is still a "ring" left in each of the primary clusters.
1.50.2. Recomputing the limits
To recompute the limits we run
./runLimits \ --path ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/ \ --outpath ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/limits_fix_sigEff_noiseFilter \ --prefix lhood_c18_R2_crAll \ --nmc 1000
Note the adjusted output directory.
We start with the following processed.txt file already in the output
directory. That way we skip the "no vetoes" cases completely.
/home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.85_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.8_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.99_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5
Generate the expected limits table:
./generateExpectedLimitsTable \ -p ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/limits_fix_sigEff_noiseFilter/ \ --prefix mc_limit_lkMCMC_skInterpBackground_nmc_1000
| εlnL | MLP | MLPeff | Scinti | FADC | εFADC | Septem | Line | eccLineCut | εSeptem | εLine | εSeptemLine | Total eff. | Limit no signal | Expected Limit |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.8 | 0.95 | 0.9107 | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7677 | 3.638e-21 | 7.7467e-23 |
| 0.8 | 0.99 | 0.9718 | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.8192 | 3.3403e-21 | 7.8596e-23 |
| 0.8 | 0.9 | 0.8474 | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7143 | 3.8192e-21 | 7.8876e-23 |
| 0.8 | 0.85 | 0.7926 | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6681 | 4.3209e-21 | 8.1569e-23 |
| 0.8 | 0.8 | 0.7398 | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6237 | 4.6466e-21 | 8.1907e-23 |
| 0.8 | 0.95 | 0.9107 | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6538 | 4.464e-21 | 8.2924e-23 |
| 0.8 | 0.9 | 0.8474 | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6083 | 4.7344e-21 | 8.4169e-23 |
| 0.8 | 0.99 | 0.9718 | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6976 | 4.0383e-21 | 8.4416e-23 |
| 0.8 | 0.99 | 0.9718 | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7468 | 3.867e-21 | 8.4691e-23 |
| 0.8 | 0.95 | 0.9107 | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6998 | 3.9627e-21 | 8.5747e-23 |
| 0.8 | 0.9 | 0.8474 | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6511 | 4.3262e-21 | 8.6508e-23 |
| 0.8 | 0.85 | 0.7926 | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.569 | 4.8645e-21 | 8.7205e-23 |
| 0.8 | 0.85 | 0.7926 | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.609 | 4.7441e-21 | 8.8143e-23 |
| 0.8 | 0.8 | 0.7398 | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.5685 | 5.0982e-21 | 8.8271e-23 |
| 0.8 | 0.8 | 0.7398 | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.5311 | 5.1353e-21 | 8.8536e-23 |
1.51.
[ ]IMPORTANT: When using MLP in classification, how do we deal with inputs that have NaN values??? Do we filter them out, i.e. reject them?
let data = h5f.readValidDsets(grp, filterNan = false)
-> How do we deal with them? Ah, from likelihood:
(classify(nnPred[ind]) == fcNaN or # some clusters are NaN due to certain bad geometry, kick those out! # -> clusters of sparks on edge of chip
However, is this actually enough? Maybe there are cases where the result is not NaN for some bizarre reason? Shouldn't be allowed to happen, because NaN "infects" everything.
Still: check the mapping of input NaN values to output NaN values! Any weirdness? What do outputs of all those many clusters in the corners look like? Maybe there is something to learn there? Same with very low energy behavior.
1.52.
I noticed that there seems to be a bug in the sorted_seq based k-d
tree implementation of arraymancer! Some test cases do not pass!
[ ]INVESTIGATE
This might be essential for the limit calculation!
Also: Yesterday evening in discussion with Cris she told me that their group thinks the focal spot of the telescope is actually in the center of the detector chamber and not the focal plane as we always assumed!
[ ]FIND OUT -> I wrote a message to Johanna to ask if she knows anything more up to date
1.53.
[ ]recompile MCMC limit program with different leaf sizes of the k-d tree (try 64)[ ]Redo limit calculations for the best 3 cases of MLP & LnL with 10000 toys[ ]compute the axion image again in the actual focal spot, then use that input to generate the limit for the best case of the above![X]I moved the data from ./../CastData/data/ to ./../../../mnt/1TB/Uni/DataFromHomeCastData/ to make more space on the home partition!
1.53.1. Expected limits with more toys
Let's get the table from the limit method talk ./Talks/LimitMethod/limit_method.html
| Method | \(ε_S\) | FADC | \(ε_{\text{FADC}}\) | Septem | Line | Total eff. | Limit no signal | Expected Limit |
|---|---|---|---|---|---|---|---|---|
| MLP | 0.9107 | true | 0.98 | false | true | 0.7677 | 6.0315e-23 | 7.7467e-23 |
| MLP | 0.9718 | true | 0.98 | false | true | 0.8192 | 5.7795e-23 | 7.8596e-23 |
| MLP | 0.8474 | true | 0.98 | false | true | 0.7143 | 6.1799e-23 | 7.8876e-23 |
| LnL | 0.9 | true | 0.98 | false | true | 0.7587 | 6.1524e-23 | 7.9443e-23 |
| LnL | 0.9 | false | 0.98 | false | true | 0.7742 | 6.0733e-23 | 8.0335e-23 |
| MLP | 0.7926 | true | 0.98 | false | true | 0.6681 | 6.5733e-23 | 8.1569e-23 |
| MLP | 0.7398 | true | 0.98 | false | true | 0.6237 | 6.8165e-23 | 8.1907e-23 |
We'll do the first 3 MLP rows and the two LnL rows.
Instead of using the tool that runs limit calcs for all input files, we'll manually call the limit calc.
We'll put the output into ./resources/lhood_limits_12_06_23_10k_toys
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --nmc 1000 \ --suffix=_sEff_0.95_mlp_mlp_tanh300_mse_epoch_485000_loss_0.0055_acc_0.9933 \ --path "" \ --outpath /home/basti/org/resources/lhood_limits_12_06_23_10k_toys
NOTE : The file used here was the wrong one. Didn't include any vetoes..
1.54.
Yesterday I sent a mail to multiple people from CAST who might know about the beamline design behind the LLNL telescope to find out the intended idea for the focal spot. The question is where in the detector the focal spot was intended to be.
I was always under the impression the focal spot was on the readout plane, but in a discussion with Cristina she mentioned that she thinks it's in the center of the gas volume.
See the mail titled "CAST detectors behind LLNL telescope" (sent from uni bonn address via fastmail).
Turns out from Igor's and Juan's answer the idea was indeed to place the focal spot in the center of the volume!
This is massive and means we need to recompute the raytracing image!
[X]recompute the axion image! -> DONE![ ]Rerun the correct expected limit calculations incl vetoes! What we ran yesterday didn't include vetoes, hence so slow![ ]Rerun the same case (best case) with the correct axion image![ ]Think about whether we ideally really should have a systematic for the z position of the detector. I.e. varying it change the size of the axion image
1.54.1. DONE Updating the axion image
Regenerate the DF for the solar flux:
cd ~/CastData/ExternCode/AxionElectronLimit/src
./readOpacityFile
Note that the config.toml file contains the output path (out
directory) and output file name solar_model_dataframe.csv.
That file should then be moved to resources and set in the config
file as the DF to use.
In the raytracer we now set the distance correctly using the config file:
[DetectorInstallation] useConfig = true # sets whether to read these values here. Can be overriden here or useng flag `--detectorInstall` # Note: 1500mm is LLNL focal length. That corresponds to center of the chamber! distanceDetectorXRT = 1497.2 # mm distanceWindowFocalPlane = 0.0 # mm lateralShift = 0.0 # mm lateral ofset of the detector in repect to the beamline transversalShift = 0.0 # mm transversal ofset of the detector in repect to the beamline #0.0.mm #
\(\SI{1497}{mm}\) comes from the mean conversion point being at \(\SI{12.2}{mm}\) behind the detector window. If the focal point at \(\SI{1500}{mm}\) is in th center of the chamber, the point to compute the image for is at \(1500 - (15 - 12.2) = 1500 - 2.8 = 1497\).
We will also compare it for sanity with the old axion image we've been using in the limit calculation, namely at \(1470 + 12.2 = 1482.2\).
We generated the following files:
with the data files in
- ./resources/axion_images/axion_image_2018_1000.csv
- ./resources/axion_images/axion_image_2018_1470_12.2mm.csv
- ./resources/axion_images/axion_image_2018_1497.2mm.csv
- ./resources/axion_images/axion_image_2018_1500.csv
First of all the 1000mm case shows us that reading from the config file actually works. Then we can compare to the actual center, the old used value and the new. The difference between the old and new is quite profound!
1.54.2. Expected limit calculations
The calculation we started yesterday didn't use the correct input files…
We used the version without any vetoes instead of the MLP + FADC + Line veto case! Hence it was also so slow!
[X]Case 1:
| MLP | 0.9107 | true | 0.98 | false | true | 0.7677 | 6.0315e-23 | 7.7467e-23 |
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --nmc 10000 \ --suffix=_sEff_0.95_scinti_fadc_line_mlp_tanh300_mse_epoch_485000_loss_0.0055_acc_0.9933 \ --path "" \ --outpath /home/basti/org/resources/lhood_limits_12_06_23_10k_toys
-> This seems to take about 20s per chain. 10000 might still be too many until the meeting at 4pm if we want to update the plots. NOTE: for now I restarted it with only 1000 toys! Yielded:
Expected limit: 6.001089083451825e-21
which is \(g_{ae} g_{aγ} = \SI{7.746e-23}{GeV^{-1}}\).
[ ]Case 2:
| MLP | 0.9718 | true | 0.98 | false | true | 0.8192 | 5.7795e-23 | 7.8596e-23 |
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.99_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.99_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --nmc 1000 \ --suffix=_sEff_0.99_scinti_fadc_line_mlp_tanh300_mse_epoch_485000_loss_0.0055_acc_0.9933 \ --path "" \ --outpath /home/basti/org/resources/lhood_limits_12_06_23_10k_toys
- Rerunning with new axion image
- Recompile the limit code with the new axion image, namely: ./resources/axion_images/axion_image_2018_1497.2mm.csv
Now run the calculation again with a new suffix! (sigh, I had started with the full 10k samples :( )
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --nmc 10000 \ --suffix=_sEff_0.95_scinti_fadc_line_mlp_tanh300_mse_epoch_485000_loss_0.0055_acc_0.9933_fixed_axion_image_1497.2 \ --path "" \ --outpath /home/basti/org/resources/lhood_limits_12_06_23_10k_toys
The result:
Expected limit: 5.789608342190943e-21
implying \(g_{ae} g_{aγ} = \SI{7.609e-11}{GeV^{-1}}\)
In order to redo one of the plots we can follow the following from when we ran the 100k toy sets:
./mcmc_limit_calculation \ limit --plotFile \ ~/org/resources/mc_limit_lkMCMC_skInterpBackground_nmc_100000_uncertainty_ukUncertain_σs_0.0469_σb_0.0028_posUncertain_puUncertain_σp_0.0500.csv \ --xLow 2.5e-21 \ --xHigh 1.5e-20 \ --limitKind lkMCMC \ --yHigh 3000 \ --bins 100 \ --linesTo 2000 \ --xLabel "Limit [g_ae² @ g_aγ = 1e-12 GeV⁻¹]" \ --yLabel "MC toy count" \ --nmc 100000
1.54.3. Limit talk for my group at
Today at 4pm I gave the talk about the limit method to my colleagues.
The talk as of right now: ./Talks/LimitMethod/limit_method.html (see the commit from today adding the sneak preview to see the talk as given today).
Some takeaways:
- it would have been good to also have some background rate plots & improvements of the vetoes etc
- show the background cluster plot when talking about background rate being position dependent. Unclear otherwise why needed
- better explain how the position nuisance parameter and generally the parameters works
[ ]Show the likelihood function with nuisance parameters without modification[ ]Finish the slide about background interpolation w/ normal distr. weighting[ ]fix numbers for scintillator veto[ ]remove "in practice" section of candidate sampling for expected limits. Move that to actual candidate sampling thing[ ]better explain what the likelihood function looks like when talking about the limit at 95% CDF (seeing the histogram there made it a bit confusing!)[ ]Update plot of expected limit w/ many candidates![ ]better explain no candidates in sensitive region?[ ]Klaus said I should add information about the time varying uncertainty stuff into the talk
Discussions:
- discussion about the estimate of the efficiencies of septem & line veto. Tobi thinks it should be improved by sampling not from all outer chip data, because events with center chip X-ray like cluster typically are shorter than 2.2 s and therefore they should see slightly less background! -> Discussed with Klaus, it could be improved, but it's not trivial
- Johanna asked about axion image & where the position comes from etc. -> Ideally make energy dependent and even compute the axion flux by sampling from the absorption position distribution
- Jochen wondered about the "feature" in 3 keV interpolated background in the top right (but not quite corner) -> Already there in the data, likely statistics
- Klaus thinks we shouldn't include systematics for the uncertainty of the solar model!
- We discussed improving systematics by taking into account the average position of the Sun over the data taking period!
- Markus asked about simulated events for MLP training
[X]Send Johanna my notes about how I compute the mean absorption position!
1.55.
[X]Send Johanna my notes about how I compute the mean absorption position![ ]Potentially take out systematic uncertainty for solar model[ ]Think about systematic of Sun ⇔ Earth, better estimate number by using real value as mean.[ ]SEE TODOs FROM YESTERDAY
UPDATE: In the end I spent the majority of the day working on the notes for Johanna for the mean conversion point of solar X-rays from axions in the gas. Turns out my previous assumption was wrong after all. Not 1.22 cm but rather about 0.55 cm for the mean and 0.3 cm are realistic numbers. :)
1.56.
We started writing notes on the LLNL telescope for the REST raytracer yesterday and finished them today, here: ./Doc/LLNL_def_REST_format/llnl_def_rest_format.html
In addition, let's now quickly try to generate the binary files
required by REST in nio format.
Our LLNL file is generated from ./../CastData/ExternCode/AxionElectronLimit/tools/llnl_layer_reflectivity.nim and lives here: ./../CastData/ExternCode/AxionElectronLimit/resources/llnl_layer_reflectivities.h5
Let's look into it:
import nimhdf5 const path = "/home/basti/CastData/ExternCode/AxionElectronLimit/resources/llnl_layer_reflectivities.h5" let h5f = H5open(path, "r") h5f.visit_file() let grp = h5f["/".grp_str] for dset in grp: echo dset.name, " of shape ", dset.shape discard h5f.close()
From the raytracing code or the code generating the file we can remind ourselves about the layout of he reflectivity datasets:
let energies = h5f["/Energy", float] let angles = h5f["/Angles", float] var reflectivities = newSeq[Interpolator2DType[float]]() for i in 0 ..< numCoatings: let reflDset = h5f[("Reflectivity" & $i).dset_str] let data = reflDset[float].toTensor.reshape(reflDset.shape) reflectivities.add newBilinearSpline( data, (angles.min, angles.max), (energies.min, energies.max) )
newBilinearSpline takes the x limits first and then the y limits,
meaning the first dimension is the angles and the second the energy.
Given that we have 4 different coatings:
let m1 = drp.Multilayer(MultilayerType="DepthGraded", SubstrateMaterial="SiO2", D_min = 11.5, D_max = 22.5, Gamma = 0.45, C = 1.0, LayerMaterial=["Pt", "C"], Repetition=2, SigmaValues=[1.0]) let m2 = drp.Multilayer(MultilayerType="DepthGraded", SubstrateMaterial="SiO2", D_min = 7.0, D_max = 19.0, Gamma = 0.45, C = 1.0, LayerMaterial=["Pt", "C"], Repetition=3, SigmaValues=[1.0]) let m3 = drp.Multilayer(MultilayerType="DepthGraded", SubstrateMaterial="SiO2", D_min = 5.5, D_max = 16.0, Gamma = 0.4, C = 1.0, LayerMaterial=["Pt", "C"], Repetition=4, SigmaValues=[1.0]) let m4 = drp.Multilayer(MultilayerType="DepthGraded", SubstrateMaterial="SiO2", D_min = 5.0, D_max = 14.0, Gamma = 0.4, C = 1.0, LayerMaterial=["Pt", "C"], Repetition=5, SigmaValues=[1.0])
we need to generate 4 different binary files for REST.
Our data has 1000x1000 elements, meaning our filename will be
.N1000f.
First let's download a file from REST though and open it with nio.
cd /t
wget https://github.com/rest-for-physics/axionlib-data/raw/master/opticsMirror/Reflectivity_Single_Au_250_Ni_0.4.N901f
import nio const Size = 1000 #901 #const path = "/tmp/Reflectivity_Single_Au_250_Ni_0.4.N901f" const path = "/tmp/R1.N1000f" #let fa = initFileArray[float32](path) #echo fa #let mm = mOpen(path) #echo mm let fa = load[array[Size, float32]](path) #echo fa[0] echo fa import ggplotnim, sequtils block Angle: let df = toDf({"x" : toSeq(0 ..< Size), "y" : fa[0].mapIt(it.float)}) ggplot(df, aes("x", "y")) + geom_line() + ggsave("/tmp/test_angle.pdf") block Refl: var refl = newSeq[float]() var i = 0 for row in fa: refl.add row[400] echo "I = ", i inc i echo fa.len echo refl.len let df = toDf({"x" : toSeq(0 ..< fa.len), "y" : refl}) echo df ggplot(df, aes("x", "y")) + geom_line() + ggsave("/tmp/test_refl.pdf") for j, row in fa: echo "I = ", j echo row[0 ..< 100] for x in countup(0, 10, 1): echo x
What we saw from the above:
- it's the transposed of our data, each row is all energies for one angle whereas ours is all angles for one energy
Now let's save each reflectivity file using nio. We read it,
transform it into a arrays of the right size and write.
import nimhdf5, strutils, sequtils, arraymancer import nio const path = "/home/basti/CastData/ExternCode/AxionElectronLimit/resources/llnl_layer_reflectivities.h5" let h5f = H5open(path, "r") h5f.visit_file() let grp = h5f["/".grp_str] for dset in grp: echo dset.name, " of shape ", dset.shape if "Reflectivity" in dset.name: let data = toTensor(h5f[dset.name, float]).asType(float32) # convert to float32 .reshape(dset.shape) .transpose # transpose our data echo data.shape let dataS = data.toSeq2D echo dataS.shape # convert to seq[array] var dataA = newSeq[array[1000, float32]](1000) for i in 0 ..< dataS.len: copyMem(dataA[i][0].addr, dataS[i][0].addr, 1000 * sizeof(float32)) dataA.save "/tmp/R1" discard h5f.close()
The above is enough to correctly generate the data files. However, the range of the files used by REST does not match what our data needs.
The README for REST about the data files says:
The following directory contains data files with X-ray reflectivity pre-downloaded data from the https://henke.lbl.gov/ database. These files will be generated by the TRestAxionOpticsMirror metadata class. The files will be used by that class to quickly load reflectivity data in memory, in case the requested optics properties are already available a this database.
See TRestAxionOpticsMirror documentation for further details on how to generate or load these datasets.
The file is basically a table with 501 rows, each row corresponding to an energy, starting at 30eV in increments of 30eV. The last row corresponds with 15keV. The number of columns is 901, describing the data as a function of the angle of incidence in the range between 0 and 9 degrees with 0.01 degree precision
which tells us in what range we need to generate the data.
Our data:
- θ = (0, 1.5)°
- E = (0.03, 15) keV
- 1000x1000
REST:
- θ = (0, 9)°
- E = (0.03, 15) keV
- 901x500
So let's adjust our data generation script and rerun it with the correct ranges and number of points:
I've changed the generator code such that it accepts arguments:
Usage:
main [optional-params]
Options:
-h, --help print this
cligen-erated help
--help-syntax advanced:
prepend,plurals,..
-e=, --energyMin= keV 0.03 keV set energyMin
--energyMax= keV 15 keV set energyMax
-n=, --numEnergy= int 1000 set numEnergy
-a=, --angleMin= ° 0 ° set angleMin
--angleMax= ° 1.5 ° set angleMax
--numAngle= int 1000 set numAngle
-o=, --outfile= string "llnl_layer_reflectivities.h5" set outfile
--outpath= string "../resources/" set outpath
./llnl_layer_reflectivity \ --numEnergy 500 \ --angleMax 9.0.° \ --numAngle 901 \ --outfile llnl_layer_reflectivities_rest.h5 \ --outpath /home/basti/org/resources/
Having generated the correct file we can now use the above snippet to
construct the binary files using nio.
import nimhdf5, strutils, sequtils, arraymancer import nio const path = "/home/basti/org/resources/llnl_layer_reflectivities_rest.h5" let h5f = H5open(path, "r") h5f.visit_file() let grp = h5f["/".grp_str] for dset in grp: echo dset.name, " of shape ", dset.shape if "Reflectivity" in dset.name: let data = toTensor(h5f[dset.name, float]).asType(float32) # convert to float32 .reshape(dset.shape) .transpose # transpose our data echo data.shape let dataS = data.toSeq2D echo dataS.shape # convert to seq[array] var dataA = newSeq[array[901, float32]](500) for i in 0 ..< dataS.len: copyMem(dataA[i][0].addr, dataS[i][0].addr, 901 * sizeof(float32)) let name = dset.name dataA.save "/tmp/" & name discard h5f.close()
Let's read one of the files using nio again to check:
import nio const Size = 901 const path = "/tmp/Reflectivity0.N901f" let fa = load[array[Size, float32]](path) echo fa import ggplotnim, sequtils block Angle: let df = toDf({"x" : toSeq(0 ..< Size), "y" : fa[0].mapIt(it.float)}) ggplot(df, aes("x", "y")) + geom_line() + ggsave("/tmp/test_angle.pdf") block Refl: var refl = newSeq[float]() var i = 0 for row in fa: refl.add row[50] inc i let df = toDf({"x" : toSeq(0 ..< fa.len), "y" : refl}) ggplot(df, aes("x", "y")) + geom_line() + ggsave("/tmp/test_refl.pdf")
1.57.
Let's finally pick up where we left off, namely finishing up the limit talk for the CAST collaboration.
UPDATE: : While explaining things to Cristina I noticed that our assumption of the LLNL multilayer was flawed. The thesis and paper always talk about a Pt/C coating. But this actually means the carbon is at the top and not at the bottom (see fig. 4.11 in the thesis).
So now I'll regenerate all the files for REST as well as the one we use and update our code.
Updated the layer in code and time to rerun:
./llnl_layer_reflectivity \ --numEnergy 500 \ --angleMax 9.0.° \ --numAngle 901 \ --outfile llnl_layer_reflectivities_rest.h5 \ --outpath /tmp/
and now to regenerate the files:
import nimhdf5, strutils, sequtils, arraymancer import nio const path = "/home/basti/org/resources/llnl_layer_reflectivities_rest.h5" let h5f = H5open(path, "r") h5f.visit_file() let grp = h5f["/".grp_str] for dset in grp: echo dset.name, " of shape ", dset.shape if "Reflectivity" in dset.name: let data = toTensor(h5f[dset.name, float]).asType(float32) # convert to float32 .reshape(dset.shape) .transpose # transpose our data echo data.shape let dataS = data.toSeq2D echo dataS.shape # convert to seq[array] var dataA = newSeq[array[901, float32]](500) for i in 0 ..< dataS.len: copyMem(dataA[i][0].addr, dataS[i][0].addr, 901 * sizeof(float32)) let name = dset.name dataA.save "/tmp/" & name discard h5f.close()
I renamed them
basti at void in /t λ mv Reflectivity0.N901f Reflectivity_Multilayer_Pt_C_LLNL_layers_1,2,3.N901f basti at void in /t λ mv Reflectivity1.N901f Reflectivity_Multilayer_Pt_C_LLNL_layers_4,5,6.N901f basti at void in /t λ mv Reflectivity2.N901f Reflectivity_Multilayer_Pt_C_LLNL_layers_7,8,9,10.N901f basti at void in /t λ mv Reflectivity3.N901f Reflectivity_Multilayer_Pt_C_LLNL_layers_11,12.N901f basti at void in /t λ cp Reflectivity_Multilayer_Pt_C_LLNL_layers_* ~/src/axionlib-data/opticsMirror/
and time to update the PR.
1.58.
For the talk about the limit calculation method I need two new plots:
- background rate with only LnL & MLP
- background rate with LnL + each different veto
and the latest numbers for the background rate achieved.
For that we need the latest MLP, from ./resources/lhood_limits_10_05_23_mlp_sEff_0.99/ (on desktop!)
First the background rate of MLP @ 99% (97% on real data) and LnL without any vetoes:
NOTE: In the naming below we use the efficiencies more closely matching the real efficiencies based on the CDL data instead of the target based on simulated events!
plotBackgroundRate \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crGold_sEff_0.99_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crGold_sEff_0.99_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ --names "MLP@97" --names "MLP@97" --names "LnL@80" --names "LnL@80" \ --centerChip 3 \ --title "Background rate CAST, LnL@80%, SGD tanh300 MLE MLP@97% + vetoes" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.2 \ --outfile background_rate_run2_3_mlp_0.99_no_vetoes.pdf \ --outpath ~/Sync/limitMethod/ \ --quiet
[INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.2795e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 1.8996e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@97 [INFO]: Integrated background rate in range: 0.0 .. 12.0: 2.6735e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 12.0: 2.2279e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@97 [INFO]: Integrated background rate in range: 0.5 .. 2.5: 4.9952e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 2.4976e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@97 [INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1661e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5914e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 8.4954e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.3982e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@97 [INFO]: Integrated background rate in range: 0.0 .. 2.5: 9.1110e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 2.5: 3.6444e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@97 [INFO]: Integrated background rate in range: 4.0 .. 8.0: 3.2012e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 8.0029e-06 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.6076e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.0095e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@97 [INFO]: Integrated background rate in range: 0.0 .. 8.0: 1.8310e-04 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 0.0 .. 8.0: 2.2887e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: LnL@80 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹ [INFO]:Dataset: MLP@97 [INFO]: Integrated background rate in range: 2.0 .. 8.0: 9.6563e-05 cm⁻²·s⁻¹ [INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.6094e-05 keV⁻¹·cm⁻²·s⁻¹
results in the plot:
and including all vetoes for both, as a reference:
plotBackgroundRate \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crGold_sEff_0.99_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crGold_sEff_0.99_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crGold_sEff_0.99_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crGold_sEff_0.99_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold_scinti_fadc_line_vetoPercentile_0.99.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold_scinti_fadc_line_vetoPercentile_0.99.h5 \ --names "MLP@97" --names "MLP@97" --names "MLP@97+V" --names "MLP@97+V" --names "LnL@80" --names "LnL@80" --names "LnL@80+V" --names "LnL@80+V" \ --centerChip 3 \ --title "Background rate CAST, LnL@80%, SGD tanh300 MLE MLP@97% + vetoes" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.2 \ --outfile background_rate_run2_3_mlp_0.99_plus_vetoes.pdf \ --outpath ~/Sync/limitMethod/ \ --quiet
[INFO]:Dataset: LnL@80 [29/1134]
[INFO]: Integrated background rate in range: 0.2 .. 12.0: 2.2566e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 12.0: 1.9124e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 0.2 .. 12.0: 1.1239e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 12.0: 9.5248e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97
[INFO]: Integrated background rate in range: 0.2 .. 12.0: 2.5838e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 12.0: 2.1897e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97+V
[INFO]: Integrated background rate in range: 0.2 .. 12.0: 1.5302e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 12.0: 1.2968e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80
[INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 0.5 .. 2.5: 1.4071e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 7.0355e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97
[INFO]: Integrated background rate in range: 0.5 .. 2.5: 4.9952e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 2.4976e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97+V
[INFO]: Integrated background rate in range: 0.5 .. 2.5: 1.6182e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 8.0909e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80
[INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 0.5 .. 5.0: 4.4500e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 9.8888e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97
[INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1661e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5914e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97+V
[INFO]: Integrated background rate in range: 0.5 .. 5.0: 6.0154e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 1.3367e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80
[INFO]: Integrated background rate in range: 0.2 .. 2.5: 8.2667e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 2.5: 3.5942e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 0.2 .. 2.5: 2.0051e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 2.5: 8.7179e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97
[INFO]: Integrated background rate in range: 0.2 .. 2.5: 8.2140e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 2.5: 3.5713e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97+V
[INFO]: Integrated background rate in range: 0.2 .. 2.5: 2.9725e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 2.5: 1.2924e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80
[INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 4.0 .. 8.0: 1.3543e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 3.3858e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97
[INFO]: Integrated background rate in range: 4.0 .. 8.0: 3.2012e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 8.0029e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97+V
[INFO]: Integrated background rate in range: 4.0 .. 8.0: 1.9524e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 4.8809e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80
[INFO]: Integrated background rate in range: 0.2 .. 8.0: 1.5848e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 8.0: 2.0317e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 0.2 .. 8.0: 6.2264e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 8.0: 7.9826e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97
[INFO]: Integrated background rate in range: 0.2 .. 8.0: 1.7413e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 8.0: 2.2324e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97+V
[INFO]: Integrated background rate in range: 0.2 .. 8.0: 8.8823e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 8.0: 1.1388e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80
[INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 2.0 .. 8.0: 4.4324e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 7.3873e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97
[INFO]: Integrated background rate in range: 2.0 .. 8.0: 9.6563e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.6094e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@97+V
[INFO]: Integrated background rate in range: 2.0 .. 8.0: 6.1385e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.0231e-05 keV⁻¹·cm⁻²·s⁻¹
And further for reference the MLP at 85%, which equals about the 80% of the LnL:
plotBackgroundRate \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crGold_sEff_0.85_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crGold_sEff_0.85_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crGold_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crGold_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run2_crGold_scinti_fadc_line_vetoPercentile_0.99.h5 \ ~/org/resources/lhood_limits_automation_correct_duration/likelihood_cdl2018_Run3_crGold_scinti_fadc_line_vetoPercentile_0.99.h5 \ --names "MLP@80" --names "MLP@80" --names "MLP@80+V" --names "MLP@80+V" --names "LnL@80" --names "LnL@80" --names "LnL@80+V" --names "LnL@80+V" \ --centerChip 3 \ --title "Background rate CAST, LnL@80%, SGD tanh300 MLE MLP@80% + vetoes" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.2 \ --outfile background_rate_run2_3_mlp_0.85_plus_vetoes.pdf \ --outpath ~/Sync/limitMethod/ \ --quiet
[INFO]:Dataset: LnL@80 [29/1264]
[INFO]: Integrated background rate in range: 0.2 .. 12.0: 2.2566e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 12.0: 1.9124e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 0.2 .. 12.0: 1.1239e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 12.0: 9.5248e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80
[INFO]: Integrated background rate in range: 0.2 .. 12.0: 1.5408e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 12.0: 1.3057e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80+V
[INFO]: Integrated background rate in range: 0.2 .. 12.0: 1.0747e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 12.0: 9.1074e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80
[INFO]: Integrated background rate in range: 0.5 .. 2.5: 6.2088e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 3.1044e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 0.5 .. 2.5: 1.4071e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 7.0355e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80
[INFO]: Integrated background rate in range: 0.5 .. 2.5: 1.9348e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 9.6738e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80+V
[INFO]: Integrated background rate in range: 0.5 .. 2.5: 1.0026e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 2.5: 5.0128e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80
[INFO]: Integrated background rate in range: 0.5 .. 5.0: 1.1626e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 2.5836e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 0.5 .. 5.0: 4.4500e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 9.8888e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80
[INFO]: Integrated background rate in range: 0.5 .. 5.0: 6.2968e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 1.3993e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80+V
[INFO]: Integrated background rate in range: 0.5 .. 5.0: 4.0454e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.5 .. 5.0: 8.9898e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80
[INFO]: Integrated background rate in range: 0.2 .. 2.5: 8.2667e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 2.5: 3.5942e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 0.2 .. 2.5: 2.0051e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 2.5: 8.7179e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80
[INFO]: Integrated background rate in range: 0.2 .. 2.5: 3.1132e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 2.5: 1.3536e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80+V
[INFO]: Integrated background rate in range: 0.2 .. 2.5: 1.5478e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 2.5: 6.7296e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80
[INFO]: Integrated background rate in range: 4.0 .. 8.0: 2.6383e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 6.5958e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 4.0 .. 8.0: 1.3543e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 3.3858e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80
[INFO]: Integrated background rate in range: 4.0 .. 8.0: 1.8292e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 4.5731e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80+V
[INFO]: Integrated background rate in range: 4.0 .. 8.0: 1.1960e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 4.0 .. 8.0: 2.9901e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80
[INFO]: Integrated background rate in range: 0.2 .. 8.0: 1.5848e-04 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 8.0: 2.0317e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 0.2 .. 8.0: 6.2264e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 8.0: 7.9826e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80
[INFO]: Integrated background rate in range: 0.2 .. 8.0: 9.0055e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 8.0: 1.1545e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80+V
[INFO]: Integrated background rate in range: 0.2 .. 8.0: 5.5932e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 0.2 .. 8.0: 7.1708e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80
[INFO]: Integrated background rate in range: 2.0 .. 8.0: 8.1788e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.3631e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: LnL@80+V
[INFO]: Integrated background rate in range: 2.0 .. 8.0: 4.4324e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 7.3873e-06 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80
[INFO]: Integrated background rate in range: 2.0 .. 8.0: 6.1913e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 1.0319e-05 keV⁻¹·cm⁻²·s⁻¹
[INFO]:Dataset: MLP@80+V
[INFO]: Integrated background rate in range: 2.0 .. 8.0: 4.2037e-05 cm⁻²·s⁻¹
[INFO]: Integrated background rate/keV in range: 2.0 .. 8.0: 7.0062e-06 keV⁻¹·cm⁻²·s⁻¹
which then is actually slightly better than the LnL method.
The plots are also found here: ./Figs/statusAndProgress/backgroundRates/limitMethodTalk/
1.59.
Things left TODO for the limit talk:
[ ]update raytracing image with correct new location based on median of simulation result[ ]update systematic uncertainties?[ ]rerun new limits with raytracing & systematics updated[X]remove "sneak preview"[ ]clarify likelihood space & relation to plot of likelihood histogram
1.60.
Left over todos from yesterday:
[ ]update raytracing image with correct new location based on median of simulation result[ ]update systematic uncertainties?[ ]rerun new limits with raytracing & systematics updated[ ]clarify likelihood space & relation to plot of likelihood histogram
To get started on the systematic uncertainties: The biggest one for sure is the Sun ⇔ Earth distance one at 3.3%.
The idea is to get the distance during each solar tracking, then compute the weighted mean of the distances. From that we can compute a new uncertainty based on the variation visible in the data. That should reduce the uncertainty to ~1% or so.
So first we need to get information about Sun ⇔ Earth distance at different dates. Maybe we can get a CSV file with distances for each date in the past?
NASA's Horizon system: https://ssd.jpl.nasa.gov/horizons/
The correct query is the following:
except we want it for 1 minute intervals instead of 1 hour (to have multiple data points per tracking).
If one selects not just one setting, the request contains the following things:
w: 1 format: json input: !$$SOF MAKE_EPHEM=YES COMMAND=10 EPHEM_TYPE=OBSERVER CENTER='coord@399' COORD_TYPE=GEODETIC SITE_COORD='+6.06670,+46.23330,0' START_TIME='2017-01-01' STOP_TIME='2019-12-31' STEP_SIZE='1 HOURS' QUANTITIES='3,6,10,11,13,16,20,27,30,35' REF_SYSTEM='ICRF' CAL_FORMAT='CAL' CAL_TYPE='M' TIME_DIGITS='SECONDS' ANG_FORMAT='HMS' APPARENT='AIRLESS' RANGE_UNITS='AU' SUPPRESS_RANGE_RATE='NO' SKIP_DAYLT='NO' SOLAR_ELONG='0,180' EXTRA_PREC='NO' R_T_S_ONLY='NO' CSV_FORMAT='NO' OBJ_DATA='YES'
The full API documentation can be found at: https://ssd-api.jpl.nasa.gov/doc/horizons.html
The example from the documentation:
https://ssd.jpl.nasa.gov/api/horizons.api?format=text&COMMAND='499'&OBJ_DATA='YES'&MAKE_EPHEM='YES'&EPHEM_TYPE='OBSERVER'&CENTER='500@399'&START_TIME='2006-01-01'&STOP_TIME='2006-01-20'&STEP_SIZE='1%20d'&QUANTITIES='1,9,20,23,24,29'
i.e. we simply make a GET request to https://ssd.jpl.nasa.gov/api/horizons.api with all the parameters added in key-value format.
Let's write a simple API library:
-> Now lives here: ./../CastData/ExternCode/horizonsAPI/horizonsapi.nim
import std / [strutils, strformat, httpclient, asyncdispatch, sequtils, parseutils, os, json, tables, uri] const basePath = "https://ssd.jpl.nasa.gov/api/horizons.api?" const outPath = currentSourcePath().parentDir.parentDir / "resources/" when not defined(ssl): {.error: "This module must be compiled with `-d:ssl`.".} ## See the Horizons manual for a deeper understanding of all parameters: ## https://ssd.jpl.nasa.gov/horizons/manual.html ## And the API reference: ## https://ssd-api.jpl.nasa.gov/doc/horizons.html type CommonOptionsKind = enum coFormat = "format" ## 'json', 'text' coCommand = "COMMAND" ## defines the target body! '10' = Sun, 'MB' to get a list of available targets coObjData = "OBJ_DATA" ## 'YES', 'NO' coMakeEphem = "MAKE_EPHEM" ## 'YES', 'NO' coEphemType = "EPHEM_TYPE" ## 'OBSERVER', 'VECTORS', 'ELEMENTS', 'SPK', 'APPROACH' coEmailAddr = "EMAIL_ADDR" ## Available for 'O' = 'OBSERVER', 'V' = 'VECTOR', 'E' = 'ELEMENTS' EphemerisOptionsKind = enum ## O V E eoCenter = "CENTER" ## x x x 'coord@399' = coordinate from `SiteCoord' on earth (399) eoRefPlane = "REF_PLANE" ## x x eoCoordType = "COORD_TYPE" ## x x x 'GEODETIC', 'CYLINDRICAL' eoSiteCoord = "SITE_COORD" ## x x x if GEODETIC: 'E-long, lat, h': e.g. Geneva: '+6.06670,+46.23330,0' eoStartTime = "START_TIME" ## x x x Date as 'YYYY-MM-dd' eoStopTime = "STOP_TIME" ## x x x eoStepSize = "STEP_SIZE" ## x x x '60 min', '1 HOURS', ... eoTList = "TLIST" ## x x x eoTListType = "TLIST_TYPE" ## x x x eoQuantities = "QUANTITIES" ## x !!! These are the data fields you want to get !!! eoRefSystem = "REF_SYSTEM" ## x x x eoOutUnits = "OUT_UNITS " ## x x 'KM-S', 'AU-D', 'KM-D' (length & time, D = days) eoVecTable = "VEC_TABLE " ## x eoVecCorr = "VEC_CORR " ## x eoCalFormat = "CAL_FORMAT" ## x eoCalType = "CAL_TYPE" ## x x x eoAngFormat = "ANG_FORMAT" ## x eoApparent = "APPARENT" ## x eoTimeDigits = "TIME_DIGITS" ## x x x eoTimeZone = "TIME_ZONE" ## x eoRangeUnits = "RANGE_UNITS" ## x 'AU', 'KM' eoSuppressRangeRate = "SUPPRESS_RANGE_RATE" ## x eoElevCut = "ELEV_CUT" ## x eoSkipDayLT = "SKIP_DAYLT" ## x eoSolarELong = "SOLAR_ELONG" ## x eoAirmass = "AIRMASS" ## x eoLHACutoff = "LHA_CUTOFF" ## x eoAngRateCutoff = "ANG_RATE_CUTOFF" ## x eoExtraPrec = "EXTRA_PREC" ## x eoCSVFormat = "CSV_FORMAT" ## x x x eoVecLabels = "VEC_LABELS" ## x eoVecDeltaT = "VEC_DELTA_T" ## x eoELMLabels = "ELM_LABELS " ## x eoTPType = "TP_TYPE" ## x eoRTSOnly = "R_T_S_ONLY" ## x Quantities = set[1 .. 48] ## 1. Astrometric RA & DEC ## * 2. Apparent RA & DEC ## 3. Rates; RA & DEC ## ,* 4. Apparent AZ & EL ## 5. Rates; AZ & EL ## 6. Satellite X & Y, position angle ## 7. Local apparent sidereal time ## 8. Airmass and Visual Magnitude Extinction ## 9. Visual magnitude & surface Brightness ## 10. Illuminated fraction ## 11. Defect of illumination ## 12. Satellite angle of separation/visibility code ## 13. Target angular diameter ## 14. Observer sub-longitude & sub-latitude ## 15. Sun sub-longitude & sub-latitude ## 16. Sub-Sun position angle & distance from disc center ## 17. North pole position angle & sistance from disc center ## 18. Heliocentric ecliptic longitude & latitude ## 19. Heliocentric range & range-rate ## 20. Observer range & range-rate ## 21. One-way down-leg light-time ## 22. Speed of target with respect to Sun & observer ## 23. Sun-Observer-Targ ELONGATION angle ## 24. Sun-Target-Observer ~PHASE angle ## 25. Target-Observer-Moon/Illumination% ## 26. Observer-Primary-Target angle ## 27. Position Angles; radius & -velocity ## 28. Orbit plane angle ## 29. Constellation Name ## 30. Delta-T (TDB - UT) ##,* 31. Observer-centered Earth ecliptic longitude & latitude ## 32. North pole RA & DEC ## 33. Galactic longitude and latitude ## 34. Local apparent SOLAR time ## 35. Earth->Site light-time ## > 36. RA & DEC uncertainty ## > 37. Plane-of-sky (POS) error ellipse ## > 38. Plane-of-sky (POS) uncertainty (RSS) ## > 39. Range & range-rate sigma ## > 40. Doppler/delay sigmas ## 41. True anomaly angle ##,* 42. Local apparent hour angle ## 43. PHASE angle & bisector ## 44. Apparent target-centered longitude of Sun (L_s) ##,* 45. Inertial frame apparent RA & DEC ## 46. Rates: Inertial RA & DEC ##,* 47. Sky motion: angular rate & angles ## 48. Lunar sky brightness & target visual SNR CommonOptions* = Table[CommonOptionsKind, string] EphemerisOptions* = Table[EphemerisOptionsKind, string] ## Example URL: ## https://ssd.jpl.nasa.gov/api/horizons.api?format=text&COMMAND='499'&OBJ_DATA='YES'&MAKE_EPHEM='YES'&EPHEM_TYPE='OBSERVER'&CENTER='500@399'&START_TIME='2006-01-01'&STOP_TIME='2006-01-20'&STEP_SIZE='1%20d'&QUANTITIES='1,9,20,23,24,29' proc serialize*[T: CommonOptions | EphemerisOptions](opts: T): string = # turn into seq[(string, string)] and encase values in `'` let opts = toSeq(opts.pairs).mapIt(($it[0], &"'{it[1]}'")) result = opts.encodeQuery() proc serialize*(q: Quantities): string = result = "QUANTITIES='" var i = 0 for x in q: result.add &"{x}" if i < q.card - 1: result.add "," inc i result.add "'" proc request*(cOpt: CommonOptions, eOpt: EphemerisOptions, q: Quantities): Future[string] {.async.} = var req = basePath req.add serialize(cOpt) & "&" req.add serialize(eOpt) & "&" req.add serialize(q) echo "Performing request to: ", req var client = newAsyncHttpClient() return await client.getContent(req) # let's try a simple request let comOpt = { #coFormat : "text", coMakeEphem : "YES", coCommand : "10", coEphemType : "OBSERVER" }.toTable let ephOpt = { eoCenter : "coord@399", eoStartTime : "2017-01-01", eoStopTime : "2019-12-31", eoStepSize : "1 HOURS", eoCoordType : "GEODETIC", eoSiteCoord : "+6.06670,+46.23330,0", eoCSVFormat : "YES" }.toTable var q: Quantities q.incl 20 ## Observer range! let fut = request(comOpt, ephOpt, q) ## If multiple we would `poll`! let res = fut.waitFor() echo res.parseJson.pretty() ## TODO: construct time ranges such that 1 min yields less than 90k elements ## then cover whole range # 1. iterate all elements and download files when false: var futs = newSeq[Future[string]]() for element in 1 ..< 92: futs.add downloadFile(element) echo "INFO: Downloading all files..." while futs.anyIt(not it.finished()): poll() echo "INFO: Downloading done! Writing to ", outpath var files = newSeq[string]() for fut in futs: files.add waitFor(fut) for f in files: f.extractData.writeData()
The common parameters:
Parameter Default Allowable Values/Format Description Manual format json json, text specify output format: json for JSON or text for plain-text COMMAND none see details below target search, selection, or enter user-input object mode link OBJ_DATA YES NO, YES toggles return of object summary data MAKE_EPHEM YES NO, YES toggles generation of ephemeris, if possible EPHEM_TYPE OBSERVER OBSERVER, VECTORS, ELEMENTS, SPK, APPROACH selects type of ephemeris to generate (see details below) EMAIL_ADDR none any valid email address optional; used only in the event of highly unlikely problems needing follow-up
The ephemeris parameters:
Parameter O V E Default Allowable Values/Format Description Manual CENTER x x x Geocentric see details below selects coordinate origin (observing site) link REF_PLANE x x ECLIPTIC ECLIPTIC, FRAME, BODY EQUATOR Ephemeris reference plane (can be abbreviated E, F, B, respectively) ) COORD_TYPE x x x GEODETIC GEODETIC, CYLINDRICAL selects type of user coordinates link SITE_COORD x x x '0,0,0' set coordinate triplets for COORD_TYPE link START_TIME x x x none specifies ephemeris start time link STOP_TIME x x x none specifies ephemeris stop time link STEP_SIZE x x x '60 min' see details below ephemeris output print step. Can be fixed time, uniform interval (unitless), calendar steps, or plane-of-sky angular change steps. See also TLIST alternative. link TLIST x x x none see details below list of up to 10,000 of discrete output times. Either Julian Day numbers (JD), Modified JD (MJD), or calendar dates TLIST_TYPE x x x none JD, MJD, CAL optional specification of type of time in TLIST QUANTITIES x 'A' list of desired output quantity option codes link, link REF_SYSTEM x x x ICRF ICRF, B1950 specifies reference frame for any geometric and astrometric quantities link OUT_UNITS x x KM-S KM-S, AU-D, KM-D selects output units for distance and time; for example, AU-D selects astronomical units (au) and days (d) VEC_TABLE x 3 see details below selects vector table format link VEC_CORR x NONE NONE, LT, LT+S selects level of correction to output vectors; NONE (geometric states), LT (astrometric light-time corrected states) or LT+S (astrometric states corrected for stellar aberration) CAL_FORMAT x CAL CAL, JD, BOTH selects type of date output; CAL for calendar date/time, JD for Julian Day numbers, or BOTH for both CAL and JD CAL_TYPE x x x MIXED MIXED, GREGORIAN Selects Gregorian-only calendar input/output, or mixed Julian/Gregorian, switching on 1582-Oct-5. Recognized for close-approach tables also. ANG_FORMAT x HMS HMS, DEG selects RA/DEC output format APPARENT x AIRLESS AIRLESS, REFRACTED toggles refraction correction of apparent coordinates (Earth topocentric only) TIME_DIGITS x x x MINUTES MINUTES, SECONDS, FRACSEC controls output time precision TIME_ZONE x '+00:00' specifies local civil time offset relative to UT RANGE_UNITS x AU AU, KM sets the units on range quantities output SUPPRESS_RANGE_RATE x NO NO, YES turns off output of delta-dot and rdot (range-rate) ELEV_CUT x '-90' integer [-90:90] skip output when object elevation is less than specified SKIP_DAYLT x NO NO, YES toggles skipping of print-out when daylight at CENTER SOLAR_ELONG x '0,180' sets bounds on output based on solar elongation angle AIRMASS x 38.0 select airmass cutoff; output is skipped if relative optical airmass is greater than the single decimal value specified. Note than 1.0=zenith, 38.0 ~= local-horizon. If value is set >= 38.0, this turns OFF the filtering effect. LHA_CUTOFF x 0.0 skip output when local hour angle exceeds a specified value in the domain 0.0 < X < 12.0. To restore output (turn OFF the cut-off behavior), set X to 0.0 or 12.0. For example, a cut-off value of 1.5 will output table data only when the LHA is within +/- 1.5 angular hours of zenith meridian. ANG_RATE_CUTOFF x 0.0 skip output when the total plane-of-sky angular rate exceeds a specified value EXTRA_PREC x NO NO, YES toggles additional output digits on some angles such as RA/DEC CSV_FORMAT x x x NO NO, YES toggles output of table in comma-separated value format VEC_LABELS x YES NO, YES toggles labeling of each vector component VEC_DELTA_T x NO NO, YES toggles output of the time-varying delta-T difference TDB-UT ELM_LABELS x YES NO, YES toggles labeling of each osculating element TP_TYPE x ABSOLUTE ABSOLUTE, RELATIVE determines what type of periapsis time (Tp) is returned R_T_S_ONLY x NO NO, YES toggles output only at target rise/transit/set
The quantities documentation: https://ssd.jpl.nasa.gov/horizons/manual.html#output
UPDATE: We've now turned this into a small nimble package: https://github.com/SciNim/horizonsAPI
We can now use that easily to construct the requests we need for the CAST trackings:
import horizonsapi, datamancer, times let startDate = initDateTime(01, mJan, 2017, 00, 00, 00, 00, local()) let stopDate = initDateTime(31, mDec, 2019, 23, 59, 59, 00, local()) let nMins = (stopDate - startDate).inMinutes() const blockSize = 85_000 # max line number somewhere above 90k. Do less to have some buffer let numBlocks = ceil(nMins.float / blockSize.float).int # we end up at a later date than `stopDate`, but that's fine echo numBlocks let blockDur = initDuration(minutes = blockSize) let comOpt = { #coFormat : "json", # data returned as "fake" JSON coMakeEphem : "YES", coCommand : "10", # our target is the Sun, index 10 coEphemType : "OBSERVER" }.toTable # observational parameters var ephOpt = { eoCenter : "coord@399", # observational point is a coordinate on Earth (Earth idx 399) eoStartTime : startDate.format("yyyy-MM-dd"), eoStopTime : (startDate + blockDur).format("yyyy-MM-dd"), eoStepSize : "1 MIN", # in 1 hour steps eoCoordType : "GEODETIC", eoSiteCoord : "+6.06670,+46.23330,0", # Geneva eoCSVFormat : "YES" }.toTable # data as CSV within the JSON (yes, really) var q: Quantities q.incl 20 ## Observer range! In this case range between our coordinates on Earth and target var reqs = newSeq[HorizonsRequest]() for i in 0 ..< numBlocks: # modify the start and end dates ephOpt[eoStartTime] = (startDate + i * blockDur).format("yyyy-MM-dd") ephOpt[eoStopTime] = (startDate + (i+1) * blockDur).format("yyyy-MM-dd") echo "From : ", ephOpt[eoStartTime], " to ", ephOpt[eoStopTime] reqs.add initHorizonsRequest(comOpt, ephOpt, q) let res = getResponsesSync(reqs) proc convertToDf(res: seq[HorizonsResponse]): DataFrame = result = newDataFrame() for r in res: result.add parseCsvString(r.csvData) let df = res.convertToDf().unique("Date__(UT)__HR:MN") .select(["Date__(UT)__HR:MN", "delta", "deldot"]) echo df df.writeCsv("/home/basti/org/resources/sun_earth_distance_cast_datataking.csv", precision = 16)
import ggplotnim, sequtils # 2017-Jan-01 00:00 const Format = "yyyy-MMM-dd HH:mm" var df = readCsv("/home/basti/org/resources/sun_earth_distance_cast_datataking.csv") df["min since 2017"] = toSeq(0 ..< df.len) ggplot(df, aes("min since 2017", "delta")) + geom_line() + ggtitle("Distance in AU Sun ⇔ Earth") + ggsave("/tmp/distance_sun_earth_cast_datataking.pdf")
1.61.
Continuing from yesterday (Horizons API).
With the API constructed and data available for the Sun ⇔ Earth distance for each minute during the CAST data taking period, we now need the actual start and end times of the CAST data taking campaign.
[X]modifycast_log_readerto output CSV / Org file of table with start / stop times & their runs
Running
./cast_log_reader \ tracking \ -p ../resources/LogFiles/tracking-logs \ --startTime 2017/01/01 \ --endTime 2018/05/01 \ --h5out ~/CastData/data/DataRuns2017_Reco.h5
and
./cast_log_reader \ tracking \ -p ../resources/LogFiles/tracking-logs \ --startTime 2018/05/01 \ --endTime 2018/12/31 \ --h5out ~/CastData/data/DataRuns2018_Reco.h5
(on voidRipper) now produces the following two files for each H5 file:
- ./../CastData/ExternCode/TimepixAnalysis/resources/DataRuns2017_Reco_tracking_times.csv
- ./../CastData/ExternCode/TimepixAnalysis/resources/DataRuns2017_Reco_tracking_times.html
and
- ./../CastData/ExternCode/TimepixAnalysis/resources/DataRuns2018_Reco_tracking_times.csv
- ./../CastData/ExternCode/TimepixAnalysis/resources/DataRuns2018_Reco_tracking_times.html
which are the following Org table combined:
| Tracking start | Tracking stop | Run |
|---|---|---|
| 76 | ||
| 77 | ||
| 78 | ||
| 79 | ||
| 80 | ||
| 81 | ||
| 82 | ||
| 82 | ||
| 84 | ||
| 86 | ||
| 87 | ||
| 87 | ||
| 89 | ||
| 90 | ||
| 91 | ||
| 92 | ||
| 94 | ||
| 95 | ||
| 97 | ||
| 98 | ||
| 99 | ||
| 100 | ||
| 101 | ||
| 103 | ||
| 104 | ||
| 106 | ||
| 105 | ||
| 107 | ||
| 109 | ||
| 112 | ||
| 112 | ||
| 114 | ||
| 113 | ||
| 115 | ||
| 117 | ||
| 119 | ||
| 121 | ||
| 123 | ||
| 124 | ||
| 124 | ||
| 125 | ||
| 127 | ||
| 146 | ||
| 150 | ||
| 148 | ||
| 152 | ||
| 154 | ||
| 156 | ||
| 158 | ||
| 160 | ||
| 162 | ||
| 162 | ||
| 162 | ||
| 164 | ||
| 164 | ||
| 166 | ||
| 170 | ||
| 172 | ||
| 174 | ||
| 176 | ||
| 178 | ||
| 178 | ||
| 178 | ||
| 178 | ||
| 178 | ||
| 180 | ||
| 182 | ||
| 182 | ||
| -1 | ||
| -1 | ||
| 240 | ||
| 242 | ||
| 244 | ||
| 246 | ||
| 248 | ||
| 250 | ||
| 254 | ||
| 256 | ||
| 258 | ||
| 261 | ||
| 261 | ||
| 261 | ||
| 263 | ||
| 265 | ||
| 268 | ||
| 270 | ||
| 270 | ||
| 272 | ||
| 272 | ||
| 272 | ||
| 274 | ||
| 274 | ||
| 274 | ||
| 276 | ||
| 276 | ||
| 279 | ||
| 279 | ||
| 281 | ||
| 283 | ||
| 283 | ||
| 283 | ||
| 285 | ||
| 285 | ||
| 287 | ||
| 289 | ||
| 291 | ||
| 291 | ||
| 293 | ||
| 295 | ||
| 297 | ||
| 297 | ||
| 298 | ||
| 299 | ||
| 301 | ||
| 301 | ||
| 303 | ||
| 306 | ||
| -1 | ||
| -1 | ||
| -1 |
1.61.1. TODO
[ ]Update the systematics code in the limit calculation!
1.62.
Let's combine the tracking start/stop information with the Horizons API data about the Sun's location to compute:
[ ]a plot showing trackings in the plot for the distances[ ]the mean value of the positions during trackings and their variance / std
import ggplotnim, sequtils, times, strutils # 2017-Jan-01 00:00 const Format = "yyyy-MMM-dd HH:mm" const OrgFormat = "'<'yyyy-MM-dd ddd H:mm'>'" const p2017 = "~/CastData/ExternCode/TimepixAnalysis/resources/DataRuns2017_Reco_tracking_times.csv" const p2018 = "~/CastData/ExternCode/TimepixAnalysis/resources/DataRuns2018_Reco_tracking_times.csv" var df = readCsv("/home/basti/org/resources/sun_earth_distance_cast_datataking.csv") .mutate(f{string -> int: "Timestamp" ~ parseTime(idx("Date__(UT)__HR:MN").strip, Format, local()).toUnix.int}) proc readRuns(f: string): DataFrame = result = readCsv(f) echo result.pretty(-1) result = result .gather(["Tracking start", "Tracking stop"], "Type", "Time") echo result.pretty(-1) result = result .mutate(f{Value -> int: "Timestamp" ~ parseTime(idx("Time").toStr, OrgFormat, local()).toUnix.int}) result["delta"] = 0.0 var dfR = readRuns(p2017) dfR.add readRuns(p2018) echo dfR ggplot(df, aes("Timestamp", "delta")) + geom_line() + geom_linerange(data = dfR, aes = aes("Timestamp", y = "", yMin = 0.98, yMax = 1.02)) + ggtitle("Distance in AU Sun ⇔ Earth") + ggsave("/tmp/distance_sun_earth_with_cast_datataking.pdf")
import ggplotnim, sequtils, times, strutils, strformat # 2017-Jan-01 00:00 const Format = "yyyy-MMM-dd HH:mm" const OrgFormat = "'<'yyyy-MM-dd ddd H:mm'>'" const p2017 = "~/CastData/ExternCode/TimepixAnalysis/resources/DataRuns2017_Reco_tracking_times.csv" const p2018 = "~/CastData/ExternCode/TimepixAnalysis/resources/DataRuns2018_Reco_tracking_times.csv" var df = readCsv("/home/basti/org/resources/sun_earth_distance_cast_datataking.csv") .mutate(f{string -> int: "Timestamp" ~ parseTime(idx("Date__(UT)__HR:MN").strip, Format, local()).toUnix.int}) proc readRuns(f: string): DataFrame = result = readCsv(f) .mutate(f{string -> int: "TimestampStart" ~ parseTime(idx("Tracking start"), OrgFormat, local()).toUnix.int}) .mutate(f{string -> int: "TimestampStop" ~ parseTime(idx("Tracking stop"), OrgFormat, local()).toUnix.int}) var dfR = readRuns(p2017) dfR.add readRuns(p2018) var dfHT = newDataFrame() for tracking in dfR: let start = tracking["TimestampStart"].toInt let stop = tracking["TimestampStop"].toInt dfHT.add df.filter(f{int: `Timestamp` >= start and `Timestamp` <= stop}) dfHT["Type"] = "Trackings" df["Type"] = "HorizonsAPI" df.add dfHT let deltas = dfHT["delta", float] let meanD = deltas.mean let varD = deltas.variance let stdD = deltas.std echo "Mean distance during trackings = ", meanD echo "Variance of distance during trackings = ", varD echo "Std of distance during trackings = ", stdD # and write back the DF of the tracking positions dfHT.writeCsv("/home/basti/org/resources/sun_earth_distance_cast_solar_trackings.csv") ggplot(df, aes("Timestamp", "delta", color = "Type")) + geom_line(data = df.filter(f{`Type` == "HorizonsAPI"})) + geom_point(data = df.filter(f{`Type` == "Trackings"}), size = 1.0) + scale_x_date(isTimestamp = true, formatString = "yyyy-MM", dateSpacing = initDuration(days = 60)) + xlab("Date", rotate = -45.0, alignTo = "right", margin = 1.5) + annotate(text = &"Mean distance during trackings = {meanD:.4f}", x = 1.52e9, y = 1.0175) + annotate(text = &"Variance distance during trackings = {varD:.4g}", x = 1.52e9, y = 1.015) + annotate(text = &"Std distance during trackings = {stdD:.4f}", x = 1.52e9, y = 1.0125) + margin(bottom = 2.0) + ggtitle("Distance in AU Sun ⇔ Earth") + ggsave("/home/basti/org/Figs/statusAndProgress/systematics/sun_earth_distance_cast_solar_tracking.pdf")
Which produces the plot
and yields the output:
Mean distance during trackings = 0.9891144450781392 Variance of distance during trackings = 1.399449924353128e-05 Std of distance during trackings = 0.003740922245052853
so the real mean position is about 1.1% closer than 1 AU! The standard deviation is much smaller at 0.37% especially.
The relevant section about the systematic calculation of the distance
is in sec. [BROKEN LINK: statusAndProgress.org#sec:uncertain:distance_earth_sun] in
statusAndProgress.org.
The file ./resources/sun_earth_distance_cast_solar_trackings.csv contains the subset of the input CSV file for the actual solar trackings.
See the new subsection of the linked section in statusAndProgress
for the final numbers we now need to use for our systematics.
1.63.
[X]push newunchainedunits & tag new version[X]mergenimhdf5RP & tag new version[X]Need to incorporate the new systematics[X]update table of systematics instatusAndProgress[X]change default systematic
σ_svalue inmcmc_limitOld line:σ_sig = 0.04244936953654317, ## <- is the value *without* uncertainty on signal efficiency! # 0.04692492913207222 <- incl 2%
new line:
σ_sig = 0.02724743263827172, ## <- is the value *without* uncertainty on signal efficiency!
[X]change usage of 2% for LnL software efficiency to 1.71% inmcmc_limit
[-]Need to adjust the flux according to the new absolute distance! -> Should we add a command line argument tomcmc_limitthat gives the distance to use in AU? NO. Done using CSV files, as there would be changes to the axion image too, which flux scaling does not reproduce! -> use 0.989AU differential solar flux CSV file![X]Make the differential axion flux input CSV file a command line option[X]calculate a new differential flux withreadOpacityFileand compare it to our direct approach of scaling by1/r²[X]Implement AU as CL argument inreadOpacityFile[X]run with 1 AU as reference (storing files in
/org/resources/differential_flux_sun_earth_distanceand/org/Figs/statusAndProgress/differential_flux_sun_earth_distance)./readOpacityFile --suffix "_1AU" --distanceSunEarth 1.0.AU[X]run with the correct mean distance ~0.989 AU:
./readOpacityFile --suffix "_0.989AU" --distanceSunEarth 0.9891144450781392.AU[X]update solar radius to correct value inreadOpacityFile[X]Compare the result to 1/r² expectation. We'll read the CSV files generated by
readOpacityFileand compare the maxima:import ggplotnim let df1 = readCsv("~/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_1AU.csv") .filter(f{`type` == "Total flux"}) let df2 = readCsv("~/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv") .filter(f{`type` == "Total flux"}) let max1AU = df1["diffFlux", float].max let max0989AU = df2["diffFlux", float].max echo "Ratio of 1 AU to 0.989 AU = ", max0989AU / max1AU
Bang on reproduction of our 2.2% increase!
[-]Implement X AU as command line argument intomcmc_limitFor the limit calculation we will use the 1 AU "reference flux". From there a CL argument can be used to adjust the distance. The raytraced image should not change, as only the amount of flux changes. -> Well, we do expect a small change. Because if the Sun is closer, its angular size is larger too! In that sense maybe it is better after all to just handle this by the axion image + differential flux file? -> YES, we won't implement AU scaling intomcmc_limit
[X]add differential solar fluxes toTimepixAnalysis/resourcesdirectory- update axion image using
[X]correct solar radius[X]correct sun earth distance at 0.989 AU[X]correct median conversion point as computed numerically, namely
Mean conversion position = 0.556813 cm Median conversion position = 0.292802 cm Variance of conversion position = 0.424726 cm
from ./Doc/SolarAxionConversionPoint/axion_conversion_point.html. This corresponds to a position of:
import unchained let f = 1500.mm let xp = 0.292802.cm let d = 3.cm # detector volume height let fromFocal = (d / 2.0) - xp let imageAt = f - fromFocal echo "Image at = ", imageAt.to(mm) echo "From focal = ", fromFocal.to(mm)
which is ~1.23 cm in front of the actual focal spot.
We could use the mean, but that would be disingenuous.
Note though: compared to our original calculation of being 1.22 cm behind the window but believing the focal spot is in the readout plane, we now still gain about 5 mm towards the focal spot! That old number was
1482.2 mm. Before runningraytracer, make sure the config file contains:distanceDetectorXRT = 1487.93 # mm
Then run:
./raytracer \ --ignoreDetWindow \ --ignoreGasAbs \ --suffix "_1487_93_0.989AU" \ --distanceSunEarth 0.9891144450781392.AU
The produced files are found in ./resources/axion_image_2018_1487_93_0.989AU.csv and the plot:
(comparing it with
shows that indeed it is quite a bit smaller than our "old" input!)
With all of the above finally done, we can finally compute some expected limits for the new input files, i.e. axion image and solar flux corresponding to:
- correct conversion point based on numerical median
- correct solar radius based on SOHO
- correct mean distance to Sun
So let's run the limit calculation for the best case scenario:
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --nmc 1000 \ --suffix=_sEff_0.95_scinti_fadc_line_mlp_tanh300_mse_epoch_485000_loss_0.0055_acc_0.9933_axion_image_1487.9_0.989AU \ --path "" \ --outpath /home/basti/org/resources/lhood_limits_03_07_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel /home/basti/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv
For notes on the meeting with Klaus, see the next point.
1.63.1. Understanding slowness of mcmc_limit
For some reason the mcmc_limit code is much slower now than it was
in the past. I don't understand why. In my notes further up, from
I mention that a command using the same files
as in the above snippet only takes about 20s to build the chains. Now
it takes 200-260s.
[X]check ifsorted_seqdifference -> no[X]check if somearraymancerdifference -> no[X]run withseptemveto in addition -> also as slow[X]is it the noisy pixels? -> it seems to be using the up to date list, incl the "Deich"[X]Numbers of noisy pixel removal logic:
Number of elements before noise filter: 25731 Number of elements after noise filter: 24418 Number of elements before noise filter: 10549 Number of elements after noise filter: 10305 [INFO]: Read a total of 34723 input clusters. And after energy filter: 20231
-> From running with septem+line veto MLP case.
Further: the HDF5 files for the Septem + Line veto case are essentially the same size as the pure Line veto case. How does that make any sense?
Let's look at the background cluster plot of the septem & line and only line veto case for MLP@95%.
plotBackgroundClusters \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters CAST MLP@95+scinti+fadc+line" \ --outpath ~/Sync/mcmc_limit_very_slow/ \ --suffix "03_07_23_mlp_0.95_scinti_fadc_line_mlp" \ --energyMax 12.0 --energyMin 0.2 \ --filterNoisyPixels
UPDATE: Could it be related to our new axion model? Does it have more energies or something, which slows down the interpolation? Seems stupid, but who knows? At least try with old axion model and see. -> No, they seem to have the same binning. Just our new one goes to 15 keV.
I let the code run over night and it yielded:
Acceptance rate: 0.2324666666666667 with last two states of chain: @[@[6.492996449371051e-22, -0.01196348404464549, -0.002164366481936349, -0.02809605322316696, -0.007979752246365442], @[1.046326715495785e-21, -0.02434126786591704, 0.0008550422211706134, -0.04539491720412565, -0.003574795727520216]]
Limit at 3.453757576271354e-21
Number of candidates: 0
INFO: The integer column `Hist` has been automatically determined to be continuous. To overwrite this behavior add a `+ scale_x/y_discrete()` call to the plotting chain. Choose `x` or `y` depending on which axis this column refers to. Or apply a `factor` to the column name in the `aes` call, i.e. `aes(..., factor("Hist"), ...)`.
Expected limit: 1.378909932139855e-20
85728
Generating group /ctx/axionModel
datasets.nim(849) write
Error: unhandled exception: Wrong input shape of data to write in `[]=` while accessing `/ctx/axionModel/type`. Given shape `@[1500]`, dataset has shape `@[1000]` [ValueError]
So outside of the fact that the code didn't even manage to save the freaking file, the limit is also completely bonkers. 1.37e-20 corresponds to something like 1e-10·1e-12, so absolutely horrible.
Something is fucked, which also will explain the slowness.
1.64.
Main priority today: understand and fix the slowness of the limit calculation.
[X]Check how fast it is if we use the old differential solar flux:
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --nmc 1000 \ --suffix=_sEff_0.95_scinti_fadc_line_mlp_tanh300_mse_epoch_485000_loss_0.0055_acc_0.9933_axion_image_1487.9_0.989AU \ --path "" \ --outpath /home/basti/org/resources/lhood_limits_03_07_23/ \ --energyMin 0.2 --energyMax 12.0
i.e. the same command as yesterday without the
axionModelargument (i.e. using the default which is the old file). -> It was so ridiculously slow that I stopped after 10 minutes. What the fuck is going on.
NOTE: Just discovered something. I wanted to reboot the computer,
because maybe something is messed up. I found a candidates.pdf in
/tmp/ that I smartly produce before starting the limit
calculation. The plot is:
As we can see the number of candidates is HUMONGOUS!!!
Is something broken with the tracking / background times?
First reboot though.
[X]Do another start after reboot of the same command as yesterday. -> I expect this to be the same slowness. The issue will be a regression introduced for the background / tracking time logic we refactored? -> Still seems to be as slow.[X]Checking thecandidates.pdffor the new data run: -> Looks the same. So something is broken in that logic.[X]Checking the background and tracking time that is assigned to
Context:Background time = 3158.57 h Tracking time = 161.111 h
-> That also looks reasonable.
[X]Investigate candidate drawing -> the drawing looks fine[X]Background interpolation -> Found the culprit! We handed the inputbackgroundTimeandtracknigTimeparameters to thesetupBackgroundInterpolationfunction instead of the local modified parametersbackTimeandtrackTime! That lead these values to be-1 Hourinside of that function causing fun side effects for the expected number of counts. That then lead to bad candidate sampling in the candidate drawing procedure (which itself looked fine).
The candidates after the fix:
Freaking hell.
Still getting things like:
Building chain of 150000 elements took 127.3677394390106 s
Acceptance rate: 0.30088 with last two states of chain: @[@[9.311633190740021e-22, 0.01744901674235642, 0.00349084434202456, -0.06634240340482739, 0.03999664726123401], @[9.311633190740021e-22, 0.01744901674235642, 0.00349084434202456, -0.06634240340482739, 0.03999664726123401]]
Initial chain state: @[4.668196570108809e-21, -0.3120389306029943, 0.3543889354717579, 0.286701390433319, 0.1226804125360241]
Building chain of 150000 elements took 128.6130454540253 s
Acceptance rate: 0.3034866666666667 with last two states of chain: @[@[3.731887947371716e-21, 0.02452035569228822, 0.000773644639561432, -0.08992991789316797, -0.0382258117838525], @[3.731887947371716e-21, 0.02452035569228822, 0.000773644639561432, -0.08992991789316797, -0.0382258117838525]]
Initial chain state: @[2.660442796473178e-22, -0.2011569539539821, -0.2836544777277811, 0.02919490998034624, 0.4127775646701672]
Building chain of 150000 elements took 128.8146977424622 s
Acceptance rate: 0.2591533333333333 with last two states of chain: @[@[3.636435825606668e-22, -0.009764842941003157, -0.0007353516663395031, 0.03297060483409234, -0.04076920903469726], @[6.506720970027227e-22, -0.0107001279962231, -6.017950416918778e-05, 0.04780628462897407, -0.04483761760499658]]
Initial chain state: @[9.722845479265146e-22, 0.3584189020390509, -0.1514954111305945, -0.03343978579815121, -0.2637922163333362]
Building chain of 150000 elements took 138.6971650123596 s
Acceptance rate: 0.2639666666666667 with last two states of chain: @[@[3.438289368883349e-22, -0.01304748715057187, 0.004184991829399071, -0.04636487615831818, 0.0302346566894824], @[1.541274009225669e-21, -0.02093515375501852, 0.003417056328213522, -0.04313773041382048, 0.02677047733100371]]
Initial chain state: @[2.436881668011995e-21, 0.3695082702072843, 0.04051624101632562, -0.458195482427621, -0.07043128904485663]
Building chain of 150000 elements took 144.5500540733337 s
Acceptance rate: 0.2609733333333333 with last two states of chain: @[@[4.194422598229001e-22, -0.01096894725308242, -0.001059399554620779, -0.04838608283669801, 0.005199899235731185], @[4.194422598229001e-22, -0.01096894725308242, -0.001059399554620779, -0.04838608283669801, 0.005199899235731185]]
Initial chain state: @[1.025364583527896e-21, 0.3425107036102778, 0.26050622555894, -0.1392108662060235, -0.3609805077820832]
Building chain of 150000 elements took 145.2971315383911 s
Acceptance rate: 0.28042 with last two states of chain: @[@[6.825664933417517
running on
mcmc_limit_calculation limit -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 -f /home/basti/
org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 --years 2017 --years 2018 --σ_p 0.05 --limitKind lkMCMC --nmc 1000 --suffix=_sEff_0.95_scinti_fadc_septem_line_mlp_tanh300_mse_epoch_485000
_loss_0.0055_acc_0.9933_axion_image_1487.9_0.989AU --path "" --outpath /home/basti/org/resources/lhood_limits_03_07_23/ --energyMin 0.2 --energyMax 12.0 --axionModel /home/basti/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv
(i.e. mlp+scinti+fadc+septem+line @ 95%!)
Still way too slow!!!
[X]Checking again with the old differential axion flux (without
--axionModel): -> SameBuilding chain of 150000 elements took 126.3729739189148 s Acceptance rate: 0.2716533333333334 with last two states of chain: @[@[1.270974433734064e-21, -0.008762536914031409, -0.0009393362144718906, 0.08807054442679391, 0.06807056108511295], @[1.270974433734064e-21, -0.008762536914031409, -0.0009393362144718906, 0.08807054442679391, 0.06807056108511295]] Initial chain state: @[4.161681061397676e-21, 0.03937891262715859, -0.2687772585382085, 0.4510828436114304, 0.4645657545530211] Building chain of 150000 elements took 125.4626288414001 s Acceptance rate: 0.2654533333333333 with last two states of chain: @[@[6.984109549639046e-23, 0.02177393681219079, -0.0009694252520926414, -0.01536573917383219, 0.06357336308703909], @[6.984109549639046e-23, 0.02177393681219079, -0.0009694252520926414, -0.01536573917383219, 0.06357336308703909]] Initial chain state: @[2.436881668011995e-21, 0.3695082702072843, 0.04051624101632562, -0.458195482427621, -0.07043128904485663] Building chain of 150000 elements took 145.4075906276703 s Acceptance rate: 0.2648733333333333 with last two states of chain: @[@[1.854805063479706e-21, 0.02828329851122759, -0.001409250857040086, -0.07848092399906945, -0.01008439148632219], @[4.68305467951519e-22, 0.03745033276146833, 0.002680253359587353, -0.09263439093421814, -0.02574252887010509]] Initial chain state: @[1.025364583527896e-21, 0.3425107036102778, 0.26050622555894, -0.1392108662060235, -0.3609805077820832] Building chain of 150000 elements took 146.0146522521973 s Acceptance rate: 0.28422 with last two states of chain: @[@[3.023581967426795e-21, -0.09020389993493418, -0.005375722700108269, -0.009890672103045093, 0.03342292616291231], @[2.466627578743573e-21, -0.0871729066832931, 0.005329262454946779, -0.0002405123197451453, 0.03887706119504662]] Initial chain state: @[2.98188342976756e-21, 0.0[X]Check with old systematic value (not that I expect this to change anything):
mcmc_limit_calculation limit -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 --years 2017 --years 2018 --σ_p 0.05 --limitKind lkMCMC --nmc 1000 --suffix=_sEff_0.95_scinti_fadc_septem_line_mlp_tanh300_mse_epoch_485000_loss_0.0055_acc_0.9933_axion_image_1487.9_0.989AU --path "" --outpath /home/basti/org/resources/lhood_limits_03_07_23/ --energyMin 0.2 --energyMax 12.0 --σ_sig 0.04244936953654317-> Same
Building chain of 150000 elements took 109.3231236934662 s Acceptance rate: 0.2735866666666666 with last two states of chain: @[@[5.061099862447965e-22, -0.02857602726297297, -0.00130806717688539, 0.03063698419159643, 0.08021558103217649], @[5.061099862447965e-22, -0.02857602726297297, -0.00130806717688539, 0.03063698419159643, 0.08021558103217649]] Initial chain state: @[4.161681061397676e-21, 0.03937891262715859, -0.2687772585382085, 0.4510828436114304, 0.4645657545530211] Building chain of 150000 elements took 141.4028820991516 s Acceptance rate: 0.2680066666666667 with last two states of chain: @[@[2.116498657020219e-22, 0.00157812420011463, -0.001191578637594618, -0.03903883316617535, 0.001184257609417868], @[2.116498657020219e-22, 0.00157812420011463, -0.001191578637594618, -0.03903883316617535, 0.001184257609417868]] Initial chain state: @[2.436881668011995e-21, 0.3695082702072843, 0.04051624101632562, -0.458195482427621, -0.07043128904485663] Building chain of 150000 elements took 142.4829633235931 s
What else:
[ ]Run with LnL with all vetoes and see what we get:
-> Need to regenerate likelihood files to work with them in limit code due to veto config missing in old files.
1.64.1. Regenerate likelihood output files
We'll only generate a single case for now:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkLogL, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto}" \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.8 \ --out ~/org/resources/lhood_lnL_04_07_23/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 12
After an extremely painful amount of work to get likelihood compiled
again to fix a few small issues (i.e. depending on the cacheTab
files even when using no MLP), the files are finally there.
BUUUUUUUUUUUUUUUT I ran with the default clustering algorithm,
instead of dbscan…
Rerunning again using dbscan.
This brings up the question on the efficiency of the septem veto in case of the MLP though…
Renamed the files to have a _default_cluster suffix.
Let's plot the cluster centers for the default case files:
plotBackgroundClusters \ ~/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99_default_cluster.h5 \ --zMax 30 \ --title "LnL+FADC+Scinti+Septem+Line default cluster algo" \ --outpath ~/Sync/lnL_04_07_23/ \ --suffix "04_07_23_lnL_scinti_fadc_septem_line_default_cluster_R2" \ --energyMax 12.0 --energyMin 0.0 --filterNoisyPixels
plotBackgroundClusters \ ~/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99_default_cluster.h5 \ --zMax 30 \ --title "LnL+FADC+Scinti+Septem+Line default cluster algo" \ --outpath ~/Sync/lnL_04_07_23/ \ --suffix "04_07_23_lnL_scinti_fadc_septem_line_default_cluster_R3" \ --energyMax 12.0 --energyMin 0.0 --filterNoisyPixels
yield
-> 6420 clusters
-> 3471 clusters
So 9891 clusters instead of the roughly 8900 we expect for DBSCAN. Let's check though if we reproduce those.
: Finished the dbscan run, let's look at clusters:
plotBackgroundClusters \ ~/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ --zMax 30 \ --title "LnL+FADC+Scinti+Septem+Line default cluster algo" \ --outpath ~/Sync/lnL_04_07_23/ \ --suffix "04_07_23_lnL_scinti_fadc_septem_line_dbscan_R2" \ --energyMax 12.0 --energyMin 0.0 --filterNoisyPixels
plotBackgroundClusters \ ~/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ --zMax 30 \ --title "LnL+FADC+Scinti+Septem+Line default cluster algo" \ --outpath ~/Sync/lnL_04_07_23/ \ --suffix "04_07_23_lnL_scinti_fadc_septem_line_dbscan_R3" \ --energyMax 12.0 --energyMin 0.0 --filterNoisyPixels
yield
-> 6242 clusters
-> 3388 clusters
-> Less, 9630, still more than initially assumed. Maybe due to the binning changes of the histograms going into the LnL method? Anyhow.
In the meantime let's check how slow mcmc_limit is with the default
cluster files:
mcmc_limit_calculation \ limit \ -f ~/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99_default_cluster.h5 \ -f ~/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99_default_cluster.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 --σ_sig 0.04244936953654317 \ --limitKind lkMCMC \ --nmc 1000 \ --suffix=_lnL_scinti_fadc_septem_line_axion_image_1487.9_0.989AU_default_cluster \ --path "" \ --outpath /home/basti/org/resources/lhood_limits_03_07_23/ \ --energyMin 0.2 --energyMax 12.0
results in times like:
Building chain of 150000 elements took 59.16278481483459 s
Acceptance rate: 0.3301133333333333 with last two states of chain: @[@[3.221218754153123e-21, 0.1001582746475284, -0.0006750750804661032, -0.01909225821269168, -0.01271736094489837], @[2.557915589131322e-21, 0.09303695310023562, 0.00136396883333453, -0.00593985734664134, -0.001857904842659075]]
Initial chain state: @[2.568718767500517e-21, -0.3503092502559793, 0.02507620318499509, -0.3212106439381629, -0.1823391110232517]
Building chain of 150000 elements took 59.59976840019226 s
Acceptance rate: 0.27792 with last two states of chain: @[@[2.43150416951269e-22, 0.04679422926871175, 0.003060880706465222, 0.005988965372613596, -0.1462321981756096], @[2.43150416951269e-22, 0.04679422926871175, 0.003060880706465222, 0.005988965372613596, -0.1462321981756096]]
Building chain of 150000 elements took 59.39858341217041 s
Acceptance rate: 0.2637866666666667 with last two states of chain: @[@[4.009161808514372e-21, 0.02699119698256826, 0.0008468364946590864, 0.00313360442843261, 0.03944583054445015], @[4.009161808514372e-21, 0.02699119698256826, 0.0008468364946590864, 0.00313360442843261, 0.03944583054445015]]
Initial chain state: @[1.551956111345227e-21, -0.02085127777101975, 0.2274015842900468, -0.3652020071376869, 0.06496986846631414]
Initial chain state: @[1.404278364950456e-22, 0.1851804887591793, -0.23513445609526, -0.4396648010325593, -0.328970832476948]
Building chain of 150000 elements took 59.805743932724 s
Acceptance rate: 0.2787866666666667 with last two states of chain: @[@[2.818591563704671e-21, 0.01977281003326564, -0.001144346574617646, 0.06980766970784988, -0.05324435377403436], @[2.818591563704671e-21, 0.01977281003326564, -0.001144346574617646, 0.06980766970784988, -0.05324435377403436]]
Initial chain state: @[3.436712853810167e-21, 0.3576059646653303, -0.3810145277979216, -0.01900304799919095, -0.3084630290908293]
Building chain of 150000 elements took 60.12974739074707 s
Acceptance rate: 0.2755866666666666 with last two states of chain: @[@[1.139005379041843e-22, -0.02549423683147078, -0.0004239605850902325, -0.008100179554892915, 0.07260243062580041], @[1.139005379041843e-22, -0.02549423683147078, -0.0004239605850902325, -0.008100179554892915, 0.07260243062580041]]
Initial chain state: @[3.891438297177983e-21, 0.1823988603616008, 0.06236190504128392, 0.2538882767591366, 0.3203063266792117]
Building chain of 150000 elements took 61.13999462127686 s
so something is CLEARLY still amiss!
What's going on? :(
mcmc_limit_calculation \ limit \ -f ~/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ -f ~/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 --σ_sig 0.04244936953654317 \ --limitKind lkMCMC \ --nmc 1000 \ --suffix=_lnL_scinti_fadc_septem_line_axion_image_1487.9_0.989AU_dbscan \ --path "" \ --outpath /home/basti/org/resources/lhood_limits_03_07_23/ \ --energyMin 0.2 --energyMax 12.0
Building chain of 150000 elements took 41.53342080116272 s
Acceptance rate: 0.3049866666666667 with last two states of chain: @[@[7.036224223613161e-21, 0.01614397518543687, 8.574973310970443e-05, 0.0872665054520058, 0.0804204777465774], @[7.036224223613161e-21, 0.01614397518543687, 8.574973310970443e-05, 0.0872665054520058, 0.0804204777465774]]
Initial chain state: @[3.59043282379205e-21, 0.2925015742273372, -0.3931338424871418, 0.4063058330665388, 0.4222861762129114]
Building chain of 150000 elements took 44.86023664474487 s
Acceptance rate: 0.3131333333333333 with last two states of chain: @[@[4.971679231139565e-21, -0.03200714694510957, 0.000626967579541237, 0.06151432017642863, -0.07064431496540197], @[4.971679231139565e-21, -0.03200714694510957, 0.000626967579541237, 0.06151432017642863, -0.07064431496540197]]
Initial chain state: @[4.515550746128827e-21, -0.09548612273662183, 0.2106540833085406, -0.1093334950239145, 0.3220710095688022]
Building chain of 150000 elements took 53.00375294685364 s
Acceptance rate: 0.4512 with last two states of chain: @[@[3.749539171666764e-21, -0.03673449807793086, -0.001626297352381822, 0.00590080323259861, 0.07538790528734959], @[3.749539171666764e-21, -0.03673449807793086, -0.001626297352381822, 0.00590080323259861, 0.07538790528734959]]
Initial chain state: @[3.574065128929081e-21, 0.0482956327541732, -0.1815499308190825, -0.1561039982914719, -0.4663740396633153]
Building chain of 150000 elements took 57.14369440078735 s
which is also clearly slower. This is using the OLD AXION IMAGE and old differential flux.
1.64.2. DONE Found the "bug" root cause
UPDATE
OH MY SWEET FUCKING JESUS
I simply didn't compile in -d:danger mode! I want to cry.
After compiling correctly we get numbers like:
Building chain of 150000 elements took 2.175987958908081 s
Acceptance rate: 0.3000066666666666 with last two states of chain: @[@[1.180885247697067e-21, 0.08481352589490687, 0.001453176163411386, -0.02849094252852952, -0.07246502908793442], @[1.180885247697067e-21, 0.08481352589490687, 0.001453176163411386, -0.02849094252852952, -0.07246502908793442]]
Initial chain state: @[3.135081713854781e-21, -0.3830730177637535, 0.1014735248650233, -0.3582165398036626, -0.07658294956061662]
Building chain of 150000 elements took 1.937663078308105 s
which is what I like to see.
This was for:
mcmc_limit_calculation \ limit \ -f ~/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ -f ~/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 --σ_sig 0.04244936953654317 \ --limitKind lkMCMC \ --nmc 1000 \ --suffix=_lnL_scinti_fadc_septem_line_axion_image_dbscan_old_defaults \ --path "" \ --outpath /home/basti/org/resources/lhood_limits_03_07_23/ \ --energyMin 0.2 --energyMax 12.0
which means:
- old systematics
- old axion image
- old axion flux
- lnL80+fadc+scinti+septem+line
and yielded:
Expected limit: 7.474765424923508e-21
A limit of: 8.64567257356e-23
The corresponding number from the bigger table in statusAndProgress:
| 0.8 | true | true | 0.98 | true | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.5743 | 4.9249e-21 | 8.7699e-23 |
So that seems at least more or less in line with expectations. The improvement may be from our accidental energy cut?
Let's now recompile with the correct axion image and run with correct systematics and flux.
mcmc_limit_calculation \ limit \ -f ~/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ -f ~/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --nmc 1000 \ --suffix=_lnL_scinti_fadc_septem_line_axion_1487.93_0989AU_new_syst_dbscan \ --path "" \ --outpath /home/basti/org/resources/lhood_limits_03_07_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel /home/basti/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv
This yields:
Expected limit: 7.336461324602653e-21
which comes out to: 8.56531454449e-23 for the limit. That's a decent improvement for not actually changing anything fundamentally!
So time to run it on the MLP:
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --nmc 1000 \ --suffix=_sEff_0.95_scinti_fadc_line_mlp_tanh300_mse_epoch_485000_loss_0.0055_acc_0.9933_axion_image_1487.9_0.989AU \ --path "" \ --outpath /home/basti/org/resources/lhood_limits_03_07_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel /home/basti/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv
Expected limit: 5.592200029700092e-21
Yields: 7.47810138317e-23 !!
That's a pretty good number! The last number for this setup was
7.74e-23 (see big table in statusAndProgress).
1.64.3. Starting all limit calculations
We can now start the limit calculations again for all the settings of LnL & MLP of interest. Essentially the best of the previous limit calculation table.
Let's check. We have the MLP files in:
However, for the LnL approach we are still lacking a large number of HDF5 files that have the "correct" NN support, i.e. have the veto settings a part of the HDF5 files for easier reading etc.
So let's first regenerate all the likelihood combinations that we actually care about and then run the limits after.
Let's first consider the setups we actually want to reproduce and then about the correct calls.
The top part of the expected limits result table in
sec. ./Doc/StatusAndProgress.html of
statusAndProgress is:
| εlnL | Scinti | FADC | εFADC | Septem | Line | eccLineCut | εSeptem | εLine | εSeptemLine | Total eff. | Limit no signal | Expected Limit |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.9 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.7587 | 3.7853e-21 | 7.9443e-23 |
| 0.9 | true | false | 0.98 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.7742 | 3.6886e-21 | 8.0335e-23 |
| 0.9 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.8794 | 0.7415 | 0.7757 | 3.6079e-21 | 8.1694e-23 |
| 0.8 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.6744 | 4.0556e-21 | 8.1916e-23 |
| 0.9 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.8946 | 0.7482 | 0.7891 | 3.5829e-21 | 8.3198e-23 |
| 0.8 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.8794 | 0.7415 | 0.6895 | 3.9764e-21 | 8.3545e-23 |
| 0.8 | true | true | 0.9 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.6193 | 4.4551e-21 | 8.4936e-23 |
| 0.9 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.9076 | 0.754 | 0.8005 | 3.6208e-21 | 8.5169e-23 |
| 0.8 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.8946 | 0.7482 | 0.7014 | 3.9491e-21 | 8.6022e-23 |
| 0.8 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.9076 | 0.754 | 0.7115 | 3.9686e-21 | 8.6462e-23 |
| 0.9 | true | false | 0.98 | true | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.6593 | 4.2012e-21 | 8.6684e-23 |
| 0.7 | true | true | 0.98 | false | true | 0 | 0.7841 | 0.8602 | 0.7325 | 0.5901 | 4.7365e-21 | 8.67e-23 |
NOTE: These have different rows for different ε line veto cutoffs, but the table does not highlight that fact! 0.8602 corresponds to ε = 1.0, i.e. disable the cutoff. As a result we will only c
which tells us the following:
- FADC either at 99% or off
- scinti on always
- line veto always
- ε line veto cutoff disabled is best
- lnL efficiency 0.7 only without
So effectively we just want the septem & line veto combinations with 0.7, 0.8, and 0.9 software efficiency.
Also make sure the config.toml file contains the DBSCAN algo for
the likelihood method!
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkLogL, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.7 --signalEfficiency 0.8 --signalEfficiency 0.9 \ --out ~/org/resources/lhood_lnL_04_07_23/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 12
This should produce all the combinations we really care about.
--dryRun yields:
Command: (fname: "/home/basti/CastData/data/DataRuns2017_Reco.h5", calib: "/home/basti/CastData/data/CalibrationRuns2017_Reco.h5", mlpPath: "", settings: (year: 2017, region: crAll, signalEff: 0.7, eccentricityCutoff: 1.0, vetoes: {fkLogL, fkScinti, fkFadc, fkSeptem, fkLineVeto}, vetoPercentile: 0.99))
As filename: /home/basti/org/resources/lhood_lnL_04_07_23//lhood_c18_R2_crAll_sEff_0.7_lnL_scinti_fadc_septem_line_vQ_0.99.h5
Command: (fname: "/home/basti/CastData/data/DataRuns2017_Reco.h5", calib: "/home/basti/CastData/data/CalibrationRuns2017_Reco.h5", mlpPath: "", settings: (year: 2017, region: crAll, signalEff: 0.7, eccentricityCutoff: 1.0, vetoes: {fkLogL, fkScinti, fkFadc, fkExclusiveLineVeto}, vetoPercentile: 0.99))
As filename: /home/basti/org/resources/lhood_lnL_04_07_23//lhood_c18_R2_crAll_sEff_0.7_lnL_scinti_fadc_line_vQ_0.99.h5
Command: (fname: "/home/basti/CastData/data/DataRuns2017_Reco.h5", calib: "/home/basti/CastData/data/CalibrationRuns2017_Reco.h5", mlpPath: "", settings: (year: 2017, region: crAll, signalEff: 0.8, eccentricityCutoff: 1.0, vetoes: {fkLogL, fkScinti, fkFadc, fkSeptem, fkLineVeto}, vetoPercentile: 0.99))
As filename: /home/basti/org/resources/lhood_lnL_04_07_23//lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5
Command: (fname: "/home/basti/CastData/data/DataRuns2017_Reco.h5", calib: "/home/basti/CastData/data/CalibrationRuns2017_Reco.h5", mlpPath: "", settings: (year: 2017, region: crAll, signalEff: 0.8, eccentricityCutoff: 1.0, vetoes: {fkLogL, fkScinti, fkFadc, fkExclusiveLineVeto}, vetoPercentile: 0.99))
As filename: /home/basti/org/resources/lhood_lnL_04_07_23//lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5
Command: (fname: "/home/basti/CastData/data/DataRuns2017_Reco.h5", calib: "/home/basti/CastData/data/CalibrationRuns2017_Reco.h5", mlpPath: "", settings: (year: 2017, region: crAll, signalEff: 0.9, eccentricityCutoff: 1.0, vetoes: {fkLogL, fkScinti, fkFadc, fkSeptem, fkLineVeto}, vetoPercentile: 0.99))
As filename: /home/basti/org/resources/lhood_lnL_04_07_23//lhood_c18_R2_crAll_sEff_0.9_lnL_scinti_fadc_septem_line_vQ_0.99.h5
Command: (fname: "/home/basti/CastData/data/DataRuns2017_Reco.h5", calib: "/home/basti/CastData/data/CalibrationRuns2017_Reco.h5", mlpPath: "", settings: (year: 2017, region: crAll, signalEff: 0.9, eccentricityCutoff: 1.0, vetoes: {fkLogL, fkScinti, fkFadc, fkExclusiveLineVeto}, vetoPercentile: 0.99))
As filename: /home/basti/org/resources/lhood_lnL_04_07_23//lhood_c18_R2_crAll_sEff_0.9_lnL_scinti_fadc_line_vQ_0.99.h5
Command: (fname: "/home/basti/CastData/data/DataRuns2018_Reco.h5", calib: "/home/basti/CastData/data/CalibrationRuns2018_Reco.h5", mlpPath: "", settings: (year: 2018, region: crAll, signalEff: 0.7, eccentricityCutoff: 1.0, vetoes: {fkLogL, fkScinti, fkFadc, fkSeptem, fkLineVeto}, vetoPercentile: 0.99))
As filename: /home/basti/org/resources/lhood_lnL_04_07_23//lhood_c18_R3_crAll_sEff_0.7_lnL_scinti_fadc_septem_line_vQ_0.99.h5
Command: (fname: "/home/basti/CastData/data/DataRuns2018_Reco.h5", calib: "/home/basti/CastData/data/CalibrationRuns2018_Reco.h5", mlpPath: "", settings: (year: 2018, region: crAll, signalEff: 0.7, eccentricityCutoff: 1.0, vetoes: {fkLogL, fkScinti, fkFadc, fkExclusiveLineVeto}, vetoPercentile: 0.99))
As filename: /home/basti/org/resources/lhood_lnL_04_07_23//lhood_c18_R3_crAll_sEff_0.7_lnL_scinti_fadc_line_vQ_0.99.h5
Command: (fname: "/home/basti/CastData/data/DataRuns2018_Reco.h5", calib: "/home/basti/CastData/data/CalibrationRuns2018_Reco.h5", mlpPath: "", settings: (year: 2018, region: crAll, signalEff: 0.8, eccentricityCutoff: 1.0, vetoes: {fkLogL, fkScinti, fkFadc, fkSeptem, fkLineVeto}, vetoPercentile: 0.99))
As filename: /home/basti/org/resources/lhood_lnL_04_07_23//lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5
Command: (fname: "/home/basti/CastData/data/DataRuns2018_Reco.h5", calib: "/home/basti/CastData/data/CalibrationRuns2018_Reco.h5", mlpPath: "", settings: (year: 2018, region: crAll, signalEff: 0.8, eccentricityCutoff: 1.0, vetoes: {fkLogL, fkScinti, fkFadc, fkExclusiveLineVeto}, vetoPercentile: 0.99))
As filename: /home/basti/org/resources/lhood_lnL_04_07_23//lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5
Command: (fname: "/home/basti/CastData/data/DataRuns2018_Reco.h5", calib: "/home/basti/CastData/data/CalibrationRuns2018_Reco.h5", mlpPath: "", settings: (year: 2018, region: crAll, signalEff: 0.9, eccentricityCutoff: 1.0, vetoes: {fkLogL, fkScinti, fkFadc, fkSeptem, fkLineVeto}, vetoPercentile: 0.99))
As filename: /home/basti/org/resources/lhood_lnL_04_07_23//lhood_c18_R3_crAll_sEff_0.9_lnL_scinti_fadc_septem_line_vQ_0.99.h5
Command: (fname: "/home/basti/CastData/data/DataRuns2018_Reco.h5", calib: "/home/basti/CastData/data/CalibrationRuns2018_Reco.h5", mlpPath: "", settings: (year: 2018, region: crAll, signalEff: 0.9, eccentricityCutoff: 1.0, vetoes: {fkLogL, fkScinti, fkFadc, fkExclusiveLineVeto}, vetoPercentile: 0.99))
As filename: /home/basti/org/resources/lhood_lnL_04_07_23//lhood_c18_R3_crAll_sEff_0.9_lnL_scinti_fadc_line_vQ_0.99.h5
which looks fine.
It finished with:
Running all likelihood combinations took 1571.099282264709 s
Finally, let's run the expected limits for the full directory:
./runLimits \ --path ~/org/resources/lhood_lnL_04_07_23/ \ --outpath ~/org/resources/lhood_lnL_04_07_23/limits \ --prefix lhood_c18_R2_crAll \ --energyMin 0.0 --energyMax 12.0 \ --axionModel /home/basti/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --nmc 1000
1.65.
Last nights run of runLimits crashed due to us compiling with
seqmath from latest tag instead of the version that uses stop as
the final value in linspace if endpoint is true.
Rerunning now.
Output from the end:
shell> Expected limit: 6.936205119829989e-21 shell> 40980 shell> Generating group /ctx/axionModel shell> Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl shell> Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl shell> Serializing Interpolator by evaluating 0.0 to 12.0 of name: backgroundSpl shell> Generating group /ctx/backgroundDf shell> Wrote outfile /home/basti/org/resources/lhood_lnL_04_07_23/limits/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0323_σb_0.0028_posUncertain_puUncertain_σp_0.0500_sEff_0.9_lnL_scinti_fadc_septem_line_vQ_0.99.h5 Computing single limit took 568.60129737854 s Computing all limits took 3136.202635526657 s
Looking good!
Time to run the MLP limits. To avoid the limits taking forever to run, we will exclude the MLP only limits from ./resources/lhood_limits_10_05_23_mlp_sEff_0.99 and instead only run the combinations that have at least the line veto.
Unfortunately, the input files have the efficiency before the used
vetoes. So we cannot go by a prefix. Can we update the runLimits to
allow a standard glob?
-> See below, glob already works for the "prefix"!
./runLimits \ --path ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/ \ --prefix "lhood_c18_R2_crAll_*_scinti_fadc_" \ --outpath ~/org/resources/lhood_MLP_05_07_23/limits \ --energyMin 0.0 --energyMax 12.0 \ --axionModel /home/basti/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --nmc 1000
A --dryRun yields:
Limit calculation will be performed for the following files: /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.8_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.8_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.8_scinti_fadc_septem_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.99_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.99_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.99_scinti_fadc_septem_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_septem_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5
which looks good!
[X]Check if we can extend
runLimitsto allow glob to select files -> AHH I think because we usewalkFilesthat might already be supported! The main code isfor file in walkFiles(path / prefix & "*.h5"): if file.extractFilename notin alreadyProcessed: echo file else: echo "Already processed: ", file
Let's test it quickly:
import os, strutils const path = "/home/basti/org/resources/*_xray_*" for file in walkFiles(path): echo file
Yup, works perfectly!
1.65.1. MLP limit output
The limits are done
shell> Expected limit: 6.969709361359805e-21 shell> 160362 shell> Generating group /ctx/axionModel shell> Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl shell> Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl shell> Serializing Interpolator by evaluating 0.0 to 12.0 of name: backgroundSpl shell> Generating group /ctx/backgroundDf shell> Wrote outfile /home/basti/org/resources/lhood_MLP_05_07_23/limits/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0374_σb_0.0028_posUncertain_puUncertain_σp_0.0500lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_septem_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 Computing single limit took 2208.284529924393 s Computing all limits took 27139.7917163372 s
1.66.
[X]Generate the expected limit table! -> UPDATED the path to the MLP files to indicate thatenergyMinwas set to 0.0!
./generateExpectedLimitsTable \ --path ~/org/resources/lhood_lnL_04_07_23/limits/ \ --prefix mc_limit_lkMCMC_skInterpBackground_nmc_1000 \ --path ~/org/resources/lhood_MLP_05_07_23_energyMin_0.0/limits/ \ --prefix mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty
[X]WHY is the limit a 7.64e-23 instead of 7.46e-23?? That's what we got when we ran that case manually, no? -> RERUN the case manually. Then rerun withrunLimitsand check! UPDATE: -> I found the reason. I forgot to update theenergyMinto0.2and left it at 0.0!!! -> Moved the limits to ./resources/lhood_MLP_05_07_23_energyMin_0.0 from its original directory to make clear what we used!
1.67.
[X]ImplementaxionModelstring field with filename toContextor whatever to have it in output H5 files[X]make used axion image a CL parameter for the limit and store the used file in the output!
Before we start rerunning the limits again with the correct minimum energy, let's implement the two TODOs.
Both implemented, time to run the limits again, this time with correct minimum energy.
./runLimits \ --path ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/ \ --prefix "lhood_c18_R2_crAll_*_scinti_fadc_" \ --outpath ~/org/resources/lhood_MLP_06_07_23/limits \ --energyMin 0.2 --energyMax 12.0 \ --axionModel /home/basti/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --nmc 1000
Code is running .
Back to the limit talk for now.
Limits finished :
shell> Expected limit: 6.952194554128882e-21 shell> 103176 shell> Generating group /ctx/axionModel shell> Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl shell> Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl shell> Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl shell> Generating group /ctx/backgroundDf shell> Wrote outfile /home/basti/org/resources/lhood_MLP_06_07_23/limits/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0374_σb_0.0028_posUncertain_puUncertain_σp_0.0500lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_septem_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 Computing single limit took 1537.849467039108 s Computing all limits took 17801.93424248695 s
Let's generate the expected result table again:
./generateExpectedLimitsTable \ --path ~/org/resources/lhood_lnL_04_07_23/limits/ \ --prefix mc_limit_lkMCMC_skInterpBackground_nmc_1000 \ --path ~/org/resources/lhood_MLP_06_07_23/limits/ \ --prefix mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty
1.67.1. Run the best case limit with more statistics
Let's run the best case expected limit with more statistics so that we can generate the plot of expected limits again with up to date data.
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --nmc 30000 \ --suffix=_sEff_0.95_scinti_fadc_line_mlp_tanh300_mse_epoch_485000_loss_0.0055_acc_0.9933_axion_image_1487.9_0.989AU \ --path "" \ --outpath /home/basti/org/resources/lhood_MLP_06_07_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel /home/basti/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv
1.68.
[X]Check the expected limits with 30k nmc! ->
Expected limit: 5.749270497358374e-21 85728 Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_MLP_06_07_23/limits/mc_limit_lkMCMC_skInterpBackground_nmc_30000_uncertainty_ukUncertain_σs_0.0328_σb_0.0028_posUncertain_puUncertain_σp_0.0500_sEff_0.95_scinti_fadc_line_mlp_tanh300_mse_epoch_485000_loss_0.0055_acc_0.9933_axion_image_1487.9_0.989AU.h5
So indeed we do lose a bit more using more statistics! Unfortunate, but it is what it is I guess.
[ ]Because of the "degradation" from 1000 to 30k toys in the 'best case' scenario, I should also rerun the next 2-3 options with more statistics to see if those expected limits might actually improve and thus give better results. As such let's rerun these other cases as well:
0.8474 MLP true true 0.98 false true 1 0.7841 0.8602 0.7325 0.7143 6.1381e-23 7.643e-23 0.9718 MLP true true 0.98 false true 1 0.7841 0.8602 0.7325 0.8192 5.8374e-23 7.6619e-23 0.9 LnL true true 0.98 false true 1 0.7841 0.8602 0.7325 0.7587 6.0434e-23 7.7375e-23 0.7926 MLP true true 0.98 false true 1 0.7841 0.8602 0.7325 0.6681 6.2843e-23 7.8575e-23 0.7398 MLP true true 0.98 false true 1 0.7841 0.8602 0.7325 0.6237 6.5704e-23 7.941e-23 i.e. the best case scenario for LnL and the other MLP cases without septem veto. I think for the MLP we can use
runLimitsto rerun all of them with more statistics. Given that the 30k for91% eff. took at least several hours (not sure how many exactly, forgot to ~time), maybe 15k? Keeping in mind that only the ~97% case should be slower. The following command:./runLimits \ --path ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/ \ --prefix "lhood_c18_R2_crAll_*_scinti_fadc_line_" \ --outpath ~/org/resources/lhood_MLP_06_07_23/limits \ --energyMin 0.2 --energyMax 12.0 \ --axionModel /home/basti/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --nmc 15000
matches all efficiencies (note the addition of the
_lineto the prefix!). So in order to exclude running the MLP@95% case again, we'll add it to theprocessed.txtfile in the output. Note that running the above to the same output file (using--dryRun) tells us (currently) correctly that it wouldn't do anything, because theprocessed.txtfile still contains all files.Limit calculation will be performed for the following files: Already processed: /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 Already processed: /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.8_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 Already processed: /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 Already processed: /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.99_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 Already processed: /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5
-> i.e. it wouldn't do anything. So we remove all the listed files with the exception of the
*_0.95_*file and rerun. which now yields on a--dryRun:Limit calculation will be performed for the following files: /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.8_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 Already processed: /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.99_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5
which looks correct! So let's run it (same command as above).
[ ]Let's create the plot for the expected limits of 30k toy samples. We'll use the old command as a reference (from
voidRipper.zsh_history):: 1659807499:0;./mcmc_limit_testing limit --plotFile ~/org/resources/mc_limit_lkMCMC_skInterpBackground_nmc_100000_uncertainty_ukUncertain_\317\203\243s_0.0469_\317\203\243b_0.0028_posUncertain_puUncertain_\317\203\243p_0.0500.csv --xLow 2.5e-21 --xHigh 1.5e-20 --limitKind lkMCMC --yHigh 3000 --bins 100 --linesTo 200\ 0 --xLabel "Limit [g_ae\302\262 @ g_a\316\263 = 1e-12 GeV\342\201\273\302\271]" --yLabel "MC toy count" --nmc 100000
to construct:
NOTE: I added the option
as_gae_gaγto plot the histogram in theg_ae · g_aγspace!mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --path "" \ --years 2017 --years 2018 \ --σ_p 0.05 \ --energyMin 0.2 --energyMax 12.0 \ --plotFile "mc_limit_lkMCMC_skInterpBackground_nmc_30000_uncertainty_ukUncertain_σs_0.0328_σb_0.0028_posUncertain_puUncertain_σp_0.0500_sEff_0.95_scinti_fadc_line_mlp_tanh300_mse_epoch_485000_loss_0.0055_acc_0.9933_axion_image_1487.9_0.989AU.csv" \ --xLow 2.5e-21 \ --xHigh 1.5e-20 \ --limitKind lkMCMC \ --yHigh 600 \ --bins 100 \ --linesTo 400 \ --as_gae_gaγ \ --xLabel "Limit g_ae·g_aγ [GeV⁻¹]" \ --yLabel "MC toy count" \ --outpath "/tmp/" \ --suffix "nmc_30k_pretty" \ --nmc 30000
The resulting plot
One striking feature is the fact that there is non-zero contribution
to the region in gae² below the "limit w/o signal, only RT" case!
As it turns out this is really just due to the variation on the MCMC
method on the limit calculation! Even the no candidates case varies by
quite a bit.
This can be verified by running multiple lkMCMC limit calculations
of the "no candidates" case. The variations are non negligible.
1.68.1. Estimating the variance of the median
[X]Add standard deviation to expected limits table ingenerateExpectedLimitTable!
That is the equivalent of our uncertainty on the expected limit.
-> Oh! It is not the equivalent of that. The issue with the
variance and standard deviation is that they are measures like the
mean, i.e. they take into account the absolute numbers of the
individual limits, which we don't care about.
Googling led me to:
https://en.wikipedia.org/wiki/Median_absolute_deviation
the 'Median Absolute Deviation', which is a measure for the
variability based on the median. However, to use it as a consistent
estimator of the median, we would need a scale factor \(k\)
\[
\hat{σ} = k · \text{MAD}
\]
which is distribution dependent. For generally well defined
distributions this can be looked up / computed, but our limits don't
follow a simple distribution.
Talking with BingChat then reminded me I could use bootstrapping for
this!
See [BROKEN LINK: sec:expected_limits:bootstrapping] in statusAndProgress for
our approach.
UPDATE: The numbers seem much smaller than the change from 1k to 30k implies.
The output after the change to the exp. limit table tool:
| ε | Type | Scinti | FADC | εFADC | Septem | Line | eccLineCut | εSeptem | εLine | εSeptemLine | Total eff. | Limit no signal [GeV⁻¹] | Expected limit [GeV⁻¹] | Exp. limit variance [GeV⁻¹] | Exp. limit σ [GeV⁻¹] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.9107 | MLP | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7677 | 5.9559e-23 | 7.4781e-23 | 1.6962e-49 | 4.1185e-25 |
| 0.8474 | MLP | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7143 | 6.1381e-23 | 7.643e-23 | 2.4612e-49 | 4.9611e-25 |
| 0.9718 | MLP | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.8192 | 5.8374e-23 | 7.6619e-23 | 2.1702e-49 | 4.6586e-25 |
| 0.9 | LnL | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7587 | 6.0434e-23 | 7.7375e-23 | 2.5765e-49 | 5.0759e-25 |
| 0.7926 | MLP | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6681 | 6.2843e-23 | 7.8575e-23 | 1.8431e-49 | 4.2932e-25 |
| 0.7398 | MLP | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6237 | 6.5704e-23 | 7.941e-23 | 1.5265e-49 | 3.907e-25 |
| 0.8 | LnL | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6744 | 6.3147e-23 | 8.0226e-23 | 4.4364e-49 | 6.6606e-25 |
| 0.9718 | MLP | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6976 | 6.2431e-23 | 8.0646e-23 | 2.0055e-49 | 4.4783e-25 |
| 0.9107 | MLP | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6538 | 6.432e-23 | 8.0878e-23 | 2.1584e-49 | 4.6459e-25 |
| 0.9718 | MLP | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7468 | 5.9835e-23 | 8.1654e-23 | 3.2514e-49 | 5.7021e-25 |
| 0.9107 | MLP | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6998 | 6.2605e-23 | 8.2216e-23 | 1.7728e-49 | 4.2104e-25 |
| 0.8474 | MLP | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6083 | 6.6739e-23 | 8.2488e-23 | 2.4405e-49 | 4.9401e-25 |
| 0.9 | LnL | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6461 | 6.4725e-23 | 8.3284e-23 | 1.5889e-49 | 3.9861e-25 |
| 0.8474 | MLP | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6511 | 6.4585e-23 | 8.338e-23 | 1.771e-49 | 4.2083e-25 |
| 0.7926 | MLP | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.569 | 6.8883e-23 | 8.3784e-23 | 1.7535e-49 | 4.1875e-25 |
| 0.7926 | MLP | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.609 | 6.6309e-23 | 8.4116e-23 | 2.132e-49 | 4.6174e-25 |
| 0.8 | LnL | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.5743 | 6.8431e-23 | 8.5315e-23 | 2.8029e-49 | 5.2942e-25 |
| 0.8 | LnL | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.5743 | 6.875e-23 | 8.5437e-23 | 2.4348e-49 | 4.9344e-25 |
| 0.7398 | MLP | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.5311 | 7.1279e-23 | 8.5511e-23 | 3.5032e-49 | 5.9188e-25 |
| 0.7398 | MLP | true | true | 0.98 | true | false | 1 | 0.7841 | 0.8602 | 0.7325 | 0.5685 | 6.9024e-23 | 8.6142e-23 | 2.9235e-49 | 5.4069e-25 |
| 0.7 | LnL | true | true | 0.98 | true | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.5025 | 7.2853e-23 | 8.9271e-23 | 2.8981e-49 | 5.3834e-25 |
Let's test locally using the CSV file of the 1k sample case:
import datamancer, stats, sequtils, seqmath, ggplotnim import random template withBootstrap(rnd: var Rand, samples: seq[float], num: int, body: untyped): untyped = let N = samples.len for i in 0 ..< num: # resample var newSamples {.inject.} = newSeq[float](N) for j in 0 ..< N: newSamples[j] = samples[rnd.rand(0 ..< N)] # get an index and take its value # compute our statistics body proc expLimitVarStd(limits: seq[float], plotname: string): (float, float) = var rnd = initRand(12312) let limits = limits.mapIt(sqrt(it) * 1e-12) # rescale limits const num = 1000 var medians = newSeqOfCap[float](num) withBootstrap(rnd, limits, num): medians.add median(newSamples, 50) if plotname.len > 0: ggplot(toDf(medians), aes("medians")) + geom_histogram() + ggsave(plotname) result = (variance(medians), standardDeviation(medians)) proc expLimit(limits: seq[float]): float = let limits = limits.mapIt(sqrt(it) * 1e-12) # rescale limits result = limits.median(50) proc slice30k(limits: seq[float]) = let N = limits.len let M = N div 1000 for i in 0 ..< M: let stop = min(limits.high, (i+1) * 1000) let start = i * 1000 echo "start ", start , " to ", stop echo "Exp limit for 1k at i ", i, " = ", limits[start ..< stop].expLimit() let df1k = readCsv("~/Sync/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0328_σb_0.0028_posUncertain_puUncertain_σp_0.0500lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.csv") let df30k = readCsv("~/Sync/mc_limit_lkMCMC_skInterpBackground_nmc_30000_uncertainty_ukUncertain_σs_0.0328_σb_0.0028_posUncertain_puUncertain_σp_0.0500_sEff_0.95_scinti_fadc_line_mlp_tanh300_mse_epoch_485000_loss_0.0055_acc_0.9933_axion_image_1487.9_0.989AU.csv") let limits30k = df30k["limits", float].toSeq1D echo "30k samples = ", expLimit(limits30k), " and std = ", expLimitVarStd(limits30k, "/tmp/medians_30k.pdf") let limits1k = df1k["limits", float].toSeq1D echo "1k samples = ", expLimit(limits1k), " and std = ", expLimitVarStd(limits1k, "/tmp/medians_1k.pdf") echo limits1k.median(50) + expLimitVarStd(limits1k, "")[1] slice30k(limits30k) let df = bind_rows([("1k", df1k), ("30k", df30k)], "Type") .filter(f{`limits` < 3e-20}) echo df["limits", float].percentile(50) ggplot(df, aes("limits", fill = "Type")) + geom_histogram(bins = 100, density = true, hdKind = hdOutline, alpha = 0.5, position = "identity") + ggsave("/tmp/histo_limits_compare_1k_30k.pdf") ggplot(df, aes("limits", fill = "Type")) + geom_density(alpha = 0.5, normalize = true) + ggsave("/tmp/kde_limits_compare_1k_30k.pdf")
30k samples = 7.582394407375754e-23 and std = (6.629501098125509e-51, 8.142174831164896e-26)
1k samples = 7.478101380514423e-23 and std = (1.613156185890611e-49, 4.016411564930331e-25)
5.592601671156511e-21
start 0 to 1000
Exp limit for 1k at i 0 = 7.605583686131433e-23
start 1000 to 2000
Exp limit for 1k at i 1 = 7.561063062729676e-23
start 2000 to 3000
Exp limit for 1k at i 2 = 7.564367446420724e-23
start 3000 to 4000
Exp limit for 1k at i 3 = 7.576524753068304e-23
start 4000 to 5000
Exp limit for 1k at i 4 = 7.571433298261475e-23
start 5000 to 6000 [0/90319]
Exp limit for 1k at i 5 = 7.627270326648243e-23
start 6000 to 7000
Exp limit for 1k at i 6 = 7.564981326101799e-23
start 7000 to 8000
Exp limit for 1k at i 7 = 7.585844150790594e-23
start 8000 to 9000
Exp limit for 1k at i 8 = 7.587466858370215e-23
start 9000 to 10000
Exp limit for 1k at i 9 = 7.596984336859885e-23
start 10000 to 11000
Exp limit for 1k at i 10 = 7.62764409158667e-23
start 11000 to 12000
Exp limit for 1k at i 11 = 7.560550411740659e-23
start 12000 to 13000
Exp limit for 1k at i 12 = 7.525828942453692e-23
start 13000 to 14000
Exp limit for 1k at i 13 = 7.549498042218461e-23
start 14000 to 15000
Exp limit for 1k at i 14 = 7.54624503868307e-23
start 15000 to 16000
Exp limit for 1k at i 15 = 7.545424145628356e-23
start 16000 to 17000
Exp limit for 1k at i 16 = 7.652870644411018e-23
start 17000 to 18000
Exp limit for 1k at i 17 = 7.562933564352857e-23
start 18000 to 19000
Exp limit for 1k at i 18 = 7.6577232551744e-23
start 19000 to 20000
Exp limit for 1k at i 19 = 7.614370346235356e-23
start 20000 to 21000
Exp limit for 1k at i 20 = 7.585288632863529e-23
start 21000 to 22000
Exp limit for 1k at i 21 = 7.520098295891504e-23
start 22000 to 23000
Exp limit for 1k at i 22 = 7.627966443034063e-23
start 23000 to 24000
Exp limit for 1k at i 23 = 7.622924220295962e-23
start 24000 to 25000
Exp limit for 1k at i 24 = 7.54129310424308e-23
start 25000 to 26000
Exp limit for 1k at i 25 = 7.566466143048985e-23
start 26000 to 27000
Exp limit for 1k at i 26 = 7.615198270864553e-23
start 27000 to 28000
Exp limit for 1k at i 27 = 7.582995326700842e-23
start 28000 to 29000
Exp limit for 1k at i 28 = 7.56983868270767e-23
start 29000 to 29999
Exp limit for 1k at i 29 = 7.630222722830585e-23
(Note: the conversion from gae² to gae gaγ is not related. The 1/100 kind of std to median remains as one would expect)
Let's also look at the 30k sample case.
But first run the generateExpectedLimitsTable for the 30k sample
case:
./generateExpectedLimitsTable --path ~/org/resources/lhood_MLP_06_07_23/limits/ --prefix mc_limit_lkMCMC_skInterpBackground_nmc_30000_
| ε | Type | Scinti | FADC | εFADC | Septem | Line | eccLineCut | εSeptem | εLine | εSeptemLine | Total eff. | Limit no signal [GeV⁻¹] | Expected limit [GeV⁻¹] | Exp. limit variance [GeV⁻¹] | Exp. limit σ [GeV⁻¹] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.9107 | MLP | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7677 | 5.9559e-23 | 7.5824e-23 | 6.0632e-51 | 7.7866e-26 |
Just as a cross check: the no signal expected limit is indeed the same as in the 1k case.
Hmmm, it's a bit weird that the 30k limit doesn't remotely reproduce the 1k limit even if we slice the limits into 1k pieces. I'm still not convinced that there is some difference going on.
[X]Rerun the mcmc limit calc for the 1k case with same as before and different RNG seed! (put current limit calcs into background and run in between then!)
-> Created directory ./resources/lhood_MLP_06_07_23/limits_1k_rng_seed Let's run:
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --nmc 1000 \ --suffix=_sEff_0.95_scinti_fadc_line_mlp_tanh300_axion_image_1487.9_0.989AU_default_seed \ --path "" \ --outpath /home/basti/org/resources/lhood_MLP_06_07_23/limits_1k_rng_seed \ --energyMin 0.2 --energyMax 12.0 \ --axionModel /home/basti/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv
Currently the RNG seed is just set via:
var nJobs = if jobs > 0: jobs else: countProcessors() - 2 if nmc < nJobs: nJobs = nmc var pp = initProcPool(limitsWorker, framesLenPfx, jobs = nJobs) var work = newSeq[ProcData]() for i in 0 ..< nJobs: work.add ProcData(id: i, nmc: max(1, nmc div nJobs))
Which is used in each worker as:
var p: ProcData while i.uRd(p): echo "Starting work for ", p, " at r = ", r, " and w = ", w var rnd = wrap(initMersenneTwister(p.id.uint32))
so we simply use the ID from 0 to nJobs. Depending on which job a process receives decides which RNG to use.
-> See the subsections below.
It really seems like the default RNG is just extremely "lucky" in this case.
Let's add these 2 new RNG cases to the script that bootstraps new medians and see what we get:
block P1000: let df = readCsv("~/Sync/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0328_σb_0.0028_posUncertain_puUncertain_σp_0.0500_sEff_0.95_scinti_fadc_line_mlp_tanh300_axion_image_1487.9_0.989AU_default_plus_1000.csv") let limits = df["limits", float].toSeq1D echo "Plus 1000 = ", expLimit(limits), " and std = ", expLimitVarStd(limits, "/tmp/medians_p1000.pdf") block P500: let df = readCsv("~/Sync/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0328_σb_0.0028_posUncertain_puUncertain_σp_0.0500_sEff_0.95_scinti_fadc_line_mlp_tanh300_axion_image_1487.9_0.989AU_default_plus_500.csv") let limits = df["limits", float].toSeq1D echo "Plus 500 = ", expLimit(limits), " and std = ", expLimitVarStd(limits, "/tmp/medians_p500.pdf")
which yields:
Plus 1000 = 7.584089303320828e-23 and std = (1.679933019568643e-49, 4.098698597809606e-25) Plus 500 = 7.545710186605163e-23 and std = (3.790787496584159e-49, 6.156937141618517e-25)
so both the values as well as the variation changes. But at least with these two cases the standard deviation includes the other.
So I guess the final verdict is that the numbers are mostly sensible, even if unexpected.
NOTE: I continue running the 15k sample cases now. I recompiled the limit calculation using the default RNG again!
- Result of run with default RNG
Finished around
Expected limit: 5.592200029700092e-21 85728 Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_MLP_06_07_23/limits_1k_rng_seed/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0328_σb_0.0028_posUncertain_puUncertain_σp_0.0500_sEff_0.95_scinti_fadc_line_mlp_tanh300_axion_image_1487.9_0.989AU_default_seed.h5
- Result of run with default RNG + 1000
Now we modify the code to use
i + 1000for eachProcDataid. Running nowThe suffix used is
default_plus_1000. Finished aroundExpected limit: 5.751841473157846e-21 85728 Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_MLP_06_07_23/limits_1k_rng_seed/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0328_σb_0.0028_posUncertain_puUncertain_σp_0.0500_sEff_0.95_scinti_fadc_line_mlp_tanh300_axion_image_1487.9_0.989AU_default_seed.h5
Wow, much worse!
- Result of run with default RNG + 500
Set to
i + 500now. With suffixdefault_plus_500Finished :Expected limit: 5.693774381122e-21 85728 Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_MLP_06_07_23/limits_1k_rng_seed/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0328_σb_0.0028_posUncertain_puUncertain_σp_0.0500_sEff_0.95_scinti_fadc_line_mlp_tanh300_axion_image_1487.9_0.989AU_default_plus_500.h5
1.69.
The limits with 15k samples finished:
shell> Expected limit: 5.88262164726686e-21 shell> 65760 shell> Generating group /ctx/axionModel shell> Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl shell> Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl shell> Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl shell> Generating group /ctx/backgroundDf shell> Wrote outfile /home/basti/org/resources/lhood_MLP_06_07_23/limits/mc_limit_lkMCMC_skInterpBackground_nmc_15000_uncertainty_ukUncertain_σs_0.0374_σb_0.0028_posUncertain_puUncertain_σp_0.0500lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 Computing single limit took 14433.18434882164 s Computing all limits took 72104.13822126389 s
Let's generate the expected limit table from these:
./generateExpectedLimitsTable \ --path ~/org/resources/lhood_MLP_06_07_23/limits/ \ --prefix mc_limit_lkMCMC_skInterpBackground_nmc_15000_
| ε | Type | Scinti | FADC | εFADC | Septem | Line | eccLineCut | εSeptem | εLine | εSeptemLine | Total eff. | Limit no signal [GeV⁻¹] | Expected limit [GeV⁻¹] | Exp. limit variance [GeV⁻¹] | Exp. limit σ [GeV⁻¹] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.9718 | MLP | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.8192 | 5.8374e-23 | 7.6252e-23 | 1.6405e-50 | 1.2808e-25 |
| 0.8474 | MLP | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.7143 | 6.1381e-23 | 7.6698e-23 | 1.4081e-50 | 1.1866e-25 |
| 0.7926 | MLP | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6681 | 6.2843e-23 | 7.8222e-23 | 1.3589e-50 | 1.1657e-25 |
| 0.7398 | MLP | true | true | 0.98 | false | true | 1 | 0.7841 | 0.8602 | 0.7325 | 0.6237 | 6.5704e-23 | 7.9913e-23 | 1.6073e-50 | 1.2678e-25 |
1.70.
[ ]For the thesis need to verify how the loss function should be defined. We'll just write a mini scrip that uses our single event
predictfunction for our MLP to then look at thelosscall for that single event.Also as a file in ./Misc/inspect_mse_loss_cast.nim. Needs to be compiled with:
nim cpp -r -d:cuda inspect_mse_loss_cast.nim
and the
mlp_impl.hppfile needs to be present in the same directory!import /home/basti/CastData/ExternCode/TimepixAnalysis/Tools/NN_playground / [nn_predict, io_helpers] import flambeau / [flambeau_raw, flambeau_nn] import nimhdf5, unchained, seqmath, stats import random from xrayAttenuation import FluorescenceLine from ingridDatabase/databaseRead import initCalibInfo import ingrid / [tos_helpers, ingrid_types, gas_physics, fake_event_generator] proc getEvents(nFake: int, calibInfo: CalibInfo, gains = @[3000.0, 4000.0], diffusion = @[550.0, 650.0]): DataFrame = var fakeDesc = FakeDesc(kind: fkGainDiffusion, gasMixture: initCASTGasMixture()) var fakeEvs = newSeqOfCap[FakeEvent](nFake) var rnd = initRand(12312) var count = 0 while count < nFake: if count mod 5000 == 0: echo "Generated ", count, " events." # 1. sample an energy let energy = rnd.rand(0.1 .. 10.0).keV let lines = @[FluorescenceLine(name: "Fake", energy: energy, intensity: 1.0)] # 2. sample a gas gain let G = rnd.gauss(mu = (gains[1] + gains[0]) / 2.0, sigma = (gains[1] - gains[0]) / 4.0) let gain = GainInfo(N: 100_000.0, G: G, theta: rnd.rand(0.4 .. 2.4)) # 3. sample a diffusion let σT = rnd.gauss(mu = 660.0, sigma = (diffusion[1] - diffusion[0] / 4.0)) fakeDesc.σT = σT let fakeEv = rnd.generateAndReconstruct(fakeDesc, lines, gain, calibInfo, energy) if not fakeEv.valid: continue fakeEvs.add fakeEv inc count result = fakeToDf( fakeEvs ) const path = "/home/basti/CastData/data/DataRuns2018_Reco.h5" const mlpPath = "/home/basti/org/resources/nn_devel_mixing/10_05_23_sgd_gauss_diffusion_tanh300_mse_loss/mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.pt" proc main = let h5f = H5open(path, "r") let calibInfo = h5f.initCalibInfo() var df = newDataFrame() for num, run in runs(h5f): echo num # read a random event df = getEvents(10, calibInfo) echo df break # initiate the MLP loadModelMakeDevice(mlpPath) df["Type"] = $dtSignal template checkIt(df: DataFrame): float = let (inp, target) = toInputTensor(df) let res = model.forward(desc, inp.to(device)) echo "Outpt: ", res let loss = mse_loss(res, target.to(device)) echo "MSE = ", loss loss.item(float) var losses = newSeq[float]() for row in rows(df): echo "===============\n" losses.add checkIt(row) discard checkIt(df) echo losses.mean main()
=============Outpt: RawTensor 1.0000e+00 4.5948e-23 [ CUDAFloatType{1,2} ] MSE = RawTensor 1.4013e-45 [ CUDAFloatType{} ] Outpt: RawTensor 1.0000e+00 2.3029e-18 1.0000e+00 5.6112e-12 1.0000e+00 1.1757e-20 1.0000e+00 9.4252e-20 1.0000e+00 3.0507e-20 1.0000e+00 3.1307e-12 1.0000e+00 3.8549e-23 9.9358e-01 6.6054e-03 1.0000e+00 9.3695e-18 1.0000e+00 4.5948e-23 [ CUDAFloatType{10,2} ] MSE = RawTensor 4.24536e-06 [ CUDAFloatType{} ] 4.245364834787324e-06The above is the (manually copied) output of the last row & the and the batch loss + the manually computed batch loss (
losses.mean).So as expected this means that the MSE loss computes:
\[ l(\mathbf{y}, \mathbf{\hat{y}}) = \frac{1}{N} \sum_{i = 1}^N \left( y_i - \hat{y}_i \right)² \]
where \(\mathbf{y}\) is a vector \(∈ \mathbb{R}^N}\) of the network outputs and \(\mathbf{\hat{y}}\) the target outputs. The sum runs over all \(N\) output neurons.
1.71. TODO [0/1]
IMPORTANT
1.71.1. LLNL telescope effective area [/]
I noticed today while talking with Cris about her limit calculation that
- our limit code does not actually use the
*_parallel_light.csvfile for the LLNL telescope efficiency! - uses the
_extended.csvversion - the
_extended.csvversion is outdated, because the real efficiency actually INCREASES AGAIN below 1 keV - the
_extendedand the_parallel_lightversions both describe different settings: The_extendedversion comes from the CAST paper about the LLNL telescope and describes the effective area for solar axion emission in a 3 arcmin radius from the solar core, i.e. NOT parallel light!! The_parallel_lightof course describes parallel light. - because of 4, the effective area of the parallel version is
significantly higher than the
_extendedversion!
⇒ What this means is we need to update our telescope efficiency for the limit! The question that remains is what is the "correct" telescope efficiency? It makes sense that the efficiency is lower for non parallel light of course. But our axion emission looks different than the assumption done for the CAST paper about the telescope!
Therefore, the best thing to do would be to use the raytracer to compute the effective area! This should actually be pretty simple! Just generate axions according to the real solar emission but different energies. I.e. for each energy in 0, 10 keV send a number N axions through the telescope. At the end just compute the average efficiency of the arriving photons (incl taking into account those that are completely lost!
This should give us a correct description for the effective area. We need to make sure of course not to include any aspects like window, conversion probability, gas etc. Only telescope reflectivity!
To compute this we need:
[ ]correct reflectivity files for the telescope -> Need to add the other 3 recipes toxrayAttenuationand compute it! -> See next section[ ]add the ability to scan the effective area to theraytracer. -> This can be done equivalent to theangularScanthat we already have there. Just need an additional energy overwrite. -> the latter can be done by having some overwrite to thegetRandomEnergyFromSolarModelfunction. Maybe as an argument to thetraceAxionprocedure or similar. Or as a field toExperimentSetupthat isOption? Or alternatively merge it into thetestXraySourcebranch. Such that the X-ray source object has an overwrite for the position such that it can sample from solar model.
1.71.2. Regenerate the LLNL reflectivities using DarpanX
[X]We're currently rerunning the DarpanX based script to get the correct reflectivites for the telescope by using Ångström as inputs instead of nano meters! :DONE:
Just by running:
./llnl_layer_reflectivity
on the HEAD of the PR https://github.com/jovoy/AxionElectronLimit/pull/22.
1.71.3. Computing the LLNL telescope reflectivities with xrayAttenuation
[ ]Implement the depth graded layer to be computed automatically according to the equation in the DarpanX paper (and in the old paper of the old IDL program?)
A depth-graded multilayer is described by the equation: \[ d_i = \frac{a}{(b + i)^c} \] where \(d_i\) is the depth of layer \(i\) (out of \(N\) layers), \[ a = d_{\text{min}} (b + N)^c \] and \[ b = \frac{1 - N k}{k - 1} \] with \[ k = \left(\frac{d_{\text{min}}}{d_{\text{max}}}\right)^{\frac{1}{c}} \] where \(d_{\text{min}}\) and \(d_{\text{max}}\) are the thickness of the bottom and top most layers, respectively.
1.71.4. Computing the effective area
First attempt using the LLNL reflectivities from DarpanX after updating them to correct thicknesses & C/Pt instead Pt/C.
./raytracer \ --distanceSunEarth 0.9891144450781392.AU \ --effectiveAreaScanMin 0.03 \ --effectiveAreaScanMax 12.0 \ --numEffectiveAreaScanPoints 100 \ --xrayTest \ --suffix "_llnl"
with the config.toml file containing
[TestXraySource] useConfig = false # sets whether to read these values here. Can be overriden here or useng flag `--testXray` active = true # whether the source is active (i.e. Sun or source?) sourceKind = "sun" # whether a "classical" source or the "sun" (Sun only for position *not* for energy) parallel = true
and of course the LLNL telescope as the telescope to use (plus CAST etc):
[Setup] # settings related to the setup we raytrace through experimentSetup = "CAST" # [BabyIAXO, CAST] detectorSetup = "InGrid2018" # [InGrid2017, InGrid2018, InGridIAXO] telescopeSetup = "LLNL" stageSetup = "vacuum" # [vacuum, gas]
The resulting plot is
which when compared even with the DTU thesis plot:
import ggplotnim, math, strformat, sequtils let dfParallel = readCsv("/home/basti/org/resources/llnl_xray_telescope_cast_effective_area_parallel_light_DTU_thesis.csv") let dfCast = readCsv("/home/basti/org/resources/llnl_xray_telescope_cast_effective_area.csv") let dfJaimeNature = readCsv("/home/basti/org/resources/llnl_cast_nature_jaime_data/2016_DEC_Final_CAST_XRT/EffectiveArea.txt", sep = ' ') .rename(f{"Energy[keV]" <- "E(keV)"}, f{"EffectiveArea[cm²]" <- "Area(cm^2)"}) .select("Energy[keV]", "EffectiveArea[cm²]") echo dfJaimeNature const areaBore = 2.15*2.15 * PI proc readDf(path: string): DataFrame = result = readCsv(path) if "Energy[keV]" notin result: result = result.rename(f{"Energy[keV]" <- "Energy [keV]"}, f{"Transmission" <- "relative flux"}) result = result.mutate(f{"EffectiveArea[cm²]" ~ `Transmission` * areaBore}) proc makePlot(paths, names: seq[string], suffix: string) = var dfs: seq[(string, DataFrame)] for (p, n) in zip(paths, names): let dfM = readDf(p) dfs.add (n, dfM) let df = bind_rows(concat(@[("Thesis", dfParallel), ("CASTPaper", dfCast), ("Nature", dfJaimeNature)], dfs), "Type") ggplot(df, aes("Energy[keV]", "EffectiveArea[cm²]", color = "Type")) + geom_line() + ggtitle("Effective area LLNL comparing parallel light (thesis) and axion emission (paper)") + scale_y_continuous(secAxis = sec_axis(trans = f{1.0 / areaBore}, name = "Transmission")) + margin(top = 1.5, right = 6) + legendPosition(0.8, 0.0) + ggsave(&"~/org/Figs/statusAndProgress/effectiveAreas/llnl_effective_area_comparison_parallel_axion{suffix}.pdf") proc makePlot(path, name, suffix: string) = makePlot(@[path], @[name], suffix) makePlot("/home/basti/org/resources/effectiveAreas/llnl_effective_area_manual_attempt1.csv", "Attempt1", "_attempt1") makePlot("/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_classical_3arcmin.csv", "3Arcmin", "_3arcmin") makePlot("/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_sun_reflect_squared.csv", "Rsquared", "_reflect_squared") makePlot("/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_classical_parallel_fullbore.csv", "Parallel", "_parallel") makePlot("/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_classical_parallel_fullbore_sigma_0.45.csv", "Parallel_σ0.45", "_parallel_sigma_0.45") makePlot("/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_sigma_0.45.csv", "Sun_σ0.45", "_sun_sigma_0.45") makePlot("/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_sun_correct_shells_xrayAttenuation_fixed.csv", "xrayAtten", "_sun_xray_attenuation") makePlot(@["/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_parallel_correct_shells.csv", "/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_sun_correct_shells.csv"], @["Parallel", "Sun"], "_sun_and_parallel_correct_shells_sigma_0.45") makePlot(@["/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_sun_correct_shells_xrayAttenuation_fixed.csv", "/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_3arcmin_xrayAttenuation.csv", "/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_parallel_xrayAttenuation.csv"], @["XASun", "XA3arcmin", "XAParallel"], "_sun_and_3arcmin_and_parallel_xrayAttenuation")
Hmm!
Where do we go wrong?
Let's try with a 3 arcmin source as mentioned in the caption of the CAST paper about the LLNL telescope. We need to get a 3 arc min source described by
distance = 2000.0 # mm Distance of the X-ray source from the readout radius = 21.5 # mm Radius of the X-ray source
in the config file:
import unchained, math const size = 3.arcmin / 2.0 # (radius not diameter!) const dist = 9.26.m echo "Required radius = ", (tan(size.to(Radian)) * dist).to(mm)
The 3 arc minute case does indeed lower the reflectivity compared to our solar axion emission case. However, it is still larger than what we would expect (about the height of the DTU PhD thesis parallel)
See the figure
Again pretty bizarre that this version is relatively close to the
PhD thesis one, when that one uses parallel light.
The DTU PhD thesis mentions that the effective area uses the reflectivity squared
optic multiplied by reflectivity squared for each layer
so I tried to change the reflectivity in the raytracer to be squared (which seems ridiculous, because what I assume is meant is that he's referring to the reflectivity of the Fresnell equations, which need to be squared to get the physical reflectivity).
This yields
The really curious thing about this is though that the behavior is now almost perfect within that dip at about 2 keV compared to the CAST LLNL paper line!
But still, I think this is the wrong approach. I tried it as well using fully parallel light using the squared reflectivity and it is comparable as expected. So in particular at high energies he suppression due to squaring is just too strong.
Ahh, in sec. 1.1.1 of the thesis he states:
However, the process becomes a little more complicated considering that the reflectivity is dependent on incident angle on the reflecting surface. In an X-ray telescope consisting of concentric mirror shells, each mirror shell will reflect incoming photons at a different angle that each result in a certain reflectivity spectrum. Also to consider is the fact that Wolter I telescopes requires a double reflection, so the reflectivity spectrum should be squared.
so what he means is really the reflectivity, but not in terms of what I assumed above, but rather due to the fact that the telescope consists of 2 separate sets of mirrors!
This is of course handled in our raytracing, due to the simulated double reflection from each layer.
Maybe the reason is surface roughness after all?
The SigmaValue in DarpanX gives the surface roughness in
Ångström. We currently use 1 Å as the value, which is 0.1 nm. The PhD
thesis states that a surface roughness of 0.45 nm (page 89):
Both SPO substrates and NuSTAR glass substrates have a surface roughness of σrms ≈ 0.45 nm
Let's recompute the reflectivities using 4.5 Å!
See the generated plots:
So the results effectively don't seem to change.
But first let's rescale the parallel light case with σ = 0.45 Å to the PhD thesis data and then see if they at least follow the same curves:
proc makeRescaledPlot(path, name, suffix: string) = let dfPMax = dfParallel.filter(f{idx("Energy[keV]") > 1.0 and idx("Energy[keV]") < 2.0})["EffectiveArea[cm²]", float].max var dfManual = readDf(path) let dfMMax = dfManual.filter(f{idx("Energy[keV]") > 1.0 and idx("Energy[keV]") < 2.0})["EffectiveArea[cm²]", float].max dfManual = dfManual .mutate(f{"EffectiveArea[cm²]" ~ idx("EffectiveArea[cm²]") / dfMMax * dfPMax}) let df = bind_rows([("Thesis", dfParallel), ("CASTPaper", dfCast), ("Nature", dfJaimeNature), (name, dfManual)], "Type") ggplot(df, aes("Energy[keV]", "EffectiveArea[cm²]", color = "Type")) + geom_line() + ggtitle(&"Effective area LLNL comparing parallel light (thesis) and axion emission (paper), {name} rescaled to parallel") + scale_y_continuous(secAxis = sec_axis(trans = f{1.0 / areaBore}, name = "Transmission")) + margin(top = 1.5, right = 6) + legendPosition(0.8, 0.0) + ggsave(&"~/org/Figs/statusAndProgress/effectiveAreas/llnl_effective_area_comparison_parallel_axion_rescaled_manual{suffix}.pdf") makeRescaledPlot("/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_classical_parallel_fullbore_sigma_0.45.csv", "Parallel_σ0.45", "_parallel_sigma_0.45") ## Version from mean of all layers! makeRescaledPlot("/home/basti/org/resources/effectiveAreas/effective_area_mean_reflectivity_squared_all_layers.csv", "MeanLR²", "_mean_layers_reflectivity_squared") makeRescaledPlot("/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_parallel_correct_shells.csv", "CorrectShells", "_parallel_correct_shells") makeRescaledPlot("/home/basti/org/resources/effectiveAreas/effective_area_mean_reflectivity_squared_all_layers_weighted.csv", "LR²Weight", "_mean_layers_weighted") makeRescaledPlot("/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_sun_correct_shells_xrayAttenuation_fixed.csv", "xrayAtten", "_sun_xray_attenuation") makeRescaledPlot("/home/basti/org/resources/effectiveAreas/effective_area_scan_telescope_llnl_3arcmin_xrayAttenuation.csv", "xrayAtten3Arc", "_3_arcmin_xray_attenuation")
As we can see it doesn't even match the shape properly
Matches after rescaling at low energies, but then diverges at higher
energies.
[X]Maybe we could look at a single plot for a single reflectivity to see if it even makes a difference in DarpanX. -> Done. Look at ./../CastData/ExternCode/AxionElectronLimit/tools/understand_darpanx.py For ranges up to 4.5 Å, there is only very marginal changes. But at larger values it does change things, so the code does something.[X]Check with C/Pt coating instead of Pt/C (so Pt at top layer!) -> Generated a file ./../CastData/ExternCode/AxionElectronLimit/resources/llnl_layer_reflctivities_c_pt.h5 and ran the raytracer with that as an input. Result iswhich clearly has a very wrong shape.
So at this point I'm pretty clueless. Given that rescaling doesn't reproduce the correct results either, it seems to me that it cannot just be a problem of too many X-rays traverse (so not being completely absorbed). The fact that the shape is different implies that the reflectivities are wrong. Maybe it's only the angular aspect? Who knows.
However, given that the PhD thesis talks about squaring the reflectivity to me implies that he really assumes perfect incoming angles, i.e. such that the first and second mirrors layers are hit under exactly the same angle. If we wanted to compute it like that though, we'd need to manually compute the weighting of each layer, which is annoying. What we could do though is to compute just the mean of all layers and see how that ends up looking. Using the angles of each mirror shell as the incoming angle. Let's quickly try that
import numericalnim/interpolate import nimhdf5, ggplotnim, unchained, sequtils, algorithm import strutils, strformat type Reflectivity = object layers: seq[int] reflectivities: seq[Interpolator2DType[float]] defUnit(mm²) const areaBore = π * 2.15.cm * 2.15.cm const allAngles = @[0.579, 0.603, 0.628, 0.654, 0.680, 0.708, 0.737, 0.767, 0.798, 0.830, 0.863, 0.898, 0.933, 0.970].mapIt(it.Degree) # Opening areas of each mirror shell in `mm²` const allAreas = @[13.863, 48.175, 69.270, 86.760, 102.266, 116.172, 128.419, 138.664, 146.281, 150.267, 149.002, 139.621, 115.793, 47.648].mapIt(it.mm²) proc initRefl(path: string): Reflectivity = result = Reflectivity( #layers: @[2, 2+3, 2+3+4, 2+3+4+5] # layers of LLNL telescope layers: @[3 - 1, 3+4 - 1, 3+4+4 - 1, 3+4+4+3 - 1] # layers of LLNL telescope ) # read reflectivities from H5 file let numCoatings = result.layers.len var h5f = H5open(path, "r") let energies = h5f["/Energy", float] let angles = h5f["/Angles", float] var reflectivities = newSeq[Interpolator2DType[float]]() for i in 0 ..< numCoatings: let reflDset = h5f[("Reflectivity" & $i).dset_str] let data = reflDset[float].toTensor.reshape(reflDset.shape) reflectivities.add newBilinearSpline( data, (angles.min, angles.max), (energies.min, energies.max) ) discard h5f.close() result.reflectivities = reflectivities proc eval(refl: Reflectivity, α: Degree, E: keV, idx: int): float = # use the hit layer to know which interpolator we have to use let layerIdx = refl.layers.lowerBound(idx) echo "Hit layer: ", idx, " yields index: ", layerIdx let reflLayer = refl.reflectivities[layerIdx] let refl = reflLayer.eval(α.float, E.float) result = refl * refl const path = "/home/basti/CastData/ExternCode/AxionElectronLimit/resources/llnl_layer_reflectivities_xrayAttenuation.h5" let refl = initRefl(path) let energies = linspace(0.03, 12.0, 1000) block AllLayerMean: var effs = newSeq[float]() for E in energies: var eff = 0.0 for i, angle in allAngles: eff += refl.eval(angle, E.keV, i) effs.add (eff / allAngles.len.float) let df = toDf({"Energy[keV]" : energies, "Transmission" : effs}) df.writeCsv("/home/basti/org/resources/effectiveAreas/effective_area_mean_reflectivity_squared_all_layers.csv") ggplot(df, aes("Energy[keV]", "Transmission")) + geom_line() + ggsave("/home/basti/org/Figs/statusAndProgress/effectiveAreas/effective_area_mean_reflectivity_squared_all_layers.pdf") block AllLayerWeighted: var effs = newSeq[float]() for E in energies: var eff = 0.0 for i, angle in allAngles: let area = allAreas[i] / areaBore ## If we leave out the normlization by areaBore we compute the _actual_ effective area! eff += area.float * refl.eval(angle, E.keV, i) effs.add eff let df = toDf({"Energy[keV]" : energies, "Transmission" : effs}) df.writeCsv("/home/basti/org/resources/effectiveAreas/effective_area_mean_reflectivity_squared_all_layers_weighted.csv") ggplot(df, aes("Energy[keV]", "Transmission")) + geom_line() + ggtitle("Effective area from each mirror shell weighted by opening area") + ggsave("/home/basti/org/Figs/statusAndProgress/effectiveAreas/effective_area_mean_reflectivity_squared_all_layers_weighted.pdf") block LayerTypeIndv: var last = 0 var dfC = newDataFrame() for lIdx in 0 ..< refl.layers.len: let start = last let stop = refl.layers[lIdx] echo "start to ", start, " to ", stop var effs = newSeq[float]() for E in energies: var eff = 0.0 for j in start .. stop: eff += refl.eval(allAngles[j], E.keV, j) echo "dividing by : ", stop - start effs.add (eff / (stop - start).float) last = stop let df = toDf({"Energy[keV]" : energies, "Transmission" : effs, "Idx" : lIdx }) dfC.add df echo dfC dfC.writeCsv("/home/basti/org/resources/effectiveAreas/effective_area_mean_reflectivity_squared_layer_by_type.csv") ggplot(dfC, aes("Energy[keV]", "Transmission", color = "Idx")) + geom_line() + ggsave(&"/home/basti/org/Figs/statusAndProgress/effectiveAreas/effective_area_mean_reflectivity_squared_layer_by_type.pdf")
yielding
and
./resources/effectiveAreas/effective_area_mean_reflectivity_squared_all_layers.csv
Let's add that to the rescaled plot above!
Looking at
we see that the shape still does not match.
This means
- either our mixture of the the amount of layers is wrong
- or our reflectivities really are wrong
I think we can check which it might be by generating only the reflectivities of the different layer types!
UPDATE: I just realized that table 4.1 in the PhD thesis actually gives us the required areas that we need!
[X]Compute full effective areas using areas of each layer given in table -> Implemented in above. Not
UPDATE: I also now realized that the "number of layers" in table of fig. 4.11 (the recipes) is the number of layers of the material in the depth graded layering. NOT which layer and how many of those of the mirror exist!!
[X]make sure we use the correct number of layers (of each type)! Check if and how that affects the raytracing! -> Fig. 4.10 mentions the assignment clearly: Layer 0-3 (0 = Mandrel), recipe with N = 2 and dmin = 11.5nm Layer 4-7, recipe with N = 3 and dmin = 70 nm Layer 8-11, recipe with N = 4 and dmin = 55 nm Layer 12-14, recipe with N = 5 and dmin = 50 nm So these are from inner most to outer most (smallest radii to largest radii) -> In the current raytracing code therefore the number of layers is WRONG. -> Updated the numbers.[X]Check the ordering of the hit layers in the raytracing code as well as the order and assignment of layers. -> That means we need to check if the index of the layers from 0 starts from the shells with the smallest radii. -> Code works by walking over all R1 radii. These are ordered from inner most / smallest radii. So that seems to be correct. The hit layer is just the index \(j\) we iterate over for the radii of the closest approach of the incoming vector to the radii.[ ]On a related note I should check with what percentage each layer of the telescope is actually hit! -> We expect a not uniform distribution, but one that scales with the ratios of the areas of the table in the PhD thesis of the opening areas. -> Just insert a global? Or make the hit layer a part of the axion output? -> latter seems a good idea! -> IT ALREADY IS a property of theAxiontype! Namely theshellNumbervariable.[X]Rerun the raytracing code with the number of each layer updated. -> Also include the plots / a plot for the shell number?
./raytracer --distanceSunEarth 0.9891144450781392.AU --effectiveAreaScanMin 0.03 --effectiveAreaScanMax 12.0 --numEffectiveAreaScanPoints 100 --xrayTest --suffix "_llnl_parallel_correct_shells"
The following bar chart is the counts each shell of the telescope was
hit
we can compare this with the following table (from [BROKEN LINK: sec:llnl_telescope]
in
statusAndProgress):
| Layer | Area [mm²] | Relative area [%] | Cumulative area [mm²] | α [°] | α [mrad] | R1 [mm] | R5 [mm] |
|---|---|---|---|---|---|---|---|
| 1 | 13.863 | 0.9546 | 13.863 | 0.579 | 10.113 | 63.006 | 53.821 |
| 2 | 48.175 | 3.3173 | 62.038 | 0.603 | 10.530 | 65.606 | 56.043 |
| 3 | 69.270 | 4.7700 | 131.308 | 0.628 | 10.962 | 68.305 | 58.348 |
| 4 | 86.760 | 5.9743 | 218.068 | 0.654 | 11.411 | 71.105 | 60.741 |
| 5 | 102.266 | 7.0421 | 320.334 | 0.680 | 11.877 | 74.011 | 63.223 |
| 6 | 116.172 | 7.9997 | 436.506 | 0.708 | 12.360 | 77.027 | 65.800 |
| 7 | 128.419 | 8.8430 | 564.925 | 0.737 | 12.861 | 80.157 | 68.474 |
| 8 | 138.664 | 9.5485 | 703.589 | 0.767 | 13.382 | 83.405 | 71.249 |
| 9 | 146.281 | 10.073 | 849.87 | 0.798 | 13.921 | 86.775 | 74.129 |
| 10 | 150.267 | 10.347 | 1000.137 | 0.830 | 14.481 | 90.272 | 77.117 |
| 11 | 149.002 | 10.260 | 1149.139 | 0.863 | 15.062 | 93.902 | 80.218 |
| 12 | 139.621 | 9.6144 | 1288.76 | 0.898 | 15.665 | 97.668 | 83.436 |
| 13 | 115.793 | 7.973 | 1404.553 | 0.933 | 16.290 | 101.576 | 86.776 |
| 14 | 47.648 | 3.2810 | 1452.201 | 0.970 | 16.938 | 105.632 | 90.241 |
where we see that the ratios seem to match pretty well to the relative areas! Also it's pretty useful to see that those areas really sum up to the exact value for the bore area of π · 2.15² cm² = 14.52201 cm² we would expect!
Back to running the code…
The resulting plot individually:
and combined and rescaled with the PhD thesis plot:
./Figs/statusAndProgress/effectiveAreas/llnl_effective_area_comparison_parallel_axion_rescaled_manual_mean_layers_correct_shells.pdf
-> As we can see the shape is still off, despite apparently getting the behavior right.
Hmm. So what now? Just for reference, let's run the raytracing again with the Sun as an emission source.
1.72.
I asked Cristina for the limit code of Jaime for the gae² limit in 2013.
The code lives here: ./Misc/CAST_gae_code_2013_jaime/
and I used Google Bard to convert the Efficiency calculation in that code
to Nim in order to plot it (because it uses some pow calls to define
the efficiency in different energy ranges). That seems like it might
be the origin of the telescope efficiency in the code.
Plotted in Nim the efficiency is the following:
which seems to be (at least) the detector efficiency, given that it
goes down towards high energies (Argon absorption?) and seems to show
the Argon absorption edge near 3 keV. So this is the efficiency of the
TPC then?
The same for the CAST_new.cxx file is
Figs/statusAndProgress/efficiency_jaime_gae_code_ccd.pdf
Weirdly it shows similar behavior.
Well, the tail down to higher energies could also be the telescope efficiency!
Writing a mail to Jaime and Julia right now titled "Another LLNL telescope question".
Let's quickly make / attach two plots:
[X]Plot of thesis + paper + parallel + 3 arcmin[X]Plot of thesis + paper + parallel rescaled
The mail:
Hey,
the other day I noticed something else about the LLNL telescope that confuses me a bit. I've been using the effective area extracted from the 2015 JCAP paper (fig. 4). It tops out at about 8 cm² at ~1.5 keV.
In the PhD thesis of Anders Jakobsen he has an effective area plot (fig. 4.13), which extends down to 0 keV and peaks at near 10 cm² at the same ~1.5 keV.
According to the descriptions this might make sense, because in the thesis the effective area is (I assume) for fully parallel light (even though I cannot find it explicitly stated; given the context it seems to be computed from the reflectivity squared for each layer). In the JCAP paper the plot caption explicitly states that it is the effective area for:
``` The EA was modelled using the as-built optical prescription, assuming a half-power diameter of 75 arcsec and that the solar axion emission comes from a uniformly distributed 3 arcmin disc. ```
so a more realistic approximation. (Note: what is meant by "half-power diameter of 75 arcsec" ?)
Now my issues are the following:
- at least the plot from the thesis should be reproducible from the reflectivites I can compute based on the coating recipes and the opening areas for each mirror shell or via raytracing. However, if I do this the efficiency is too high.
- even if I rescale the absolute magnitude of my raytraced result to give the same maximum throughput as the plot from the thesis the curves don't exactly line up. I still get too much transmission at higher energies (see attached fig. llnleffectiveareacomparisonparallelaxionrescaledmanualparallelcorrectshells.pdf
- the effect of using a realistic axion emission model from the Sun compared to using parallel light is not as massive as the two different plots would indicate. According to my raytracing results that comes out to a few percent difference. (see attached fig.
llnleffectiveareacomparisonparallelaxionsunandparallelcorrectshellssigma0.45.pdf
(shows both the efficiencies being too high and parallel and Sun emission difference being smaller than the two plots make it seem)
So, I'm mainly writing to ask if one of you knows more details about where either of the two plots come from and how they are computed.
The reason is that given that my limit method should be independent of the used emission model I need to be able to reliably reproduce the efficiency based on the used axion emission. In particular if one were to consider looking at the Chameleon coupling again in the future the emission is vastly different. Using the same effective area as either of the two plots is obviously flawed.
Thanks for any help in advance! Cristina is in CC, because this will also be relevant for her limit calculation.
Cheers, Sebastian
[X]The plots that come out of the effective area scan, especially for the axion image of different energy ranges are actually really interesting. It might after all be worthwhile to include not a single axion image, but rather use multiple!
[ ]Think about computing multiple axion images for our real setup at different energies!
1.73.
And we still a have the problem that our higher energy behavior seems different than DarpanX's.
UPDATE: When constructing the H5 file of the reflectivities yesterday, we had a bug. We used only an energy range from 0 to 10 keV, but mapped it to 15 keV. Therefore everything was stretched!
UPDATE 2:
Also our usage of the layers via lowerBound was flawed. Both in the
"analytical" code above as well as in the raytracer. The layers need
to be given as N - 1 for each recipe to match the correct recipes
from the hit layers. This is fixed now.
Note that bizarrely the heatmap plots comparing the xrayAttenuation reflectivities pretty much look identical to the DarpanX version. Maybe the color is just screwing me?
1.73.1. Quick notes
- we were comparing the XRR calculation with the wrong plot of the thesis. Not fig. 4.20! But use 4.21! 4.20 is ONLY experimental data. 4.20 shows the IMD fit. And THAT one we actually perfectly reproduce.
- reproducing the XRR values seems to imply my calculation has to be mostly correct!
1.73.2. XRR - X-ray Reflectometry
The XRR (X-ray reflectrometry) measurements mentioned in the PhD thesis of Jakobsen DTU about the LLNL use an energy of 8.047 keV! Page 14 in the thesis:
After the source, along the z-axis, is placed two slits, a monochromator, an attenuator, on more slit, the sample holder and finally the detector, see figure 2.5. The first two slits ensures that only a narrow beam hits the monochromator, which then reflects only photons around the copper Kα1 emission line (8.047 keV) by reflecting the beam on two germanium crystals at an angle where Bragg reflection only allow photons of that energy.
Reproducing the XRR measurements from page 70 works almost perfectly. But the mini peak at around 1.5 keV is not being reproduced. However, neither in our code nor using DarpanX.
UPDATE:
We figured out the actual issue.
The problem was not the code for once, but just the person sitting
in front reading a thesis.
The figure we were looking at was the wrong fig.
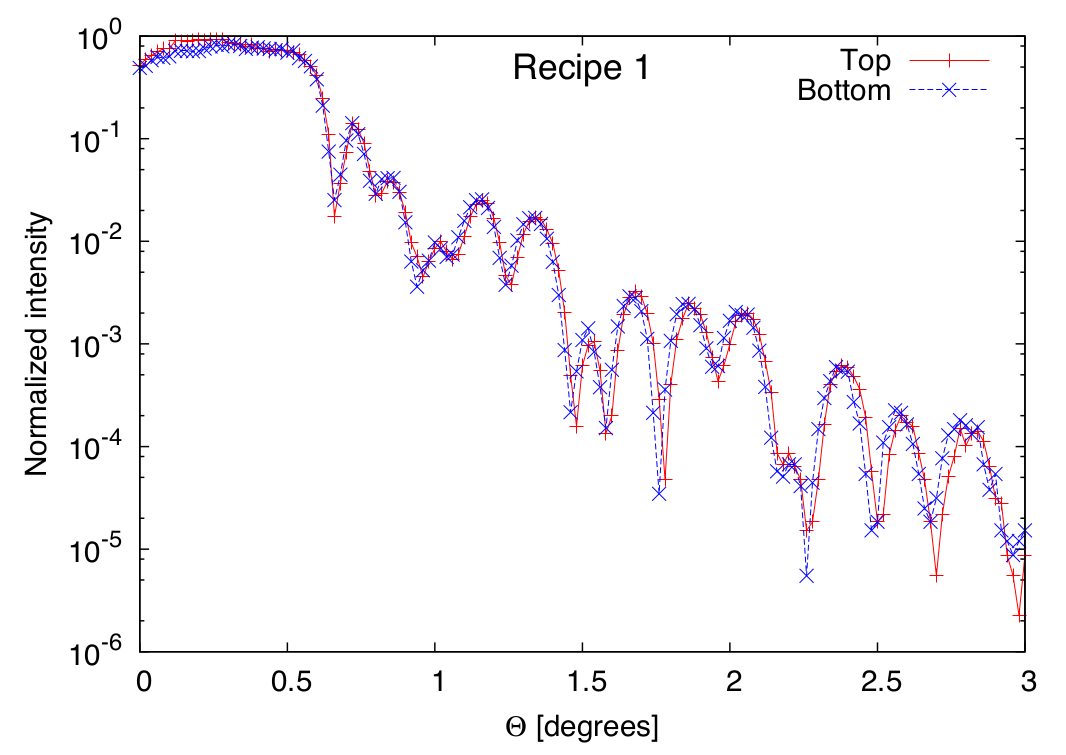 ONLY shows experimental data. The lines are just connecting the
dots.
ONLY shows experimental data. The lines are just connecting the
dots.
Fig. 4.21:
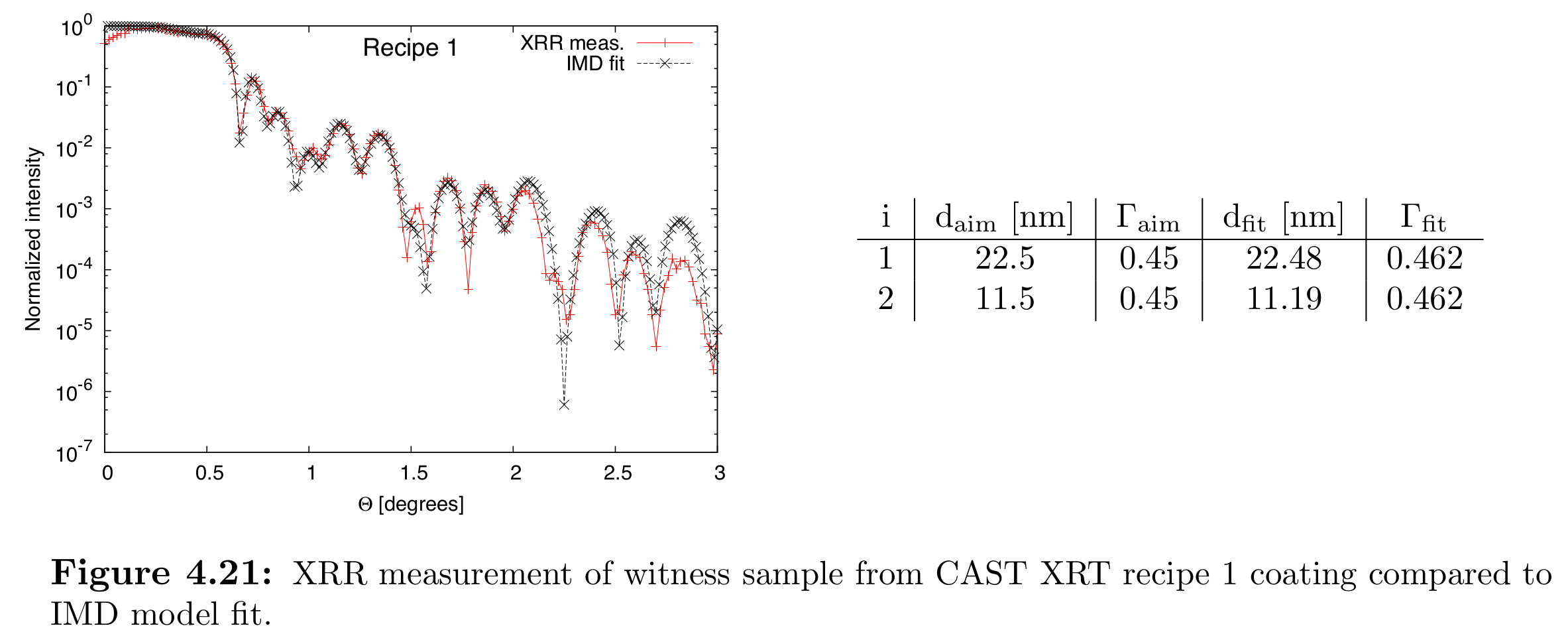 shows the fit of IMD!
shows the fit of IMD!
We included the calculation of this in ./../CastData/ExternCode/xrayAttenuation/playground/llnl_telescope_reflectivity.nim
in the automatic procedure. Note that we use the 8.047 keV as
mentioned in the quote above.
The produced figures are stored in:
where each corresponds to the recipes 1 to 4 (index 0 to 3).
Note in particular how recipe 1 (index 0) pretty much matches the values seen in fig. 4.21 of the thesis (IMD fit) perfectly!
This really means our code should be quite correct by now.
The other recipes do not match that well. But I suppose the likely reason is that the real composition of the different layers is simply the reason. IMD performs a fit after all. So if we calculate it for the fit parameters I'm pretty sure the result would look much better.
It might be a good idea to compute the XRR for the fit parameters and see what it gives us!
- DarpanX
Note that we also tried to compute the recipe 1 with DarpanX. We do indeed reproduce the numbers with it as well.
which was produced using ./../CastData/ExternCode/AxionElectronLimit/tools/understand_darpanx.py
with the important code:
import darpanx as drp import numpy as np m1 = drp.Multilayer(MultilayerType="DepthGraded", SubstrateMaterial="SiO2", D_min = 115.0, D_max = 225.0, Gamma = 0.45, C = 1.0, LayerMaterial=["C", "Pt"], Repetition=2, SigmaValues=[0.0]) θs = np.linspace(0.0, 3.0, 1000) E = [8.047] m1.get_optical_func(Theta = θs, Energy = E) m1.plot(ylog = "yes", Comp = ["Ra"], OutFile = "plots/Pt_SiC_angle_0", Struc = 'yes')
1.73.3. Effective area with xrayAttenuation
We've rerun the effective area calculations using the H5 file of the
LLNL reflectivity from xrayAttenuation.
The relevant CSV and figures are:
- ./resources/effectiveAreas/effective_area_scan_telescope_llnl_3arcmin_xrayAttenuation.csv
- ./resources/effectiveAreas/effective_area_scan_telescope_llnl_parallel_xrayAttenuation.csv
- ./resources/effectiveAreas/effective_area_scan_telescope_llnl_sun_correct_shells_xrayAttenuation_fixed.csv
In particular the last plot comparing all three cases,
- parallel light of full magnet bore (21.5 mm)
- 3 arcmin sized non parallel source
- our solar emission model
compared to the thesis plot shows a decent result.
In
we generally see that our behavior is now more or less as expected,
with the exception of that last bump (!) near about 6 keV!
An interesting aspect is that we DO indeed see the largest decrease in efficiency when going to the 3 arc minute source. However, for our realistic axion emission model the difference is much less.
So that means we really should not use the effective area from the JCAP paper, because it would give us significantly lower efficiencies than are apparently correct for our emission!
1.73.4. Reflectivity comparisons DarpanX and xrayAttenuation
Looking at the layer1.pdf produced when creating the H5 file using
./../CastData/ExternCode/AxionElectronLimit/tools/llnl_layer_reflectivity.nim
as well as for our xrayAttenuation version:
./../CastData/ExternCode/xrayAttenuation/playground/llnl_telescope_reflectivity.nim
(which will be moved at some point!!)
Compare recipe 1 (well, index 0) and at a first glance it really looks like they are the same.
However, the reflectivity for a random angle is still very different!
These are computed for θ = 0.5° using again the same code as
mentioned above.
Comparing it to the same from the code mentioned above:
./../CastData/ExternCode/AxionElectronLimit/tools/understand_darpanx.py
for DarpanX:
and we can see that the DarpanX result has a MUCH HIGHER (they are still at about 0.5 whereas ours is below 0.3!) reflectivity at values towards 10 keV!
This is questionable given that our previous effective areas computed from the DarpanX H5 file were always TOO HIGH at large values when normalizing the values to the maximum of the thesis data!
1.74. [/]
[ ]Make XRR plot using the IMD fit parameters![X]Create a reflectivity plot of only a single angle from the H5 files created from DarpanX as well as from our xrayAttenuation based file! Because I'm still confused that the layers
import numericalnim/interpolate import nimhdf5, ggplotnim, unchained, sequtils, algorithm import strutils, strformat proc initRefl(path: string, idx: int): Interpolator2DType[float] = var h5f = H5open(path, "r") let energies = h5f["/Energy", float] let angles = h5f["/Angles", float] let reflDset = h5f[("Reflectivity" & $idx).dset_str] let data = reflDset[float].toTensor.reshape(reflDset.shape) result = newBilinearSpline( data, (angles.min, angles.max), (energies.min, energies.max) ) discard h5f.close() const pathDP = "/home/basti/CastData/ExternCode/AxionElectronLimit/resources/llnl_layer_reflectivities.h5" const pathXA = "/home/basti/CastData/ExternCode/AxionElectronLimit/resources/llnl_layer_reflectivities_xrayAttenuation.h5" let reflDP = initRefl(pathDP, 0) let reflXA = initRefl(pathXA, 0) let energies = linspace(0.03, 12.0, 1000) let rDP = energies.mapIt(reflDP.eval(0.5, it)) let rXA = energies.mapIt(reflXA.eval(0.5, it)) let df = toDf(rDP, rXA, energies).gather(["rDP", "rXA"], "Type", "Refl") ggplot(df, aes("energies", "Refl", color = "Type")) + geom_line() + ggtitle("Comparison of DarpanX & xrayAttenuation at θ = 0.5° recipe 1") + ggsave("~/org/Figs/statusAndProgress/xrayReflectivities/comparison_darpanx_xrayAttenuation_reflectivity_recipe_1.pdf")
Ohhh, this is a slam dunk!
Compared to the DarpanX data our data is simply compressed along the energy axis! My assumption is that the conversion from keV in DarpanX has a linear bias and scales the numbers such that they become more and more wrong over the energy scale.
We can quickly verify this by checking the used wavelength inside the inner parts of each library at a known energy, e.g. at 9 keV.
From our point of view we expect:
import unchained proc wavelength(E: keV): Meter = result = (hp * c / E).to(Meter) echo 9.keV.wavelength().to(NanoMeter)
0.13776 nm or 1.3776 Å!
Let's check in our library first:
WAVELENGTH= == 0.13776 nm
for an input of 9 keV this is precisely what we get of course.
Now DarpanX:
ENERGY AS WAVELENGTH::: [1.54980248 1.3776022 ]
which is for 8 and 9 keV (we need to give two numbers).
Hmm, so that all looks fine. :/ And the conversion functions in DarpanX from keV to Å and back surprisingly look reasonable.
Maybe their mapping to energies from the form factor files is wrong?
[ ]Try to plot the form factors of each against one another?
1.74.1. TODO IMPORTANT [/]
- Tell Cristina she needs to include the detector window opening area!
1.75. [/]
Today I gave the limit method presentation in front of many people of the CAST collaboration.
It took 53 min 35 s with a couple of questions by Theodoros in the middle.
I think I did a pretty good job.
Generally everyone is happy with the method that we use. There are a few minor questions and comments.
Horst:
- CAST CAPP used a magnetic field of 8.8 T! We still use 9 T
[ ]Jaime used 8.8 T in the 2013 analysis! Need to tell Klaus that this is indeed the case- He would like to see the variability of measured counts over time
for all 7 chips. I.e.
[ ]Plot the count rate (in time bins) for all 7 chips before cuts[ ]…and after cuts
Igor:
[ ]He's not sure whether the unbinned likelihood approach shouldn't include a term (iiuc) of \(\exp{- \text{number of total candidates}}\) Generally he says that for the unbinned likelihood normally one would not start from the ratio of two Poissons. That makes sense because strictly speaking each candidate is not Poisson distributed! -> Read up on unbinned likelihood from scratch. I.e. how is it normally defined and how is it derived?[ ]He brought up the 'Asimov dataset'. Apparently it is the dataset that matches exactly our expectation. I.e. assuming a binned approach it would be exactly the mean used in all bins! The nice property of the 'Asimov dataset' is that it, apparently, reproduces exactly the expected limit. Which actually does make sense intuitively, no? -> The question is how does one apply this in an unbinned likelihood approach? Klaus said it might work by including fractionally weighted candidates.
Generally now:
[ ]Compute the real candidates! Using the best expected limit method.
What plots & numbers to produce?
[ ]number of total candidates in tracking over the entire chip[ ]Rate in form of "background rate plots" of the candidates in the gold region[ ]plot of s/b using the raytracing image showing the s/b for each candidadte. Maybe show thegeom_rasterwith an opacity underlying all points? That could be neat, if we can get it to work.[ ]List the number of signal sensitive candidates.[ ]Compute the real limit based on these.
1.75.1. DONE Send mail with slides to everyone there
1.75.2. Horst related TODOs
Start with plots for chip activity.
What do we want to plot?
[ ]number of clusters per time bin, raw[ ]number of clusters per time bin, after cuts
This should probably be a plot of all three run periods.
So we need a facet_wrap with mapping of the run periods, similar to
the median cluster charge plot.
Maybe we can even extend ./../CastData/ExternCode/TimepixAnalysis/Plotting/plotTotalChargeOverTime/plotTotalChargeOverTime.nim for it (the script that produces the median cluster charge plot).
1.75.3. Igor's TODOs
Cristina wrote me:
In the theses of Javi Galan and Juanan, they explain this unbinned method Javi's explanation is very similar to what I recalll Igor told me in Florence
check out these two theses!
1.76. [/]
[ ]While talking to Cristina I noticed one thing: -> Computing a simple limit using cumulative sums (or even an EDF as we do now?) might depend on the usage of the coupling constant? In our code we deal with \(g_{ae}²\). The \(g_{aγ}²\) is implicit. In the simple limit code I wrote a an explanation we scan the linear space of the coupling constant though! I believe the limit is likely wrong in that case, no? Because scaling the coupling constant to some power, will distort the likelihood function and therefore change where the limit is. The question is where is the real limit? It's at \(∫_{-∞}^g' L dg\), no?
So: Write / extend a simple limit example, pulling out the gae / gaγ contributions and compute the limit based on modifying the parameters multiplying the signal once and keeping the rest constant? -> Having done this on Cristina's computer: The limit does change when going to gaγ to gaγ⁴! It becomes worse as a matter of fact, because we sample less (right?) points near 0 on the linear scale.
[ ]Only computesimpsonin simple limit code of Cristina once!
1.76.1. Investigate impact of gaγ and gae² gaγ on limit
[X]Compute my own MCMC limit for different gaγ values. Do we recover the same limit? -> See below, the answer is YES WE DO.[ ]Compute my own MCMC limit with gae²·gaγ² as input instead of gae²! Does the limit change?
- Impact of different gaγ values
Let's start with looking at different gaγ values.
- We'll do it by extending the sanity checks first. Pick one set of
candidates, and compute limit for a few different gaγ and compare.
Run
mcmc_limitwith sanity check. Should not require any arguments.mcmc_limit_calculation sanity
The issue we encounter is that the limit goes a bit bonkers when we change the axion photon coupling to a range from
[2023-07-18 - 17:09:30] - INFO: =============== Axion-photon coupling constant =============== [2023-07-18 - 17:09:30] - INFO: Conversion probability using default g_aγ² = 9.999999999999999e-25, yields P_a↦γ = 1.70182e-21 UnitLess [2023-07-18 - 17:10:59] - INFO: Computing limits for g_aγ² from g_aγ = 1e-13 to 9.999999999999999e-12. Variation of 1e-1 normal. [2023-07-18 - 17:11:31] - INFO: Limit for g_aγ² = 1e-26, yields = 2.449779472507777e-19 and as g_ae·g_aγ = 4.949524696885326e-23 [2023-07-18 - 17:11:47] - INFO: Limit for g_aγ² = 1.112e-23, yields = 4.893620862732201e-22 and as g_ae·g_aγ = 7.376792256366047e-23 [2023-07-18 - 17:12:02] - INFO: Limit for g_aγ² = 2.223e-23, yields = 2.04984080890773e-22 and as g_ae·g_aγ = 6.750404519880186e-23 [2023-07-18 - 17:12:16] - INFO: Limit for g_aγ² = 3.334e-23, yields = 1.24410602693182e-22 and as g_ae·g_aγ = 6.440380030549973e-23 [2023-07-18 - 17:12:30] - INFO: Limit for g_aγ² = 4.445e-23, yields = 9.020510146674863e-23 and as g_ae·g_aγ = 6.332153472711299e-23 [2023-07-18 - 17:12:51] - INFO: Limit for g_aγ² = 5.555999999999999e-23, yields = 1.378284773114401e-19 and as g_ae·g_aγ = 2.767264027776101e-21 [2023-07-18 - 17:13:05] - INFO: Limit for g_aγ² = 6.666999999999999e-23, yields = 6.161123571121022e-23 and as g_ae·g_aγ = 6.409072542003551e-23 [2023-07-18 - 17:13:19] - INFO: Limit for g_aγ² = 7.778e-23, yields = 5.676375117774414e-23 and as g_ae·g_aγ = 6.644610271945932e-23 [2023-07-18 - 17:13:38] - INFO: Limit for g_aγ² = 8.888999999999999e-23, yields = 3.467061740306982e-19 and as g_ae·g_aγ = 5.551460331263185e-21 [2023-07-18 - 17:13:53] - INFO: Limit for g_aγ² = 9.999999999999999e-23, yields = 5.181832452845458e-19 and as g_ae·g_aγ = 7.198494601543753e-21
I think this is because of the starting parameters and step sizes of the MCMC.
We have (for the fully uncertain case):
2093 var totalChain = newSeq[seq[float]]() 2094 for i in 0 ..< nChains: 2095 let start = @[rnd.rand(0.0 .. 5.0) * 1e-21, # g_ae² 2096 rnd.rand(-0.4 .. 0.4), rnd.rand(-0.4 .. 0.4), # θs, θb 2097 rnd.rand(-0.5 .. 0.5), rnd.rand(-0.5 .. 0.5)] # θx, θy 2098 echo "\t\tInitial chain state: ", start 2099 let (chain, acceptanceRate) = rnd.build_MH_chain(start, @[3e-21, 0.025, 0.025, 0.05, 0.05], 150_000, fn)
where we now have to make sure to recover the equivalent of 1e-21 if gaγ is modified.
So:
import math const xsq = 1e-21 echo sqrt(xsq) const g_aγ = 1e-12 let want = xsq * g_aγ * g_aγ echo "Implicitly = ", want, " which we want to keep given changing g_aγ!" let g_aγLow = 1e-13 echo "Assume new g_aγ = ", g_aγLow let need = want / (g_aγLow * g_aγLow) echo "Value I need for g_ae² = ", need echo "Check = ", need * g_aγLow * g_aγLow
The above means that in places of hardcoded 1e-21 values (corresponding to gae²), we need to replace that by 1e-45 / gaγ²!
Implementing this reference based starting & stepsize parameter for the gae² numbers does indeed work and now the limits make sense and are independent of the axion photon coupling.
From the
sanity.logfile:[2023-07-18 - 17:44:29] - INFO: =============== Axion-photon coupling constant =============== [2023-07-18 - 17:44:29] - INFO: Conversion probability using default g_aγ² = 9.999999999999999e-25, yields P_a↦γ = 1.70182e-21 UnitLess [2023-07-18 - 17:44:52] - INFO: Limit with default g_aγ² = 9.999999999999999e-25 is = 4.22382364911193e-21, and as g_ae·g_aγ = 6.499095051706761e-23 [2023-07-18 - 17:46:00] - INFO: Computing limits for g_aγ² from g_aγ = 1e-13 to 9.999999999999999e-12. Variation of 1e-1 normal. [2023-07-18 - 17:46:22] - INFO: Limit for g_aγ² = 1e-26, yields = 4.218144490990871e-19 and as g_ae·g_aγ = 6.49472439060417e-23 [2023-07-18 - 17:46:44] - INFO: Limit for g_aγ² = 2.782559402207114e-26, yields = 2.039417518566342e-19 and as g_ae·g_aγ = 7.533127100555704e-23 [2023-07-18 - 17:47:07] - INFO: Limit for g_aγ² = 7.742636826811214e-26, yields = 5.399405788710695e-20 and as g_ae·g_aγ = 6.465727963854423e-23 [2023-07-18 - 17:47:29] - INFO: Limit for g_aγ² = 2.15443469003186e-25, yields = 1.98161971093051e-20 and as g_ae·g_aγ = 6.533965295040676e-23 [2023-07-18 - 17:47:52] - INFO: Limit for g_aγ² = 5.994842503189323e-25, yields = 7.154971209266925e-21 and as g_ae·g_aγ = 6.549269082455635e-23 [2023-07-18 - 17:48:14] - INFO: Limit for g_aγ² = 1.668100537200028e-24, yields = 2.552164701072774e-21 and as g_ae·g_aγ = 6.524773795988979e-23 [2023-07-18 - 17:48:36] - INFO: Limit for g_aγ² = 4.641588833612678e-24, yields = 9.040340406290424e-22 and as g_ae·g_aγ = 6.477773003270114e-23 [2023-07-18 - 17:48:59] - INFO: Limit for g_aγ² = 1.291549665014851e-23, yields = 3.342467000566223e-22 and as g_ae·g_aγ = 6.570359301365869e-23 [2023-07-18 - 17:49:21] - INFO: Limit for g_aγ² = 3.593813663804523e-23, yields = 1.179994862189101e-22 and as g_ae·g_aγ = 6.512051642112742e-23 [2023-07-18 - 17:49:44] - INFO: Limit for g_aγ² = 9.999999999999673e-23, yields = 4.119636934452537e-23 and as g_ae·g_aγ = 6.418439790519501e-23 [2023-07-18 - 17:51:39] - INFO: =============== Input =============== [2023-07-18 - 17:51:39] - INFO: Input path: [2023-07-18 - 17:51:39] - INFO: Input files: @[(2017, "/home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5"), (2018, "/home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.\ 99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5")] [2023-07-18 - 17:51:39] - INFO: =============== Time =============== [2023-07-18 - 17:51:39] - INFO: Total background time: -1 h [2023-07-18 - 17:51:39] - INFO: Total tracking time: -1 h [2023-07-18 - 17:51:39] - INFO: Ratio of tracking to background time: 1 UnitLess [2023-07-18 - 17:51:40] - INFO: =============== Axion-photon coupling constant =============== [2023-07-18 - 17:51:40] - INFO: Conversion probability using default g_aγ² = 9.999999999999999e-25, yields P_a↦γ = 1.70182e-21 UnitLess [2023-07-18 - 17:52:05] - INFO: Limit with default g_aγ² = 9.999999999999999e-25 is = 6.916320578920485e-21, and as g_ae·g_aγ = 8.316441894777169e-23 [2023-07-18 - 17:53:15] - INFO: Computing limits for g_aγ² from g_aγ = 1e-13 to 9.999999999999999e-12. Variation of 1e-1 normal. [2023-07-18 - 17:53:39] - INFO: Limit for g_aγ² = 1e-26, yields = 7.071860417740563e-19 and as g_ae·g_aγ = 8.409435425604125e-23 [2023-07-18 - 17:54:03] - INFO: Limit for g_aγ² = 2.782559402207114e-26, yields = 2.478176624728773e-19 and as g_ae·g_aγ = 8.30401930842465e-23 [2023-07-18 - 17:54:26] - INFO: Limit for g_aγ² = 7.742636826811214e-26, yields = 9.108023442629517e-20 and as g_ae·g_aγ = 8.397625719592601e-23 [2023-07-18 - 17:54:50] - INFO: Limit for g_aγ² = 2.15443469003186e-25, yields = 3.224445239849654e-20 and as g_ae·g_aγ = 8.334780549504706e-23 [2023-07-18 - 17:55:14] - INFO: Limit for g_aγ² = 5.994842503189323e-25, yields = 1.168884566147608e-20 and as g_ae·g_aγ = 8.37094909700429e-23 [2023-07-18 - 17:55:37] - INFO: Limit for g_aγ² = 1.668100537200028e-24, yields = 4.125379960809593e-21 and as g_ae·g_aγ = 8.295509947423794e-23 [2023-07-18 - 17:56:01] - INFO: Limit for g_aγ² = 4.641588833612678e-24, yields = 1.502288345596094e-21 and as g_ae·g_aγ = 8.35045196967523e-23 [2023-07-18 - 17:56:25] - INFO: Limit for g_aγ² = 1.291549665014851e-23, yields = 5.320321836758966e-22 and as g_ae·g_aγ = 8.289426931964138e-23 [2023-07-18 - 17:56:48] - INFO: Limit for g_aγ² = 3.593813663804523e-23, yields = 1.950497695512556e-22 and as g_ae·g_aγ = 8.372410208149297e-23 [2023-07-18 - 17:57:12] - INFO: Limit for g_aγ² = 9.999999999999673e-23, yields = 6.901452233923125e-23 and as g_ae·g_aγ = 8.30749795902647e-23
Compare the "default gaγ²" based limit and the limits for other values of gaγ².
The first entries are for an RNG seed of
0xaffeand the second for0x1337.We can conclude from these numbers that the value of gaγ is indeed irrelevant. This is of course only the case for numbers up to values for gaγ where the dominating axion flux contribution is still the axion electron coupling!
- Does limit of gae²·gaγ² change value of limit?
limit 1 = 4.223823649111892e-45
Finally managed to implement this correctly!
Still need to clean up the implementation a bit though.
Anyway though, we can now confirm that the limit remains unchanged!
[2023-07-19 - 00:46:26] - INFO: =============== Time =============== [2023-07-19 - 00:46:26] - INFO: Total background time: 3158.57 h [2023-07-19 - 00:46:26] - INFO: Total tracking time: 161.111 h [2023-07-19 - 00:46:26] - INFO: Ratio of tracking to background time: 1 UnitLess [2023-07-19 - 00:46:50] - INFO: Limit for g_ae²·g_aγ² = 4.305185213455262e-45, yields g_ae·g_aγ = 6.561391021311916e-23 [2023-07-19 - 00:47:12] - INFO: Limit for g_ae²·g_aγ² = 4.256371638039694e-45, yields g_ae·g_aγ = 6.524087398280079e-23 [2023-07-19 - 00:47:34] - INFO: Limit for g_ae²·g_aγ² = 4.147641630954775e-45, yields g_ae·g_aγ = 6.440218653861664e-23 [2023-07-19 - 00:47:56] - INFO: Limit for g_ae²·g_aγ² = 4.293718641483905e-45, yields g_ae·g_aγ = 6.552647282956642e-23 [2023-07-19 - 00:48:18] - INFO: Limit for g_ae²·g_aγ² = 4.2238512626012e-45, yields g_ae·g_aγ = 6.499116295775296e-23 [2023-07-19 - 00:48:40] - INFO: Limit for g_ae²·g_aγ² = 5.484924054642261e-45, yields g_ae·g_aγ = 7.406027312022459e-23 [2023-07-19 - 00:49:02] - INFO: Limit for g_ae²·g_aγ² = 4.223524160658419e-45, yields g_ae·g_aγ = 6.49886463981088e-23 [2023-07-19 - 00:49:24] - INFO: Limit for g_ae²·g_aγ² = 4.146771380385997e-45, yields g_ae·g_aγ = 6.439542980977762e-23 [2023-07-19 - 00:49:46] - INFO: Limit for g_ae²·g_aγ² = 4.164270234117464e-45, yields g_ae·g_aγ = 6.453115708026212e-23 [2023-07-19 - 00:50:08] - INFO: Limit for g_ae²·g_aγ² = 4.239747781864239e-45, yields g_ae·g_aγ = 6.511334565098187e-23
1.77.
1.77.1. Finishing different gaγ² and gae²·gaγ² coupling limits
I cleaned up the code from yesterday this morning to allow for different kind of limit calculations.
We introduced a CouplingKind:
CouplingKind = enum ck_g_ae² ## We vary the `g_ae²` and leave `g_aγ²` fully fixed ck_g_aγ² ## We vary the `g_aγ²` and leave `g_ae²` fully fixed (and effectively 'disabled'); for axion-photon searches ck_g_ae²·g_aγ² ## We vary the *product* of `g_ae²·g_aγ²`, i.e. direct `g⁴` proportional search. ## Note that this is equivalent in terms of the limit!
and changed the fields for the Context
g_aγ²: float # the ``reference`` g_aγ (squared) g_ae²: float # the ``reference`` g_ae value (squared) coupling: float # the ``current`` coupling constant in use. Can be a value of # `g_ae²`, `g_aγ²`, `g_ae²·g_aγ²` depending on use case! # Corresponds to first entry of MCMC chain vector! couplingKind: CouplingKind # decides which coupling to modify couplingReference: float # the full reference coupling. `g_ae²·g_aγ²` if `ck_g_ae²·g_aγ²`
where coupling now stores the actual current coupling in use (from
the MCMC vector the 0 value) and the g_ae/γ² values are the actual
reference values we normally use. Note though that g_aγ² at the
moment can be adjusted, as it is not a reference in the same way as
the g_ae² where the input is based on that.
The couplingReference is the product of the g_ae² and g_aγ²
fields. Be careful when adjusting the g_aγ² field after the fact if
you change to ck_g_ae²·g_aγ² which requires the full reference
including the g_aγ²! So initCouplingReference may need to be
called again!
Running the sanity checks now yields:
[2023-07-19 - 12:58:08] - INFO: =============== Time =============== [2023-07-19 - 12:58:08] - INFO: Total background time: 3158.57 h [2023-07-19 - 12:58:08] - INFO: Total tracking time: 161.111 h [2023-07-19 - 12:58:08] - INFO: Ratio of tracking to background time: 1 UnitLess [2023-07-19 - 12:58:08] - INFO: =============== Axion-photon coupling constant =============== [2023-07-19 - 12:58:08] - INFO: Conversion probability using default g_aγ² = 9.999999999999999e-25, yields P_a↦γ = 1.70182e-21 UnitLess [2023-07-19 - 12:58:32] - INFO: Limit with default g_aγ² = 9.999999999999999e-25 is = 4.305185213456743e-21, and as g_ae·g_aγ = 6.561391021313044e-23 [2023-07-19 - 12:59:38] - INFO: Computing limits for g_aγ² from g_aγ = 1e-13 to 9.999999999999999e-12. Variation of 1e-1 normal. [2023-07-19 - 13:00:00] - INFO: Limit for g_aγ² = 1e-26, yields = 4.223851262602536e-19 and as g_ae·g_aγ = 6.499116295776324e-23 [2023-07-19 - 13:00:21] - INFO: Limit for g_aγ² = 2.782559402207114e-26, yields = 1.971179501250353e-19 and as g_ae·g_aγ = 7.40602731202235e-23 [2023-07-19 - 13:00:43] - INFO: Limit for g_aγ² = 7.742636826811214e-26, yields = 5.454891214880666e-20 and as g_ae·g_aγ = 6.498864639810866e-23 [2023-07-19 - 13:01:05] - INFO: Limit for g_aγ² = 2.15443469003186e-25, yields = 1.924760773474564e-20 and as g_ae·g_aγ = 6.439542980977886e-23 [2023-07-19 - 13:01:27] - INFO: Limit for g_aγ² = 5.994842503189323e-25, yields = 6.94642141457768e-21 and as g_ae·g_aγ = 6.453115708026223e-23 [2023-07-19 - 13:01:49] - INFO: Limit for g_aγ² = 1.668100537200028e-24, yields = 2.541662020552458e-21 and as g_ae·g_aγ = 6.511334565098359e-23 [2023-07-19 - 13:02:11] - INFO: Limit for g_aγ² = 4.641588833612678e-24, yields = 9.034559994585504e-22 and as g_ae·g_aγ = 6.47570172162615e-23 [2023-07-19 - 13:02:32] - INFO: Limit for g_aγ² = 1.291549665014851e-23, yields = 3.335200938988032e-22 and as g_ae·g_aγ = 6.563213889175949e-23 [2023-07-19 - 13:02:53] - INFO: Limit for g_aγ² = 3.593813663804523e-23, yields = 1.186891715034903e-22 and as g_ae·g_aγ = 6.531054786899905e-23 [2023-07-19 - 13:03:15] - INFO: Limit for g_aγ² = 9.999999999999673e-23, yields = 4.15039556899915e-23 and as g_ae·g_aγ = 6.442356377133303e-23 [2023-07-19 - 13:03:15] - INFO: =============== Axion-electron · Axion-photon coupling constant limit =============== [2023-07-19 - 13:03:15] - INFO: Coupling reference to rescale by 1e-26 from g_ae² = 1e-26 and g_aγ² = 9.999999999999999e-25 [2023-07-19 - 13:03:38] - INFO: Limit for g_ae²·g_aγ² = 4.305185213456743e-21, yields g_ae·g_aγ = 6.561391021313044e-11 [2023-07-19 - 13:04:00] - INFO: Limit for g_ae²·g_aγ² = 4.256371638039793e-21, yields g_ae·g_aγ = 6.524087398280156e-11 [2023-07-19 - 13:04:22] - INFO: Limit for g_ae²·g_aγ² = 4.147641630954855e-21, yields g_ae·g_aγ = 6.440218653861727e-11 [2023-07-19 - 13:04:44] - INFO: Limit for g_ae²·g_aγ² = 4.293718641484303e-21, yields g_ae·g_aγ = 6.552647282956944e-11 [2023-07-19 - 13:05:05] - INFO: Limit for g_ae²·g_aγ² = 4.223851262602952e-21, yields g_ae·g_aγ = 6.499116295776644e-11 [2023-07-19 - 13:05:27] - INFO: Limit for g_ae²·g_aγ² = 5.484924054642192e-21, yields g_ae·g_aγ = 7.406027312022412e-11 [2023-07-19 - 13:05:49] - INFO: Limit for g_ae²·g_aγ² = 4.223524160658912e-21, yields g_ae·g_aγ = 6.498864639811259e-11 [2023-07-19 - 13:06:11] - INFO: Limit for g_ae²·g_aγ² = 4.146771380386027e-21, yields g_ae·g_aγ = 6.439542980977787e-11 [2023-07-19 - 13:06:33] - INFO: Limit for g_ae²·g_aγ² = 4.164270234117538e-21, yields g_ae·g_aγ = 6.45311570802627e-11 [2023-07-19 - 13:06:55] - INFO: Limit for g_ae²·g_aγ² = 4.239747781864322e-21, yields g_ae·g_aγ = 6.51133456509825e-11
i.e. it still works correctly and we can change the limit calculation approach at runtime!
The code is now all committed and pushed.
1.77.2. Horst related TODOs continue from yesterday
We've extended the plotTotalChargeOverTime script to produce a plot
of the number of clusters in a time window.
[ ]CHECK FOR older script about outer chip activity that we once wrote!!! ./../CastData/ExternCode/TimepixAnalysis/Tools/outerChipActivity/outerChipActivity.nim -> I think that script only produces statistics about the chips in general, but no time specific information.
To generate the plot of # of clusters in each time bin for all chips:
./plotTotalChargeOverTime \ /mnt/1TB/CAST/2017/DataRuns2017_Reco.h5 \ /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 \ --interval 90 \ --cutoffCharge 0 \ --cutoffHits 500 \ --timeSeries \ --toCount \ --readAllChips \ --outpath /tmp/overTime/ \ --ylabel "# of clusters" \ --title "Number of clusters in 90 min intervals per chip"
which yields the figure:
Lower number of clusters: likely gas gain time slices that are shorter than 90 min? Is that possible? Those at end of run.
Let's run it with an charge cutoff:
--cutoffCharge 1000
-> Changes nothing. So more likely a time interval effect?
-> Ah, of course why would it change anything? We're plotting the
number of clusters found! And some are too low.
We have done the following couple of changes to the code:
[X]change the number to be the number of clusters in each interval divided by the actual length of the current interval[X]exclude any intervals shorter than 1 hour -> This was very important to get rid of serious outliers[X]after the above two, some minor outliers to the bottom (2 cases iirc) remain to about 0.06. AcountCutoffoption was added for clarity to see a better range of the interesting data.
./plotTotalChargeOverTime \ /mnt/1TB/CAST/2017/DataRuns2017_Reco.h5 \ /mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 \ --interval 90 \ --cutoffCharge 0 \ --cutoffHits 500 \ --timeSeries \ --toCount \ --readAllChips \ --outpath /tmp/overTime/ \ --ylabel "# of clusters" \ --title "Number of clusters per second (from 90 min intervals) per chip" \ --countCutoff 0.08
The last option removes any interval entry with less than 0.08 clusters per second.
This should conclude our 'investigation' into Horst's question!
1.77.3. Unblinding the data
To unblind the data we just need to run the likelihood program with
the --tracking flage.
At least in theory.
The case we want to look at is the MLP @ 95% (91% effective) with line veto but no septem veto.
Run-2:
likelihood \ -f /home/basti/CastData/data/DataRuns2017_Reco.h5 \ --h5out ~/org/resources/lhood_tracking_data/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ --region=crAll \ --cdlYear=2018 \ --scintiveto \ --fadcveto --vetoPercentile=0.99 \ --lineveto \ --mlp ~/org/resources/nn_devel_mixing/10_05_23_sgd_gauss_diffusion_tanh300_mse_loss/mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.pt \ --nnSignalEff=0.95 \ --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --readOnly \ --calibFile=/home/basti/CastData/data/CalibrationRuns2017_Reco.h5 \ --tracking
Run-3:
likelihood \ -f /home/basti/CastData/data/DataRuns2018_Reco.h5 \ --h5out ~/org/resources/lhood_tracking_data/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --region=crAll \ --cdlYear=2018 \ --scintiveto \ --fadcveto --vetoPercentile=0.99 \ --lineveto \ --mlp ~/org/resources/nn_devel_mixing/10_05_23_sgd_gauss_diffusion_tanh300_mse_loss/mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.pt \ --nnSignalEff=0.95 \ --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --readOnly \ --calibFile=/home/basti/CastData/data/CalibrationRuns2018_Reco.h5 \ --tracking
I copied the two files to ~/Sync.
The file for Run-3 has the following duration:
totalDuration = 316176.9779 (in seconds)
-> 87.8266666667 hours
That does indeed look correct.
I also checked a random run for the number of entries in the dataset and I saw like 33. That seems a bit much.
Let's make a background cluster plot and see what's what.
plotBackgroundClusters \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --zMax 5 \ --title "MLP@95+FADC+Scinti+Line tracking clusters" \ --outpath ~/org/Figs/statusAndProgress/trackingCandidates/ \ --suffix "mlp_0.95_scinti_fadc_line_tracking_candidates" \ --energyMax 12.0 --energyMin 0.2 --filterNoisyPixels
This yields the plot
so 1610 candidates over the entire chip and few in the center.
[X]INSERT PLOT[ ]COMPUTE EXPECTED NUMBER OF CANDIDATES -> I think MCMC shows precisely about 1600 candidates expected!
1.78.
[X]ExtendplotBackgroundClustersto show:[X]Axion image with an alpha[X]Add energy of each cluster as text[-]instead of count, show all clusters with S/B -> Will be done straight inmcmc_limitbecause there we have access to S and B! Makes no sense to port this over to the plotting tool.
[ ]Make plot of background rate of clusters in gold region[ ]Make plot of S/B for all clusters[ ]Compute the real limit
We did a decently large rewrite of the plotBackgroundClusters
program to support storing the energy information of each
cluster. Using a Table[(int, int), seq[float]] now internally, which
stores the energy of all clusters that share a position. Can be
converted into the old CountTable if needed.
Can be plotted against the counts or the energy.
The axion image can be inserted with an alpha below the clusters for clarity.
The energy of each cluster can be printed above it if the
--energyText option is given and can be restricted to a radius of
energyTextRadius around the center of the chip.
Produce the cluster positions with the axion image in the center and the number of counts:
plotBackgroundClusters \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --title "MLP@95+FADC+Scinti+Line tracking clusters" \ --outpath ~/org/Figs/statusAndProgress/trackingCandidates/ \ --suffix "mlp_0.95_scinti_fadc_line_tracking_candidates_axion_image" \ --energyMin 0.2 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv
plotBackgroundClusters \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --title "MLP@95+FADC+Scinti+Line tracking clusters" \ --outpath ~/org/Figs/statusAndProgress/trackingCandidates/ \ --suffix "mlp_0.95_scinti_fadc_line_tracking_candidates_axion_image_with_energy" \ --energyMin 0.2 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy
plotBackgroundClusters \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --title "MLP@95+FADC+Scinti+Line tracking clusters" \ --outpath ~/org/Figs/statusAndProgress/trackingCandidates/ \ --suffix "mlp_0.95_scinti_fadc_line_tracking_candidates_axion_image_with_energy_radius_85" \ --energyMin 0.2 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0
1.78.1. Compute the real limit:
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --tracking /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ --tracking /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --years 2017 --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --suffix=_sEff_0.95_scinti_fadc_line_mlp_tanh300_axion_image_1487.9_0.989AU_default_seed \ --path "" \ --outpath /home/basti/org/resources/lhood_MLP_0.95_real_limit/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel /home/basti/org/resources/differential_flux_sun_earth_distance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv
-> Running this for the first time showed a total time of 770.031 h
which is obviously wrong (for the tracking)
The 2018 (Run-3) file has the totalDuration of
totalDuration = 316176.9779 (in seconds)
-> 87.8266666667 hours
and 2017 (Run-2):
totalDuration = 2455933.4681
-> 682.20 hours!! Clearly wrong.
I suppose we are counting runs that have no trackings at all in the calculation? -> Indeed, fixed.
Rerunning the likelihood with the fixed code now!
I've implemented a plot of the signal over background:
which shows only very minor contributions.
The candsInSens procedure actually counts 0 candidates as sensitive
based on the 0.5 cutoff!
Running the real limit yields some pretty crazy numbers:
[2023-07-21 - 01:27:32] - INFO: =============== Calculation of the real limit =============== [2023-07-21 - 01:27:32] - INFO: Input tracking files: @["/home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5", "/home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5"] [2023-07-21 - 01:27:32] - INFO: Number of candidates: 1604 [2023-07-21 - 01:27:32] - INFO: Total tracking time: 161.111 h [2023-07-21 - 01:27:34] - INFO: Number of candidates in sensitive region ln(1 + s/b) > 0.5 (=cutoff): 0 [2023-07-21 - 01:27:34] - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/trackingCandidates/real_candidates_signal_over_background.pdf [2023-07-21 - 01:28:14] - INFO: Real limit based on 3 150k long MCMCs: g_ae² = 4.212302074670949e-21, g_ae·g_aγ = 6.49022501510614e-23
gae·gaγ = 6.49022501510614e-23
Wow.
NOTE: This number is using a magnetic field of 9 T! Horst said CAPP used 8.8 T.
1.79. [0/1]
Let's now copy over the likelihood files that should have the correct durations and check them.
Run-2: 334086.075 s = 92.8016875
Run-3: 241223.645 s = 67.0065680556
These sum to 159.806 hours.
[ ]So we miss about 1.3 hours somewhere?[ ]Check again how we compute our normal number of 161 hours[ ]Cross check the difference between that and the likelihood way of computing the duration. Given that we only sum up the real event durations maybe there's something related to not cutting to the tracking precisely enough / having events missing at beginning or end? Or some time zone shenanigans of the tracking start and stop? -> INVESTIGATE.
Aside from the time, let's make sure it gives the same number of clusters still:
plotBackgroundClusters \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --title "MLP@95+FADC+Scinti+Line tracking clusters" \ --outpath /tmp/ \ --suffix "mlp_0.95_scinti_fadc_line_tracking_candidates_axion_image_with_energy_radius_85" \ --energyMin 0.2 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0
-> Plot looks the same, so all good, still 1610 clusters.
[ ]Make background rate plot of the clusters in gold region!
plotBackgroundRate \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --names "Candidates" --names "Candidates" \ --centerChip 3 \ --title "Rate of candidates @CAST, SGD tanh300 MLE MLP@91% + line veto" \ --showNumClusters \ --region crGold \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.2 \ --outfile rate_real_candidates_mlp_0.95_scinti_fadc_line.pdf \ --outpath ~/org/Figs/statusAndProgress/trackingCandidates/ \ --quiet
which yields
[ ]Let's recompute the real limit using a magnetic field of 8.8 T and the "correct" time of 159.8 hours.
- INFO:
=============Calculation of the real limit=============- INFO: Input tracking files: @["/home/basti/Sync/lhoodtrackingscintilinemlp0.952017.h5", "/home/basti/Sync/lhoodtrackingscintilinemlp0.952018.h5"] - INFO: Number of candidates: 1604 - INFO: Total tracking time: 159.808 h - INFO: Number of candidates in sensitive region ln(1 + s/b) > 0.5 (=cutoff): 0 - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/trackingCandidates/realcandidatessignaloverbackground.pdf - INFO: Real limit based on 3 150k long MCMCs: gae² = 4.373176811037735e-21, gae·gaγ = 6.612999932736832e-23
So even in this case the limit only decreases from 6.49 to 6.61e-23. That's acceptable. :)
We see 1604 clusters in the MCMC output, but 1610 in the background cluster plot. Why?
Energy range cuts are the same (previously there was a < vs a <=).
Without noisy filter cut:
- 1686 clusters in
plotBackgroundClusters - 1686 clusters in
mcmc_limit_calculation
So the difference comes from applying the noisy filter!
Pixel conversion from poisition in mm to pixel in
plotBackgroundCluster via:
func toPixel(s: float): int = min((255.0 * s / 14.0).round.int, 255)
and in mcmc_limit_calculation:
func toIdx*(arg: float): int = (arg / 14.0 * 256.0).round.int.clamp(0, 255)
import math func toPixel(s: float): int = min((255.0 * s / 14.0).round.int, 255) func toIdx*(arg: float): int = (arg / 14.0 * 256.0).round.int.clamp(0, 255) echo toPixel(14.0) echo toIdx(14.0) echo toPixel(13.98) echo toIdx(13.98) echo toPixel(13.96) echo toIdx(13.96)
The issue is the toIdx function. We need to multiply by 255 instead
of 256. While we have 256 pixels, we want to compute the index and
not the pixel number.
Weird changing it to use 255 we get: DataFrame with 5 columns and 1595 rows: 1595 clusters. What.
The reason is:
result = df.filter(f{not (toIdx(`centerX`) in xSet and toIdx(`centerY`) in ySet)})
compared to:
result = df.filter(f{float -> bool: (toIdx(`centerX`), toIdx(`centerY`)) notin noiseFilter.pixels})
Why do these give different results?
The former is not the same as the latter in terms of the pixels considered!
[ ]We'll show that later, for now just use the latter.[X]Check if hetoIdxfunction makes a difference in the pixels -> Using our 256 as a reference number we get DataFrame with 5 columns and 1615 rows: 1615 clusters! So use the correct 255 from now.
So the "final" limit for now based on 1610 clusters, a magnetic field of 8.8 T and our unfortunate 159.8 hours we apparently have:
- INFO:
=============Calculation of the real limit=============- INFO: Input tracking files: @["/home/basti/Sync/lhoodtrackingscintilinemlp0.952017.h5", "/home/basti/Sync/lhoodtrackingscintilinemlp0.952018.h5"] - INFO: Number of candidates: 1610 - INFO: Total tracking time: 159.808 h - INFO: Number of candidates in sensitive region ln(1 + s/b) > 0.5 (=cutoff): 0 - INFO: Saving plot: /home/basti/org/Figs/statusAndProgress/trackingCandidates/realcandidatessignaloverbackground.pdf - INFO: Real limit based on 3 150k long MCMCs: gae² = 4.298506025245605e-21, gae·gaγ = 6.556299280269018e-23
Let's combine all our plots into one PDF and send a mail to Klaus.
pdfunite \ background_cluster_centersmlp_0.95_scinti_fadc_line_tracking_candidates.pdf \ rate_real_candidates_mlp_0.95_scinti_fadc_line.pdf \ background_cluster_centersmlp_0.95_scinti_fadc_line_tracking_candidates_axion_image.pdf \ background_cluster_centersmlp_0.95_scinti_fadc_line_tracking_candidates_axion_image_with_energy.pdf \ background_cluster_centersmlp_0.95_scinti_fadc_line_tracking_candidates_axion_image_with_energy_radius_85.pdf \ real_candidates_signal_over_background.pdf \ plots_real_candidates.pdf
We forgot to make a plot of the likelihood space.
[X]Histogram of the gae² values (likelihood space) via the MCMC
We also changed the computation of the real limit to compute the limit
via g_ae²·g_aγ² and g_ae·g_aγ from the MCMC. That should give the
same limit because the transformation is monotonic in nature and thus
the quantiles of the data don't change!
That is indeed the case.
In addition we now make two histogram plots:
[X]the histogram of allg_ae²samples[X]the histogram of allg_ae·g_aγsamples. Done by converting allg_ae²MCMC elements tog_ae·g_aγfirst
We'll use the computeIntegral option that uses the numerical
integration for our real limit as well to showcase that the numbers
are indeed correct.
When comparing the gae² plot to the gae·gaγ plot the shape is
distinctly different. At first that might seem surprising, but at a
second thought it makes perfect sense:
The sqrt operation is effectively turning it into a log2 plot!
Looking at a value of the first bin, e.g. 0.5e-21 · 1e-12² = 0.5e-45 -> sqrt(0.5e-45) = 2.2360679775e-23 2e-21 · 1e-12² = 2e-45 -> sqrt(2e-45) = 4.472135955e-23
So everything in the first bin is stretched over half the plot in the
g_ae·g_aγ plot!
So we really see the distribution of the data at the very lowest edge given a much wider range! Of course keep in mind that this is because all numbers are smaller than 1 (otherwise it would do the opposite).
The following plot:
also contains 10 points of the likelihood function evaluated using
numerical integration via Romberg.
We set the computeIntegral to true for one run, but it takes over 1
hour to compute these 10 points! It's a good reference though, because
it shows our MCMC produces the correct integral.
1.80. [/]
[ ]Klaus mentioned in his mail that we should make a plot(s) to showcase the distribution of the data during background vs during tracking. i.e. before cuts make histograms / kde / plots of the distributions.
Like
but comparing background and tracking.
Those should look pretty much identical then.
-> We will likely want the ability to apply at least some cuts. I think we should extend the
plotDatasetGgplotscript to reuse theGenericCutlogic fromplotData. That way we can just apply whatever cuts we want from the command line. Just move the code to someio_helpersmodule ofingridand then build a newreadDsetsprocedure that works similar to the current one that returns a DF fromchipDsets, but also takes a variadic number of cuts and masks.
-> Extended ./../CastData/ExternCode/TimepixAnalysis/Plotting/plotDsetGgplot/plotDatasetGgplot.nim to also plot a ridgeline KDE plot of all properties. Should reproduce the above nicely.
[ ]Run it with background + tracking[ ]Run it with 55Fe + fake events & CDL + fake events -> For these need a way to filter the X-ray data a bit? We have all that filtering logic in the CDL plotting! At the heart of ./../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/cdl_spectrum_creation.nim is
for (run, grp) in tfRuns(h5f, tfKind, filename): df.add toDf({ dsetStr : h5f.readCutCDL(run, centerChip, dsetStr, tfKind, float), "run" : run })
which is defined here: ./../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/private/cdl_utils.nim -> use that.
[ ]Turn fake generation file into script to generate data and store in H5? -> Done for generation based on existing runs.[ ]Still missing for user defined energies.
[X]plotDatasetGgplotnow can plot the ridgelines similar to the CDL plot above[X]Run 83 (calibration) seems a bit off from fake generation[ ]check 241
Generate fake events based on a real run, e.g. 241 calibration run:
./fake_event_generator \ like \ -p /mnt/1TB/CAST/2018_2/CalibrationRuns2018_Reco.h5 \ --run 241 \ --outpath /tmp/test_fakegen_run241.h5 \ --outRun 241 \ --tfKind Mn-Cr-12kV \ --nmc 50000
where we specify we want X-rays like the 55Fe source (via tfKind).
Then we can compare the properties:
./plotDatasetGgplot \ -f /mnt/1TB/CAST/2018_2/CalibrationRuns2018_Reco.h5 \ -f /t/test_fakegen_run241.h5 \ --run 241
Let's automate this quickly for all calibration runs.
import shell, strutils, sequtils import nimhdf5 import ingrid / [ingrid_types, tos_helpers] const filePath = "/mnt/1TB/CAST/$#/CalibrationRuns$#_Reco.h5" const genData = """ fake_event_generator \ like \ -p $file \ --run $run \ --outpath /home/basti/org/resources/fake_events_for_runs.h5 \ --outRun $run \ --tfKind Mn-Cr-12kV \ --nmc 50000 """ const plotData = """ plotDatasetGgplot \ -f $file \ -f /home/basti/org/resources/fake_events_for_runs.h5 \ --names 55Fe --names Simulation \ --run $run \ --plotPath /home/basti/org/Figs/statusAndProgress/fakeEventSimulation/ \ --prefix ingrid_properties_run_$run \ --suffix "Run $run" """ const years = ["2017", "2018_2"] const yearFile = ["2017", "2018"] for (year, fYear) in zip(years, yearFile): #if year == "2017": continue ## skip for now, already done let file = filePath % [year, fYear] var runs = newSeq[int]() withH5(file, "r"): let fileInfo = getFileInfo(h5f) runs = fileInfo.runs for run in runs: echo "Working on run: ", run #let genCmd = genData % ["file", file, "run", $run] #shell: # ($genCmd) let plotCmd = plotData % ["file", file, "run", $run, "run", $run, "run", $run] shell: ($plotCmd)
1.81.
Received the raytracing results from Jaime (via Cristina) for the LLNL
telescope as used in the Nature CAST paper.
See more notes in statusAndProgress at
[BROKEN LINK: sec:llnl_telescope:nature_paper_raytracing].
Note that this also contains files for the effective area.
[ ]Compare the effective area file from the above data with the JCAP data.
1.82.
Finishing up the initial draft of the limit paper.
Need to generate the background rate plot for the 91% eff MLP + Line veto.
plotBackgroundRate \ /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crGold_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crGold_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --centerChip 3 \ --combName "MLP@0.91+line" \ --combYear "2018" \ --title "Background rate in center 5·5 mm², MLP@91 % + line veto," \ --showNumClusters \ --region crGold \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.2 \ --outfile background_limitPaper_rate_gold_mlp_0.95_scinti_fadc_line.pdf \ --outpath ~/org/Figs/statusAndProgress/backgroundRates/ \ --quiet
1.83.
Copied over the plots created during training of the MLP with 300 neurons on hidden layer, using MSE and tanh from
~/Sync/10_05_23_sgd_tanh300_mse/
to
~/phd/Figs/neuralNetworks/10_05_23_sgd_tanh300_mse/
to have them in the thesis.
[ ]IMPORTANT: While we set an RNG seed for the NN training, we were relying on the standard global RNG of the
randommodule to shuffle the data! Theshuffleprocedure used the standard RNG instead of our custom one. While the results are reproducible (the default seed is fixed!) this is not great and should be fixed! On current Nim the default random seed is:const DefaultRandSeed = Rand( a0: 0x69B4C98CB8530805u64, a1: 0xFED1DD3004688D67CAu64) # racy for multi-threading but good enough for now: var state = DefaultRandSeed # global for backwards compatibility
1.84.
[ ]Finish section on background rate! -> Is it really correct that MLP has such low background at low energies? Compare again with the plot from limit talk![ ]What does the axion image look like at 3 keV? UNLIKELY, but maybe there is a reason all 3 keV events are outside the center? Hehe.
1.85.
While writing the part about the real candidates in the thesis I noticed from the MCMC sanity checks:
[2023-08-07 - 17:13:37] - INFO: =============== Candidate sampling =============== [2023-08-07 - 17:13:37] - INFO: Sum of background events from candidate sampling grid (`expCounts`) = 1570.974793195856 [2023-08-07 - 17:13:37] - INFO: Expected number from background data (normalized to tracking time) = 1460.188727365468
which seems a bit off. Why such a large discrepancy? Edge correction? Probably, no? It would explain why for the old LnL+Septem+Line case we got:
[2022-07-28 - 15:09:03] - INFO: =============== Candidate sampling =============== [2022-07-28 - 15:09:03] - INFO: Sum of background events from candidate sampling grid (`expCounts`) = 426.214328925721 [2022-07-28 - 15:09:03] - INFO: Expected number from background data (normalized to tracking time) = 412.873417721519
(see statusAndProgress section about sanity.log)
Because there were less clusters in the corners / edges, the difference due to edge correction might be lower.
Plotting the probability of seeing 1610 clusters (our real number) based on 1460 is:
import math proc poisson(k: int, λ: float): float = #result = pow(λ, k.float) / fac(k).float * exp(-λ) result = exp(k.float * ln(λ) - λ - lgamma((k + 1).float)) import ggplotnim, sequtils let xs = linspace(0.0, 2000.0, 2001) let ys = xs.mapIt(poisson(it.int, 1460.0)) let df = toDf(xs, ys) ggplot(df, aes("xs", "ys")) + geom_line() + ggsave("/home/basti/org/Figs/poisson_1460.pdf")
Our 1610 is pretty much in nowhere's land….
I guess we do need to investigate a bit after all.
The most likely explanation seems to be that there is something off about the relevant times associated or we use too much of the background data or too much / little of the tracking data.
Not sure about the best way to check these things.
[X]Is the plotBackgroundCluster counting wrong? -> It seems correct to me.[X]Create timestamps of all candidates
import nimhdf5, datamancer, times import ingrid / tos_helpers const paths = ["/home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5", "/home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5"] proc readTimestamps(f: string): DataFrame = withH5(f, "r"): let df = readDsets(h5f, likelihoodBase(), commonDsets = @["timestamp"]) .mutate(f{int -> string: "Date" ~ fromUnix(`timestamp`).format("YYYY-MM-dd HH:mm")}) result = df var df = newDataFrame() for p in paths: df.add readTimestamps(p) echo df df.writeCsv("/home/basti/org/resources/candidate_cluster_dates.csv", sep = '\t')
./resources/candidate_cluster_dates.csv contains the timestamps of all candidate clusters!
[ ]Create time series plot of the combined non tracking + tracking datasets -> then we see if there's any overlap that shouldn't be there.[ ]Maybe it's related to specific energies? -> Plot background rate over entire chip
plotBackgroundRate \ /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --centerChip 3 \ --combName "MLP@0.91+line" \ --combYear "2018" \ --title "Background rate over whole chip, MLP@91 % + line veto," \ --showNumClusters \ --region crAll \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.2 \ --outfile background_limitPaper_rate_crAll_mlp_0.95_scinti_fadc_line.pdf \ --outpath ~/org/Figs/statusAndProgress/backgroundRates/ \ --quiet
[BROKEN LINK: ~org/Figs/statusAndProgress/backgroundRates/background_limitPaper_rate_crAll_mlp_0.95_scinti_fadc_line.pdf]
plotBackgroundRate \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --centerChip 3 \ --combName "MLP@0.91+line" \ --combYear "2018" \ --title "Candidate rate over whole chip, MLP@91 % + line veto," \ --showNumClusters \ --region crAll \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.2 \ --outfile rate_real_candidates_rate_crAll_mlp_0.95_scinti_fadc_line.pdf \ --outpath ~/org/Figs/statusAndProgress/trackingCandidates/ \ --quiet
Combined:
plotBackgroundRate \ /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --centerChip 3 \ --names "Background" --names "Background" --names "Candidates" --names "Candidates" \ --title "Rate over whole chip, MLP@91 % + line veto," \ --showNumClusters \ --region crAll \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.2 \ --outfile rate_real_candidates_vs_background_rate_crAll_mlp_0.95_scinti_fadc_line.pdf \ --outpath ~/org/Figs/statusAndProgress/trackingCandidates/ \ --logPlot \ --hideErrors \ --quiet
-> Hmm, it really seems like there is just a smidge more in the candidate dataset in terms of rate. Now, sure, this may still just be down to some bug in the total time. Alternatively, it may just be a real background effect due to the tilting of the magnet. The amount of e.g. orthogonal events increases.
[ ]Let's try to understand the missing time again. INTERESTING FINDING: The table we have with the active and passive times in
statusAndProgressshows:Total 180.3041 3526.36 161.0460 3157.35 3706.66 3318.38 0.89524801 so 3157.35 h for background. I just noticed that in the
plotBackgroundRateabove we have 3158.5739 hours for background and 159.8027 hours for tracking! Do we just assign the wrong numbers in the table above or in this code? The difference is: 1.2433 h for tracking 1.2239 h for background That's a pretty close MATCH![X]See where the above table comes from again! Probably one of our "number of tracking" related scripts? -> Yes, it comes fromwriteRunList./../CastData/ExternCode/TimepixAnalysis/Tools/writeRunList/writeRunList.nim[X]The major distinction is:likelihoodof course just sums up theeventDurationwriteRunListlooks atgetExtendedRunInfo, which uses the tracking fields of the H5 file
[ ]The difference is still pretty large! If anything it should amount to a few events being counted in one or the other. -> Maybe it's a time zone issue? Potentially we use the wrong indices even for different cases. :/ -> UsingDateTimefor parsing the trackings inparseTrackingforgetExtendedRunInfodoes not change the result ofwriteRunList! That's good news I guess.
[X]WHY does thesanity.logfile say tracking time of 161 hours? -> Because we ran it without the candidate files, ergo there IS no tracking. The 161.1 h are those based on the 19.x tracking ratio!
UPDATE: I may have found the "missing time"!!
The H5 output files after likelihood have less elements in the
eventNumber and timestamp datasets than there are events even in
the chip 3 dataset (let alone the other chips of course!).
Maybe this also affects how the totalDuration is computed? Leading
to a wrong number?
-> Uhh it turns out there are usually at least ~1 event that leads
to TWO clusters that pass the cuts in a single event.
Are we sure that this is not a bug? Those are the events that end up
giving "more" entries in the chip 3 datasets compared to the run
global datasets.
[X]Does the same happen in non tracking data? -> Yup it happens as well.[X]Is it possible these are noisy hits? -> Having looked at one background run comparing event numbers with thehitsdataset it seems like at least one of these clusters is always < 5 pixels. So I suppose the answer is likely yes. This certainly explains the discrepancy.
1.86.
[ ]Compare the number of expected vs real candidates for each Run-2 and Run-3 separately! Effect of scintillator?[ ]What does the "rate" of candidates look like as a function of time since start of tracking? That could be an indicator for more background under larger tilt (could even correlate to actual magnet tilt in theory)
1.86.1. 'Missing time' stuff
SUMMARY: The summary is that the determination of the tracking times was flawed. We used the total active ratio time instead of the durations of each run in terms of the eventDuration. In addition one run (89) has 10.3 hours missing due to the terminal getting stuck. The effective times in the output H5 files are correct however (so 159.8 h of solar tracking data)!
[X]I added the indices of the trackings to theRunTimeInfoobject so thatExtendedRunInfocontains these. In addition the event durations for each of these are then computed.[X]added this towriteRunInfothat it also computes the time based on theeventDurationfields
Preliminary results:
- It seems it's not that one is always larger than the other! For Run-3 the information from end - start is larger instead of smaller
- Individual trackings seem to have very weird numbers for the end-start time method!
writeRunList -b ~/CastData/data/DataRuns2017_Reco.h5 -c ~/CastData/data/CalibrationRuns2017_Reco.h5
writeRunList -b ~/CastData/data/DataRuns2018_Reco.h5 -c ~/CastData/data/CalibrationRuns2018_Reco.h5
So: for Run-2: active tracking duration: 94.12276972527778 active tracking duration from event durations: 92.80168749194445 for Run-3: active tracking duration: 66.92306679361111 active tracking duration from event durations: 67.00656808222222
But: there are very weird cases like run 89 says:
Run: 89 activeTrackingTime = 3181 Tracking: 0 duration from eventDuration = 4927.829242349919
which seems completely broken for the activeTrackingTime (end-start)
!!!
Investigating…
[X]The actual tracking end-start time seems perfectly fine!:
Run: 89 activeTrackingTime = 3181 compare tracking: 5882 Tracking: 0 duration from eventDuration = 4927.829242349919
where the 5882 s is the end-start! So the active ratio time is broken? What the.
Indeed the "active ratio" comes out to only 0.54 for run 89. Is it such a noisy run or what?
Quote from ./data_taking_2017_runlist.html
NOTE: During Run 89 the byobu buffer containing TOS got stuck on due to <F7> being pressed (which eventually pauses the thread). Was called by Cristian at roughly. I fixed the issue. Therefore the length given in the table is misleading, as it does not show the actual time of data taking of that run.
So the run is 1 day and 3 hours "long", but in reality the majority of the time the detector was NOT taking data (from 19:35 to 5:55 the next day!). For this run (and possibly others to some extent) our calculation of a "run length" is wrong!
Indeed, from the timestamps of Run 89: Index 6157 = 1510511751 Index 6158 = 1510548717 Difference = 36966, / 3600 = 10.268 h As expected, "missing" about 10.3 hours
How to compute length correctly then?
[ ]Compute time delta between successive events in all background runs. Any other cases of more than, say 10 s? -> First do this then decide what to do.
import nimhdf5, times import ingrid / tos_helpers const path = ["/home/basti/CastData/data/DataRuns2017_Reco.h5", "/home/basti/CastData/data/DataRuns2018_Reco.h5"] proc study(h5f: H5File, r: int) = let ts = h5f[recoBase() & $r & "/timestamp", int] var last = ts[0] for i in 1 ..< ts.len: if ts[i] - last > 10 or ts[i] - last < 0: echo "Difference = ", ts[i] - last, " s in run = ", r, " at ", ts[i].fromUnix last = ts[i] for p in path: withH5(p, "r"): let finfo = getFileInfo(h5f) for run in finfo.runs: h5f.study(run)
The last 3 are weird. At first I thought maybe timezone stuff, but they are all close together (so summer / winter time doesn't make sense 3 times in 2 days). 89 we know about and the others are short (but also weird of course!) Checking the logs… -> No entries in the logs about any of the other runs appearing there.
Daylight savings time came into effect on March 25 in 2018. Maybe there was something funky going on with the time of the computer? And TOS used the computer time as a reference to write the timestamps? Well, yeah. See the time stamps that are negative 1 hour roughly…
So, what does it imply? We should:
- active tracking time should be computed within the tracking. Use tracking end - start as tracking length and active as sum of duration.
- Subtract the ~10 hours in run 89. The DST stuff in run 180 and 182 we'll just ignore. The total time should come out correct regardless I hope. At most off by 1 hour.
[X]Implemented![ ]Think about fixing the DST time stuff in runs 180 and 182. It should be quite easy to change the raw data files.
writeRunList -b ~/CastData/data/DataRuns2017_Reco.h5 -c ~/CastData/data/CalibrationRuns2017_Reco.h5
writeRunList -b ~/CastData/data/DataRuns2018_Reco.h5 -c ~/CastData/data/CalibrationRuns2018_Reco.h5
NOTE:
The active non tracking time remains the same even after correcting
for the dead time of course! That is because the activeRatio
calculation already took care of 'correcting' for the missing data!
1.86.2. Expected counts and clusters by Run-2/3 separately
Let's look at the number of clusters in background / tracking for Run-2 and 3 separately now.
Maybe we find something interesting.
plotBackgroundClusters \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ --outpath ~/org/Figs/statusAndProgress/numClustersByRunPeriod/ \ --suffix "mlp_candidates_run2" \ --energyMin 0.2 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0
plotBackgroundClusters \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --outpath ~/org/Figs/statusAndProgress/numClustersByRunPeriod/ \ --suffix "mlp_candidates_run3" \ --energyMin 0.2 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0
plotBackgroundClusters \ /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --outpath ~/org/Figs/statusAndProgress/numClustersByRunPeriod/ \ --suffix "mlp_background_run2" \ --energyMin 0.2 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0
plotBackgroundClusters \ /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --outpath ~/org/Figs/statusAndProgress/numClustersByRunPeriod/ \ --suffix "mlp_background_run3" \ --energyMin 0.2 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0
Putting these clusters from the figures above into a table and computing expected vs observed numbers:
| Run | Type | Clusters | Time | Expected | Observed/Expected |
|---|---|---|---|---|---|
| Run-2 | Background | 20731 | 2144.12 | 897.27816 | 1.1144816 |
| Run-3 | Background | 7896 | 1012.68 | 522.45933 | 1.1675550 |
| Run-2 | Tracking | 1000 | 92.8017 | ||
| Run-3 | Tracking | 610 | 67.0066 |
So we are slightly closer to the expected number in Run-2 compared to Run-3. In Run-3 we have the scintillator. We see 16.7% too many in Run-3 and only 11.1% too many in Run-2. If the scintillator helped to prevent this from happening, it is not seen (it would need to be the inverse, less too many in Run-3).
1.86.3. Rate of candidates by time since start of tracking
So, then let's look at the rate of candidates seen as a function of their timing within when they happened during a tracking. If it is a systematic effect due to the tilt of the magnet that should be evident from there (most seen beginning and end of shift).
We need:
- times of all clusters
- start and end of each tracking
import nimhdf5, datamancer, times, options import ingrid / [tos_helpers, ingrid_types] const paths = ["/home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5", "/home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5"] proc readTimestamps(f: string): DataFrame = withH5(f, "r"): let fInfo = getFileInfo(h5f) let df = readDsets(h5f, likelihoodBase(), chipDsets = some((chip: 3, dsets: @["centerX", "centerY", "energyFromCharge"])), commonDsets = @["timestamp"]) .mutate(f{int -> string: "Date" ~ fromUnix(`timestamp`).format("YYYY-MM-dd HH:mm")}) # based on tracking, insert time since tracking start result = newDataFrame() for (tup, subDf) in groups(df.group_by("runNumber")): let run = tup[0][1].toInt # find tracking info for this run let rInfo = getExtendedRunInfo(h5f, run, rtBackground, basePath = likelihoodBase()) doAssert rInfo.trackings.len > 0 for tr in rInfo.trackings: echo tr, " for ", subDf proc getTracking(trs: seq[RunTimeInfo], t: int): RunTimeInfo = let tstamp = t.fromUnix for tr in trs: if tstamp >= (tr.t_start - initDuration(minutes = 1)) and tstamp <= (tr.t_end + initDuration(minutes = 1)): return tr doAssert false, "Could not find any tracking! " & $tstamp & " vs " & $trs # determine which run is the correct one for each candidate for c in rows(subDf): let t = c["timestamp"].item(int) let tr = getTracking(rInfo.trackings, t) let ss = fromUnix(t) - tr.t_start echo "Since start = ", ss var c = c c["sinceStart"] = ss.inSeconds() c["trackingProgress"] = ss.inSeconds() / (tr.t_length).inSeconds() result.add c var df = newDataFrame() for p in paths: df.add readTimestamps(p) let dfF = df.filter(f{`energyFromCharge` >= 0.2 and `energyFromCharge` <= 12.0}) import ggplotnim ggplot(dfF, aes("sinceStart")) + geom_histogram(bins = 50) + margin(top = 1.5) + ggtitle("# of candidates in time bins since start of tracking") + ggsave("/home/basti/org/Figs/statusAndProgress/trackingCandidates/count_since_start.pdf") ggplot(dfF, aes("trackingProgress")) + geom_histogram(bins = 30) + margin(top = 1.5) + ggtitle("# of candidates in time bins since start of tracking (fraction of tracking)") + ggsave("/home/basti/org/Figs/statusAndProgress/trackingCandidates/count_since_start_tracking_progress.pdf") ggplot(dfF, aes("sinceStart")) + geom_density() + margin(top = 1.5) + ggtitle("Density of candidates since start of tracking") + ggsave("/home/basti/org/Figs/statusAndProgress/trackingCandidates/density_since_start.pdf") ggplot(dfF, aes("trackingProgress")) + geom_density(adjust = 0.5) + margin(top = 1.5) + ggtitle("Density of candidates since start of tracking (fraction of tracking)") + ggsave("/home/basti/org/Figs/statusAndProgress/trackingCandidates/density_since_start_tracking_progress.pdf") df.writeCsv("/home/basti/org/resources/candidate_cluster_dates.csv")
Well, the take away is that there does not seem to be an obvious time dependence:
In particular the counts since start progress plot doesn't show anything statistically significant, even if we vary the number of bins.
So then, statistical outlier? Or do we overestimate our background time / underestimate our background rate by a factor of about 10-15%?
How would we check the latter, outside of what we currently do, namely compute the total duration of events?
Otherwise we could produce background rate plots as a function of 90 min time intervals? Given that trackings are roughly 90 min long. This would allow us to verify if there is some weird time dependent behavior.
import nimhdf5, datamancer, times, options import ingrid / [tos_helpers, ingrid_types] const paths = ["/home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5", "/home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5"] proc readTimestamps(f: string): DataFrame = withH5(f, "r"): let fInfo = getFileInfo(h5f) let df = readDsets(h5f, likelihoodBase(), chipDsets = some((chip: 3, dsets: @["centerX", "centerY", "energyFromCharge"])), commonDsets = @["timestamp"]) .mutate(f{int -> string: "Date" ~ fromUnix(`timestamp`).format("YYYY-MM-dd HH:mm")}) # based on tracking, insert time since tracking start result = df result["Hour"] = df["timestamp", int].map_inline(fromUnix(x).utc().hour().int) var df = newDataFrame() for p in paths: df.add readTimestamps(p) let dfF = df.filter(f{`energyFromCharge` >= 0.2 and `energyFromCharge` <= 12.0}) echo "Expected in tracking: ", dfF.len.float / 19.602 ggplot(dfF, aes(x = factor("Hour"))) + geom_bar() + ggtitle("Number of cluster in background data for each hour in the day") + ggsave("/home/basti/org/Figs/statusAndProgress/moreCandidatesThanExpected/cluster_per_hour_day.pdf") import ggplotnim, sequtils from ginger import transparent proc histogram(df: DataFrame): DataFrame = ## Calculates the histogam of the energy data in the `df` and returns ## a histogram of the binned data ## TODO: allow to do this by combining different `File` values let (hist, bins) = histogram(df["energyFromCharge"].toTensor(float).toSeq1D, range = (0.0, 20.0), bins = 40) result = toDf({ "Energy" : bins, "Counts" : concat(hist, @[0]) }) var dfH = newDataFrame() for (tup, subDf) in groups(dfF.group_by("Hour")): let hour = tup[0][1].toInt dfH.add histogram(subDf).mutate(f{"Hour" <- hour}) echo dfH ggplot(dfH.filter(f{`Counts` > 0.0}), aes("Energy", "Counts", color = factor("Hour"))) + geom_histogram(hdKind = hdOutline, stat = "identity", position = "identity", fillColor = transparent, lineWidth = 1.0) + scale_y_log10() + #scale_fill_discrete() + margin(top = 1.5) + ggtitle("Background counts split by hour of day") + ggsave("/home/basti/org/Figs/statusAndProgress/moreCandidatesThanExpected/rate_by_hour_of_day.pdf") #ggplot(dfF, aes("trackingProgress")) + # geom_histogram(bins = 30) + # margin(top = 1.5) + # ggtitle("# of candidates in time bins since start of tracking (fraction of tracking)") + # ggsave("/home/basti/org/Figs/statusAndProgress/trackingCandidates/count_since_start_tracking_progress.pdf")
Going by these plots, in particular
(
is too busy)
shows that there is no real difference between the number of clusters
recorded at different hours of the day.
The variation we do see is expected:
- between 5 and 8 am are the shifts (and hence less statistics there)
- in the afternoon the calibration runs typically took place, taking some statistics away too
A few more things to look at:
- candidates for a different case, e.g. lower efficiency with septem veto
- lnL with septem and line veto
- run the likelihood again with fake tracking times. I.e. replace the indices by times that are shifted by two hours forward for example. Just don't know if that won't break anything. I don't think I ever stopped a run after a tracking, but maybe I did. (checked: well, once because of a GRID measurement at 10:40am and once at around 8:50am)
[X]Add--trackingsupport tocreateAllLikelihoodCombinations
We want to run:
- LnL for [0.8, 0.9] septem + line, line only
- MLP for [0.85, 0.9] septem + line, line only
LnL cases:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkLogL, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.8 --signalEfficiency 0.9 \ --out ~/org/resources/lhood_lnL_tracking_08_08_23/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 12 \ --dryRun
^– running now :FINISHED:
MLP cases:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkMLP, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/10_05_23_sgd_gauss_diffusion_tanh300_mse_loss/mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 \ --out ~/org/resources/lhood_mlp_tanh300_mse_tracking_08_08_23/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 12 \ --dryRun
^– running now :FINISHED:
UPDATE: I forgot to add the --tracking flag!
[X]Another idea that just came to me: based on the background rate it does after all look as if the excess is mainly below 1 keV (but not entirely). How many clusters do we get if we useenergyMin = 1.5?
Background:
plotBackgroundClusters \ /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --outpath ~/org/Figs/statusAndProgress/numClustersByRunPeriod/ \ --suffix "mlp_background_run2_run3_energyMin_1.0" \ --energyMin 1.0 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0
plotBackgroundClusters \ /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ /home/basti/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ --outpath ~/org/Figs/statusAndProgress/numClustersByRunPeriod/ \ --suffix "mlp_background_run2_run3_energyMin_1.5" \ --energyMin 1.5 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0
Candidates:
plotBackgroundClusters \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --outpath ~/org/Figs/statusAndProgress/numClustersByRunPeriod/ \ --suffix "mlp_candidates_run2_run3_energyMin_1.0" \ --energyMin 1.0 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0
plotBackgroundClusters \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --outpath ~/org/Figs/statusAndProgress/numClustersByRunPeriod/ \ --suffix "mlp_candidates_run2_run3_energyMin_1.5" \ --energyMin 1.5 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0
Background: 2524 clusters @ 1.5 keV, 4145 @ 1 keV Candidates: 136 clusters @ 1.5 keV, 219 @ 1 keV
What does this mean for the ratio? Ratio of: 18.5588235294 @ 1.5 keV, 18.9269406393 @ 1 keV whereas with our real ratio of 19.7 we would have expected 128.121827411 clusters (@ 1.5 keV), 210 @ 1 keV
What does the Poisson look like for that?
import math, ggplotnim, sequtils proc poisson(k: int, λ: float): float = result = exp(k.float * ln(λ) - λ - lgamma((k + 1).float)) let xs = linspace(0.0, 150.0, 151) let ys = xs.mapIt(poisson(it.int, 128.12)) let df = toDf(xs, ys) ggplot(df, aes("xs", "ys")) + geom_line() + geom_line(aes = aes(x = 136, yMin = 0.0, yMax = 0.04), color = "red") + ggsave("/home/basti/org/Figs/poisson_128.12.pdf")
./Figs/poisson_128.12.pdf -> Which is much more in line with a statistical fluke! In particular for the 1 keV case it is pretty much as expected!
Based on this (for the time being) I'm tempted to assume the reason may be down to some sort of noise contribution (given that we know their is significant noise in the hall during tracking motor activity. Maybe enough to sometimes cause small power fluctuations and thus create activity on the chip?
1.87.
Ouch, I just realized that the likelihood combinations I've been computing were for the non tracking data………………..
Maybe run overnight?
So compute again tomorrow/today:
LnL cases:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkLogL, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.8 --signalEfficiency 0.9 \ --out ~/org/resources/lhood_lnL_tracking_08_08_23/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --tracking \ --jobs 8 \ --dryRun
^– running now :FINISHED:
MLP cases:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkMLP, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/10_05_23_sgd_gauss_diffusion_tanh300_mse_loss/mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 \ --out ~/org/resources/lhood_mlp_tanh300_mse_tracking_08_08_23/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --tracking \ --jobs 4 \ --dryRun
^– running now :FINISHED:
1.88.
Further questions about LLNL:
- why is the carbon spacer rotated in the X-ray finger measurement? How do we verify its rotation and why is it like that in the first place? -> From what I can gather from the PhD thesis it looks like it may never have been the intention to have the spacer be centered?
- why does the axion image presented in the DTU thesis on page ~80 from the raytracing of Michael P. (LLNL) look symmetric?
Our raytracer:
- reproduce X-ray finger data
- produce axion image for 3' sized source
- axion image in focal spot for our emission model
- what does sampling look like in Christophs code?
-> Uhhh, he sampled for Chameleons!
in our MSc axion limit code we just assumed:
// in case of the standard axion radiation, we use 1/100 of the solar radius as // the origin of axion radiation. In that region we assume homogeneous emission r = radius * 1e-1 * _randomGenerator->Rndm(); //r = radius * _randomGenerator->Rndm(); _randomGenerator->Sphere(x,y,z,r);
uniform sampling from inner 10%.
- where is Christoph's code?
- we should be able to sample Primakoff using
readOpacityFile, no? -> Yes, did that and works fine. Question, see below. [ ]CreatesanityCheckblock inraytracer[ ]Modify code for radius sampling plot to show as percentage of solar radius.
[ ]Where does relation to calculate Primakoff from emission come from? Redondo does not really cover that in his 2013 paper. Johanna writes Raffelt 2006 about it. -> Asked Johanna. -> She said look in her MSc thesis, Georg G. Raffelt. “Plasmon Decay Into Low Mass Bosons in Stars”. In: Phys. Rev. D 37 (1988) -> This is the same eq. as eq. (6) in.
- Images from CAST: -> Telescope can be seen rotated in fig!
1.89.
Raytracing woes with the LLNL telescope:
Turns out the bug was that I was rotating the mirrors twice in some sense. The cones DON'T have to be moved of course, because they are already angled by the required angles!!
1.89.1. 'Measurements' to find the bug
wire = -21.95418312639536 #-21.70380736885296,
top red = -14.20370513205117 for y = 0
top red = -19.13439190189884 for y = -4.631203279012007
top red for "correct" case = -24.42074328637678,
correct difference = -4.631203279012007
import math const lMirror = 225.0 const angle = 0.579 echo sin(3 * angle.degToRad) * lMirror
highest point cylinder: -23.86579369877846, lowest point cylinder: -30.86906662133978,
import math let y1 = -23.86579369877846 let y2 = -30.86906662133978 echo y2 - y1
highest point cone:, -23.65804435583882 lowest point cone: -37.66217273447373,
import math let y1 = -23.65804435583882 let y2 = -37.66217273447373 echo y2 - y1
1.90.
(Updated )
[X]Simulate LLNL telescope as "full" telescope[X]Simulate LLNL telescope as "double LLNL" telescope -> Meaning to just mirror what we have and place a "second bore" with a "second telescope" rotated by 180° and see what image looks like[ ]Create old raytracing result for all results we create with new one and compare[X]Check LLNL mirrors -> Found a few issues with them! The height of the first layers was wrong.[X]I think the better approach would be to compute the relevant mirror parameters based on:
- the known R1 values and angles for the first set of mirrors
- the known R5 values and angles for the second set of mirrors
- assume
x_sepis exactly 4 mm in the z direction
That gives a result that guarantees it will conform to the numbers of the thesis.
1.91.
[ ]Add the plots in ./Figs/statusAndProgress/rayTracing/interactiveRaytraceDevelopment/ tostatusAndProgress. The commands to generate them are inzsh_history
1.92.
./Figs/statusAndProgress/rayTracing/debugAxionImageDifference/
-> based on inner 20% of solar radius
-> using ./../CastData/ExternCode/AxionElectronLimit/resources/solar_model_dataframe_fluxKind_fkAxionPhoton_0.989AU.csv
Very different solar emission. But these should not really show such a different in the end result, I think.
Trying with 15% inner radius.
1.93.
[ ]Write short section about raytracer development[ ]Add plots in and mention what they show. ./Figs/statusAndProgress/rayTracing/debugAxionImageDifference
I've learned a lot of things this morning…
See the sections in statusAndProgress:
[BROKEN LINK: sec:llnl_telescope:info_from_nustar_phd] (and below) and the
[BROKEN LINK: sec:raytracing].
- DTU thesis about NuSTAR optic
- led to the MTRAYOR raytracer developed in Yorick
- Yorick is an interpreted language developed at LLNL in the 90s
- manual of MTRAYOR led to a public FTP server from DTU with source code -> ./../src/mt_rayor/ ./../src/yorick/ ./../Documents/ftpDTU/
- found a talk by Michael Pivovaroff about axions, CAST, IAXO
[BROKEN LINK: sec:llnl_telescope:pivovaroff_talk]
-> mentions telescope was at PANTER and raytracing image was done
for source at infinity…
- mentions Julia and Jaime were working on a paper about NuSTAR for axion/ALP limit from solar core data
1.94.
After finishing the mail ./Mails/llnlAxionImage/llnl_axion_image.html yesterday, it's time to finally continue with the actual work on the thesis.
The following things came up since then:
[ ]Potentially use the X-ray finger after all to determine the position. If we decide that:[ ]determine position of the X-ray finger[ ]determine the center position of the following simulations:
- X-ray finger without graphite spacer or detector window
- X-ray finger with graphite spacer
- X-ray finger with graphite spacer and detector window
in order to be able to gauge where the real center is, given the simulation.
[X]Fix the RNG sampling bug in the old raytracer[X]Verify what the axion image looks like
[X]Verify the uniform disk sampling is correct in the old raytracer -> The disk sampling works correctly. See ./Misc/sampling_check_random_disk_old_raytracer.nim which will be added as a sanity check in the old raytracer.[X]Implemented as sanity check.
[ ]Redo the axion image: Ideally with the new raytracer. In any case we need:[ ]correct reflectivity[ ]ImageSensorwhich does not purely count, but includes intensity[ ]also addImageSensorRGB
[ ]detector window + detector strongback
[ ]Determine the rotation angle of the graphite spacer from the X-ray finger data
1.94.1. Old raytracer sanity check
For the sanity check we ran:
./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_sun_at_1550mm" --sanity
(the main arguments don't really matter).
The files are:
And after the RNG fix:
1.94.2. Compute axion image using old raytracer after RNG bug fixed
./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_sun_at_1500mm" ./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_sun_at_1487.93mm" ./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_sun_at_1530mm"
The resulting plots for 1500mm, 1530mm and 1487.93mm. The real 1487 equivalent is still pending.
We can see that the 1530mm result is very similar to our new raytracing result (even though this includes the real reflectivities). Comparison:
Except the new raytracing result is a bit "wider" than the old one. But maybe that is due to the real reflectivity? (I don't think so), but let's compute it without reflectivity as well:
./raytracer --ignoreDetWindow --ignoreGasAbs --suffix "_sun_at_1530mm" --ignoreReflectivity
What we can clearly see though is that the center of the old raytracer is wrong.
1.95. , and
TODOs from yesterday:
[ ]Potentially use the X-ray finger after all to determine the position. If we decide that:[ ]determine position of the X-ray finger[ ]determine the center position of the following simulations:
- X-ray finger without graphite spacer or detector window
[X]window implemented!
- X-ray finger with graphite spacer
- X-ray finger with graphite spacer and detector window
in order to be able to gauge where the real center is, given the simulation.
- X-ray finger without graphite spacer or detector window
[ ]Redo the axion image: Ideally with the new raytracer. In any case we need:[ ]correct reflectivity -> Reflectivity can be implemented by: new materialXrayMetal, which mostly acts likeMetal, but its attenuation is simplycolor(reflectivity, reflectivity, reflectivity), meaning that after multiplying it with other colors (e.g. from a light), the color is simply suppressed by that amount. If input iscolor(1,1,1)from a source, it simply suppresses byreflectivityin each frequency. Only question is if it should really be fullreflecitityin each channel, or rather a fraction of it.[ ]ImageSensorwhich does not purely count, but includes intensity[ ]also addImageSensorRGB
[ ]detector window + detector strongback
[X]Determine the rotation angle of the graphite spacer from the X-ray finger data -> do now. X-ray finger run:->
 -> It comes out to 14.17°!
But for run 21 (between which detector was dismounted of course):
-> It comes out to 14.17°!
But for run 21 (between which detector was dismounted of course):
 -> Only 11.36°!
That's a huge uncertainty given the detector was only dismounted!
3°.
-> Only 11.36°!
That's a huge uncertainty given the detector was only dismounted!
3°.[X]Apply rotation of graphite spacer to axion image[X]In new raytracer
[ ]Implement detector strongback in new raytracer[X]NOTE: It seems like the -83 mm matches perfectly if we use 1530mm as the focal length. What.[ ]UNDERSTAND where / how to calculate the 83! Given that we cannot reproduce it. What is it equivalent to?
import math const x = 83.0 const bore = 21.5 let rx = x - bore const α = 0.579.degToRad const r1 = 63.006 const lM = 225.0 echo rx let z = r1 - sin(α) * lM/2 echo z ## r1 = rx + sin(α) * lM/2 #let αx = arcsin((r1 - rx)/(lM/2)).radToDeg #echo αx #echo r1 - sin(αx) * lM/2
[X]The
yLdisplacement for the telescope in theposvariable was performed "twice". Because we shift the mirror to thesetConeproc to its center to perform the rotation. That already corresponds to moving the mirror "down" to its center aligning with the magnet bore. -> The thing is: if we include the yL displacement, I noticed that a) the focal spot is not actually in the perfect center (see above about 83mm) and b) I noticed that the focus seems to be better when I move down the telescope "too far". As I think I understand now, by moving it down a bit more by a full mirror translation, I was actually putting it into the correct position, i.e. the one where we do not apply additional shifts topos.Comparing the three cases, down, none and up:
[X]Create plot with explicit displacement half down[X]Create plot with explicit displacement half up[X]Plot with new correct (?) placement- [BROKEN LINK: ~org/Figs/statusAndProgress/rayTracing/finalizeAlignment/axion_image_skParallelXrayFinger_fixed_alignment_yL_removed.pdf]
We can see that the "no displacement" aligns best with the center at 83mm displacement BUT does not actually show the best focal spot. The issue is that at the minus displacement, the lowest mirror is NOT AT ALL aligned with the magnet bore. It is completely below the bore! Do we see an effect that the telescope is actually worse at the lower layers due to their smaller radii? So if those are included the image generally becomes worse? In reality of course the total flux may go up? And light from the bottom that hits the top layers if moved up, have to hit at a larger angle causing worse focusing?
And now the
SolarEmissionaxion image for the 'correct' alignment without-yL.
NOTE: The DTU thesis mentions (page 75):
Because of the limited space at the place where the optic had to be installed and the method of which to align it, the number of layers were cut from 14 to 13 layers. With 14 layers, the freedom of movement of the optic inside the vacuum vessel would have been severely limited and possibly caused the optic to hit the vacuum vessel wall during alignment.
The issue is we don't know which layer we are removing! If it's the lowest layer 1, then our "move yL down" raytracing is actually correct.
In addition I THINK this is likely the reason for the fact that the axion image still seems to be wider and narrower than the old raytracer, but more importantly the LLNL result!
Ok:
If we subtract anything from
pos, the focus looks good. But ifwe add anything, the image becomes wide. What?
Does it somehow change the alignment between the mirrors instead of
just moving things up and down?
AHH: Nevermind the above. The effect is just not perfectly
symmetrical. In both directions the image starts to become more
and more elongated! Just by going up it was more evident. I guess
because of which layers are more inside of the bore. The top ones
are "more optimal" in a sense.
 -> Shows the alignment of the mirrors as seen from the focal point at
1530mm and 83mm. I still don't understand why it's 83mm now.
-> Shows the alignment of the mirrors as seen from the focal point at
1530mm and 83mm. I still don't understand why it's 83mm now.
-> To summarize the above: I don't understand why not using layer 0 yields a better result than using it. At least using it is not as bad as using it "fully" (i.e. full bottom layer visible). It would be nice to know which layer was not used at CAST. Probably 14 though, for obvious construction reasons. I believe we are simulating the correct thing, but the result is just not quite right due to the shape of the telescope. That will be my working assumption from here on.
[X]The Wolter equation does not yield 2.75° using 83mm!
import math const x = 83.0 const f = 1500.0 echo arctan(x / f).radToDeg
1.96.
Things I learned today about pbrt:
They actually use an approach where they propagate entire spectra by a single ray. That makes a lot of sense now that I've thought about it! It's the reason why I didn't really find anything about sampling a particular wavelength and the like.
For the attenuation of materials, I believe that's why they have a
medium pointer in the Ray, maybe? That way they keep the information
about the previous medium around.
1.97.
[X]had a big performance regression in TrAXer. Ended up being becausegetMaterialwas aprocand the compiler decided to copy the return value, so that forSolarEmissionwe would copy the CDF & radius & flux data for each ray. We turned it into a template to make sure there won't be a copy.[X]Ahh, I think the current issue is that we are copying aroundXrayMatterall the time too (e.g. inhitwhen assigning thematfield!) BAD idea. -> Made it into aref object, which fixes it :)[X]theXrayMatteris now implemented for the different layers![X]Show plots of setups -> Note: older plots from today used 10x10mm² chip![X]copy from tmp ./../CastData/ExternCode/RayTracing/tmp_31_08_23/[ ]compare both raytracers -> Partially done.[ ]Implement alternative tosSumwhere we keep the entire flux data! This would allow us to directly look at the axion image at arbitrary energies. It would need a 3DSensor3Dto model flux as 3rd. Note: when implementing that it seems a good idea to order the data such that we can copy an entire Spectrum row into the sensor. So the last dimension should be the flux, i.e.[x, y, E]. That way copying theSpectrumshould be efficient, which should be the most expensive operation on it.[ ]implement toggle between XrayMatter behavior for X-rays and light forCamera![ ]FIXCamerausage of attenuation forXrayMatterwhen callingeval! Need to convert toRGBSpectrum
Implementing reflectivity and energy emission:
UPDATE: The plots below still had a bug in the calculation of the actual angle! Therefore the reflectivity values are WRONG. Using file:///home/basti/CastData/ExternCode/xrayAttenuation/playground/llnl_telescope_reflectivity.nim we generated an HDF5 file with the reflectivities for all layer recipes from 0 to 15° in the energy range from 0.03 to 15 keV:
As a reference again the old raytracing code (without reflectivity)
and with reflectivity
THESE PLOTS WERE USING AXION PHOTON COUPLING
We can see the spot becomes a bit smaller when including the
reflectivities (but the absolute sum of flux goes down!)
After implementing the SensorKind to select if we simply count the
times the sensor was hit or take into account the incoming flux by
summing it up (sCount vs sSum), the first test run on a 10x10mm²
(!!! not 14x14) sized image sensor:
The plot above required a very lengthy run time due to some issues
with the performance. We were copying data of the
XrayMatter and
AngleInterpolator. After the fix:
i.e. the same.
Then we implemented the reflectivities for all mirror shells, as
mentioned above:
And finally the axion image on a 14x14mm² image sensor using sSum and reflectivities:
1.98.
[ ]Check the X-ray finger still looks compatible.[X]With perfect mirror and no reflectivity.
./raytracer --width 1200 --maxDepth 10 --speed 10.0 --nJobs 32 --vfov 30 --llnl --focalPoint --sourceKind skXrayFinger --rayAt 1.0 --setupRotation 0.0 --telescopeRotation 0.0 --ignoreWindow --sensorKind sCount
which looks pretty much unchanged to before.
[X]With perfect mirror and including reflectivity.
./raytracer --width 1200 --maxDepth 10 --speed 10.0 --nJobs 32 --vfov 30 --llnl --focalPoint --sourceKind skXrayFinger --rayAt 1.0 --setupRotation 0.0 --telescopeRotation 0.0 --ignoreWindow --sensorKind sSum
UPDATE:
While looking at these plots I realized that they are pretty much
identical, both for the xray fingler as well as for
skParallelXrayFinger:
It turns out I still hadn't finalized the calculation of the
angle
under which the mirrors are hit in scatter for XrayMatter!
Xray finger plot of the code with the angle fixed:
and now we can indeed see that the position of the data seems to
shift a bit between the two! As expected. The parts with the lowest
angles have the highest flux now.
And now for parallel light as well:
at least we now see a minor difference here as well!
So let's finally look at the (now hopefully correct) axion image:
./raytracer --width 1200 --maxDepth 10 --speed 10.0 --nJobs 32 --vfov 30 --llnl --focalPoint \ --sourceKind skSun --rayAt 1.0 --setupRotation 0.0 --telescopeRotation 0.0 \ --ignoreWindow \ --solarModelFile ~/CastData/ExternCode/AxionElectronLimit/resources/solar_model_dataframe_fluxKind_fkAxionPhoton_0.989AU.csv \ --sensorKind sSum # or sCount
This is FOR THE AXION PHOTON COUPLING.
And for the axion electron coupling:
./raytracer --width 1200 --maxDepth 10 --speed 10.0 --nJobs 32 --vfov 30 --llnl --focalPoint \ --sourceKind skSun --rayAt 1.0 --setupRotation 0.0 --telescopeRotation 0.0 \ --ignoreWindow \ --solarModelFile ~/CastData/ExternCode/AxionElectronLimit/resources/solar_model_dataframe.csv \ --sensorKind sSum # or sCount
So we can see differences between the two solar emissions as well as between count and sum (of course).
So comparing the correct cases with the old raytracer:
The differences in the new raytracer are much more pronounced!
NOTE:
[X]I just found where it comes from that the old raytracer seems to useR1for the second set of mirrors too. It's in the call togetMirrorAngleandgetVectoraAfterMirroras well as the hyperbolic/second cone calls!![X]I added the option to debug the differential flux that is being used in
fluxCDF. Enable thewhen falsebranch there, which creates differential fluxes for each radius. Then run:pdfunite `lc -n1 -p` /tmp/all_diff_flux_{suffix}.pdfto combine them all into a single file. We were using the axion photon flux accidentally. But they all look fine. The only thing I'm not certain about is the normalization of them. Might be missing a factor of 1/(2π).!
While trying to understand why the new raytracer produces a bigger axion image, I noticed at some point that there seem to be rays with too large incidence angles on the mirror shells. Debugging this I found a few different things:
[X]tMin -> Added the correcttMincheck in theConeandCylinderintersection tests (tShapeHit > 0->tShapeHit > tMin)[X]thickness -> We implemented the thickness of the mirrors by adding- a section of a disk at the front
- a secondary cone at the height of the thickness
[X]need 2 x thickness to block all light! See the images here for parallel light with an ImageSensor inserted in between the two telescope parts.

 These three are for parallel light.
-> Compute the real height and stack height as given from the numbers
vs as computed by hand. Can we really see that these numbers allow
some light to leak straight?
Why is this?
These three are for parallel light.
-> Compute the real height and stack height as given from the numbers
vs as computed by hand. Can we really see that these numbers allow
some light to leak straight?
Why is this?[X]INSERT PLOT It seems like adding the mirror thickness has a significant effect on the axion image. It becomes quite a bit smaller. The axion image using
SolarEmission(and axion electron flux, see above) first without a mirror thickness:which is the same image we already had before. Using the 2*expected thickness (0.4mm) yields:
(using this value to make sure no parallel rays pass through the telescope to the second set of mirrors). And finally using the NuSTAR 1 arcminute figure error:
surprisingly this is actually even closer to the LLNL result:
Still doesn't show the two side lobes we only see when we use fully parallel light! (Note: on some level their result looks like using parallel light, with a figure error and then zooming in, just saying). Running that:
./raytracer --width 1200 --maxDepth 10 \ --speed 10.0 --nJobs 32 --vfov 30 --llnl --focalPoint \ --sourceKind skParallelXrayFinger --rayAt 1.00 --setupRotation 0.0 --telescopeRotation 0.0 \ --ignoreWindow --sensorKind sSum --mirrorThickness 0.4 --usePerfectMirror=false
Hm, well. Surprisingly large change from the perfect mirror though!
[ ]Compute heights of each layer from R1, R3 and angles etc The telescope should follow the construction: R3,i+1 = R1,i + dglass. Running the following code as part of the telescope construction in the raytracer:
var lastR1 = 0.0 for i in 0 ..< tel.allR1.len: let r1 = tel.allR1[i] r5 = tel.allR5[i] angle = tel.allAngles[i] ## * 1.02 yields 1500mm focal length r2 = r1 - lMirror * sin(angle.degToRad) r3 = r2 - 0.5 * xSep * tan(angle.degToRad) r4 = r5 + lMirror * sin(3.0 * angle.degToRad) let (ySep, yL1, yL2) = calcYlYsep(angle, xSep, lMirror) echo "i = ", i, " R1 = ", r1, " R2 = ", r2, " R3 = ", r3, " lastR1+0.2 = ", lastR1 + 0.2 lastR1 = r1
i = 0 R1 = 63.006 R2 = 60.7323110157061 R3 = 60.71209941495875 lastR1+0.2 = 0.2 i = 1 R1 = 65.60599999999999 R2 = 63.23806825058707 R3 = 63.21701880264519 lastR1+0.2 = 63.206 i = 2 R1 = 68.30500000000001 R2 = 65.83889914563667 R3 = 65.81697693234052 lastR1+0.2 = 65.806 i = 3 R1 = 71.105 R2 = 68.53680377480966 R3 = 68.51397387668443 lastR1+0.2 = 68.50500000000001 i = 4 R1 = 74.011 R2 = 71.34070893282912 R3 = 71.31697134047126 lastR1+0.2 = 71.30500000000001 i = 5 R1 = 77.027 R2 = 74.24676125701171 R3 = 74.22204613684256 lastR1+0.2 = 74.211 i = 6 R1 = 80.157 R2 = 77.26288757878646 R3 = 77.23716000664587 lastR1+0.2 = 77.227 i = 7 R1 = 83.405 R2 = 80.39308800256796 R3 = 80.36631305243878 lastR1+0.2 = 80.357 i = 8 R1 = 86.77500000000001 R2 = 83.64136264166396 R3 = 83.61350538551388 lastR1+0.2 = 83.605 i = 9 R1 = 90.27200000000001 R2 = 87.01271161881355 R3 = 86.98373712642727 lastR1+0.2 = 86.97500000000001 i = 10 R1 = 93.902 R2 = 90.51313506674326 R3 = 90.48300840554474 lastR1+0.2 = 90.47200000000001 i = 11 R1 = 97.66800000000001 R2 = 94.14170661969963 R3 = 94.11035793941267 lastR1+0.2 = 94.102 i = 12 R1 = 101.576 R2 = 97.91227948851467 R3 = 97.87970876573758 lastR1+0.2 = 97.86800000000001 i = 13 R1 = 105.632 R2 = 101.823000866023 R3 = 101.7891382432993 lastR1+0.2 = 101.776
We can see that R3 on the next layer almost matches the R1 + dglass(= 0.2), but is off by about 0.01! This reminded me to check the LLNL notes I wrote ./Doc/LLNL_def_REST_format/llnl_def_rest_format.html where I compute the following numbers in the (almost) last section:
| 63.216 mm | ||||||||
| Layer | = | 0 | r1 | = | 63.006 mm | α | = | 0.603311 ° |
| Layer | = | 1 | r1 | = | 65.6062 mm | α | = | 0.603311 ° |
| Layer | = | 2 | r1 | = | 68.3046 mm | α | = | 0.628096 ° |
| Layer | = | 3 | r1 | = | 71.1049 mm | α | = | 0.653812 ° |
| Layer | = | 4 | r1 | = | 74.0109 mm | α | = | 0.680495 ° |
| Layer | = | 5 | r1 | = | 77.0266 mm | α | = | 0.70818 ° |
| Layer | = | 6 | r1 | = | 80.156 mm | α | = | 0.736904 ° |
| Layer | = | 7 | r1 | = | 83.4035 mm | α | = | 0.766706 ° |
| Layer | = | 8 | r1 | = | 86.7735 mm | α | = | 0.797625 ° |
| Layer | = | 9 | r1 | = | 90.2706 mm | α | = | 0.829702 ° |
| Layer | = | 10 | r1 | = | 93.8995 mm | α | = | 0.862981 ° |
| Layer | = | 11 | r1 | = | 97.6652 mm | α | = | 0.897504 ° |
| Layer | = | 12 | r1 | = | 101.573 mm | α | = | 0.933316 ° |
| Layer | = | 13 | r1 | = | 105.627 mm | α | = | 0.970466 ° |
where we can then see that heer the r1 is slightly off from the second layer. But looking at this I remembered that we learned the glass is not 0.2 mm thick, but 0.21! That is pretty much exactly the amount we miss. We'll change that now in the raytracer as the default and then see what we get with the image sensor in the middle..
The above numbers with 0.21 mm:
i = 0 R1 = 63.006 R2 = 60.7323110157061 R3 = 60.71209941495875 lastR1+0.2 = 0.21 i = 1 R1 = 65.60599999999999 R2 = 63.23806825058707 R3 = 63.21701880264519 lastR1+0.2 = 63.216 i = 2 R1 = 68.30500000000001 R2 = 65.83889914563667 R3 = 65.81697693234052 lastR1+0.2 = 65.81599999999999 i = 3 R1 = 71.105 R2 = 68.53680377480966 R3 = 68.51397387668443 lastR1+0.2 = 68.515 i = 4 R1 = 74.011 R2 = 71.34070893282912 R3 = 71.31697134047126 lastR1+0.2 = 71.315 i = 5 R1 = 77.027 R2 = 74.24676125701171 R3 = 74.22204613684256 lastR1+0.2 = 74.22099999999999 i = 6 R1 = 80.157 R2 = 77.26288757878646 R3 = 77.23716000664587 lastR1+0.2 = 77.23699999999999 i = 7 R1 = 83.405 R2 = 80.39308800256796 R3 = 80.36631305243878 lastR1+0.2 = 80.36699999999999 i = 8 R1 = 86.77500000000001 R2 = 83.64136264166396 R3 = 83.61350538551388 lastR1+0.2 = 83.61499999999999 i = 9 R1 = 90.27200000000001 R2 = 87.01271161881355 R3 = 86.98373712642727 lastR1+0.2 = 86.985 i = 10 R1 = 93.902 R2 = 90.51313506674326 R3 = 90.48300840554474 lastR1+0.2 = 90.482 i = 11 R1 = 97.66800000000001 R2 = 94.14170661969963 R3 = 94.11035793941267 lastR1+0.2 = 94.11199999999999 i = 12 R1 = 101.576 R2 = 97.91227948851467 R3 = 97.87970876573758 lastR1+0.2 = 97.878 i = 13 R1 = 105.632 R2 = 101.823000866023 R3 = 101.7891382432993 lastR1+0.2 = 101.786
It seems like even with the correct thickness of 0.21mm there is still a very slim line! Which is expected because even with 0.21mm there is a very minor difference of another 0.001 mm. Is the real thickness 0.211 per chance? -> I checked the papers again, but couldn't find anything along those lines. Indeed all the NuSTAR references indeed mention 0.21 mm explicitly.
See:

From here I think the next step is to simply continue with the actual work. What the code should look like / produce / etc I don't even know anymore.
1.99.
[ ]Look into
FTOAimplementation in TPAJa, hört sich gut an. Die Dateien liegen an der selben Stelle, wie auch Tobis Daten (tpc08). In allen, außer den ersten beiden Runs von heute (01.09.) ist das FToA eingeschaltet.
-> Take one of those files and try to get the data over to
raw_data_manipulation:) We should create a new datasat for the FTOA data itself.
1.100.
From a few days ago:
[ ]Potentially use the X-ray finger after all to determine the position. If we decide that:[ ]determine position of the X-ray finger[ ]determine the center position of the following simulations:
- X-ray finger without graphite spacer or detector window
[X]window implemented!
- X-ray finger with graphite spacer
- X-ray finger with graphite spacer and detector window
in order to be able to gauge where the real center is, given the simulation.
- X-ray finger without graphite spacer or detector window
X-ray finger without spacer or window:
1.100.1. Important note on compile times
After adding cacheMe to the raytracer to cache the reflectivity
calculations, the compile times took a big nose dive.
However, --profileVM:on --benchmarkVM:on still only say:
prof: µs #instr location 1017114 11944 /home/basti/CastData/ExternCode/datamancer/src/datamancer/formula.nim(901, 6)
649513 5432 /home/basti/CastData/ExternCode/units/src/unchained/define_units.nim(123, 6)
646393 37561 /home/basti/CastData/ExternCode/units/src/unchained/define_units.nim(92, 6)
644839 9477 /home/basti/CastData/ExternCode/units/src/unchained/units.nim(125, 7)
595544 476875 /home/basti/src/nim/nim_git_repo/lib/pure/collections/hashcommon.nim(72, 6)
562153 7683 /home/basti/CastData/ExternCode/units/src/unchained/define_units.nim(128, 6)
455323 44950 /home/basti/src/nim/nim_git_repo/lib/pure/hashes.nim(504, 6)
375883 69856 /home/basti/src/nim/nim_git_repo/lib/pure/hashes.nim(544, 6)
351647 134013 /home/basti/CastData/ExternCode/units/src/unchained/ct_unit_types.nim(212, 6)
317037 220781 /home/basti/src/nim/nim_git_repo/lib/pure/hashes.nim(165, 6)
297331 23030 /home/basti/src/nim/nim_git_repo/lib/pure/collections/tables.nim(375, 6)
292607 41454 /home/basti/src/nim/nim_git_repo/lib/pure/collections/tables.nim(357, 6)
291173 9005 /home/basti/CastData/ExternCode/datamancer/src/datamancer/formula.nim(1108, 7)
274084 214660 /home/basti/src/nim/nim_git_repo/lib/pure/hashes.nim(117, 6)
243133 693 /home/basti/CastData/ExternCode/datamancer/src/datamancer/formula.nim(1049, 6)
238185 61380 /home/basti/src/nim/nim_git_repo/lib/pure/hashes.nim(213, 8)
231923 1492133 /home/basti/src/nim/nim_git_repo/lib/pure/hashes.nim(100, 6)
228833 32625 /home/basti/src/nim/nim_git_repo/lib/pure/collections/tables.nim(341, 6)
213698 56861 /home/basti/CastData/ExternCode/units/src/unchained/define_units.nim(295, 6)
211817 2906 /home/basti/CastData/ExternCode/units/src/unchained/units.nim(312, 7)
180389 1556 /home/basti/CastData/ExternCode/units/src/unchained/units.nim(430, 7)
174624 6494 /home/basti/CastData/ExternCode/units/src/unchained/define_units.nim(349, 6)
153306 1962 /home/basti/CastData/ExternCode/units/src/unchained/define_units.nim(21, 6)
148289 72038 /home/basti/src/nim/nim_git_repo/lib/pure/algorithm.nim(368, 6)
147813 7368 /home/basti/CastData/ExternCode/units/src/unchained/parse_units.nim(218, 6)
136026 36404 /home/basti/src/nim/nim_git_repo/lib/pure/collections/tables.nim(286, 6)
128107 74 /home/basti/CastData/ExternCode/units/src/unchained/define_units.nim(632, 7)
126648 172942 /home/basti/CastData/ExternCode/units/src/unchained/define_units.nim(731, 7)
126216 96136 /home/basti/src/nim/nim_git_repo/lib/pure/asyncmacro.nim(66, 6)
124906 153547 /home/basti/src/nim/nim_git_repo/lib/pure/algorithm.nim(329, 6)
105664 5436 /home/basti/CastData/ExternCode/units/src/unchained/ct_unit_types.nim(188, 6)
99289 36726 /home/basti/src/nim/nim_git_repo/lib/pure/hashes.nim(272, 6)
Hint: mm: orc; threads: on; opt: speed; options: -d:danger
214299 lines; 31.554s; 2.723GiB peakmem; proj: /home/basti/CastData/ExternCode/RayTracing/raytracer; out: /home/basti/CastData/ExternCode/RayTracing/raytracer [SuccessX]
where line 901 in formula.nim points to determineTypesImpl. But
putting echoes at the beginning and end does not actually reproduce
significant chunks of time being spent in there! Unless it's really
some inner logic that takes freaking ages where we get stuck and due
to the recursive nature we "overlook" it.
1.101. and
Let's continue without cacheMe for now for the calculation.
From a few days ago:
[ ]Potentially use the X-ray finger after all to determine the position. If we decide that:
[ ]determine position of the X-ray finger[ ]determine the center position of the following simulations:
- X-ray finger without graphite spacer or detector window
[X]window implemented!
- X-ray finger with graphite spacer
- X-ray finger with graphite spacer and detector window
in order to be able to gauge where the real center is, given the simulation.
- X-ray finger without graphite spacer or detector window
-> Based on the below use a wide-ish spectrum at slightly below 4 keV and a width of 2 keV or so.
[ ]Check whether we correctly take into account the energy information in the emitted flux from the Sun! i.e. not only for the mirrors, but also for the solar emission![X]Check what the actual energy is of the X-ray finger data -> Done using the code snippet fromthesis.org. SeestatusAndProgressX-ray finger section for more. The peak seems to be at 3 keV instead of 8. I suppose they changed the target on the X-ray finger?
1.101.1. X-ray finger simulations
The real plot to compare to:
As mentioned yesterday and seen in the plot, we'll use energies in the range from 2-5 keV.
Of all the different combinations below, the most relevant plot is
likely this one:
in terms of comparison to the real data.
[X]Compute the weighted (by the flux) mean position of each of the three cases -> Seems to be negligible, < 0.03 mm.[ ]Apply gradient descent to compute the best match with the real data.
- Computing the center of each case shown below
Given that the image data is effectively a 2D tensor, we need to compute a weighted mean in x and y where the value at (x, y) is used as weight.
This also writes back a DF of the transformed coordinates.
[ ]Merge this logic intoplotBinaryas it is something that would be useful there too! In particular to see the mean position on the plot as well.
import math proc weightedMean*[T](data: seq[T], weights: seq[T]): T = ## Computes the weighted mean of the input data. doAssert data.len == weights.len, "Must have one weight per data sample." result = T(0.0) var sumW = 0.0 for i in 0 ..< data.len: result += data[i] * weights[i] sumW += weights[i] result /= sumW import std / [strutils, strscans] proc getWidthHeight(fname: string): (int, int) = const wStr = "_width_" const hStr = "_height_" let idx = fname.find(wStr) let fname = fname[idx .. ^1].replace("_", " ") # fuck this let (success, width, height) = scanTuple(fname, " width $i height $i.dat") if success: result = (width, height) else: doAssert false, "Could not parse size from name: " & $fname import std/os import ggplotnim proc getMean(data: ptr UncheckedArray[float], width, height: int, fname: string): (float, float) = ## Computes the mean x, y position let t = fromBuffer[float](data, [width, height]) var xCoords = linspace(0.0, 14.0, width) var yCoords = linspace(0.0, 14.0, height) var xs = newSeqOfCap[float](width * height) var ys = newSeqOfCap[float](width * height) var zs = newSeqOfCap[float](width * height) for x in 0 ..< width: for y in 0 ..< height: xs.add 14.0 - xCoords[x] ## <- this effectively flips `y`! ys.add yCoords[y] zs.add t[y, x].float ## because here we access in order `y, x`! let df = toDf(xs, ys, zs) let outname = fname.extractFilename.replace(".dat", "_transformed") df.writeCsv(outname & ".csv") result = (weightedMean(xs, zs), weightedMean(ys, zs)) var customInferno = inferno() customInferno.colors[0] = 0 # transparent ggplot(df, aes("xs", "ys", fill = "zs")) + geom_raster() + scale_fill_gradient(customInferno) + ggsave("/tmp/" & outname & ".pdf") proc main(fname: string) = let data = readFile(fname) let (w, h) = getWidthHeight(fname) let buf = cast[ptr UncheckedArray[float]](data[0].addr) echo "Input file: ", fname echo "\tMean position (x, y) = ", getMean(buf, w, h, fname) when isMainModule: import cligen dispatch main
cd /tmp ./calc_weighted_mean_xray_finger_pos -f ~/CastData/ExternCode/RayTracing/out/image_sensor_0_2023-09-05T16:16:21+02:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_160000_width_400_height_400.dat ./calc_weighted_mean_xray_finger_pos -f ~/CastData/ExternCode/RayTracing/out/image_sensor_0_2023-09-05T13:30:20+02:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_160000_width_400_height_400.dat ./calc_weighted_mean_xray_finger_pos -f ~/CastData/ExternCode/RayTracing/out/image_sensor_0_2023-09-05T16:32:12+02:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_160000_width_400_height_400.dat ./calc_weighted_mean_xray_finger_pos -f ~/CastData/ExternCode/RayTracing/out/image_sensor_0_2023-09-05T18:49:35+02:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_160000_width_400_height_400.datSo the center moves by less than 0.03mm, which really is pretty negligible!
- Map simulation to real data using gradient descent
- Read raw data and simulation data.
- Convert both to (x, y, z) pairs on the same grid.
- Define translation operation on the data type.
- Define rotation operation on the data type.
- Define test score via e.g. χ² of all pixels
- Define numerical gradient
- Define gradient descent
- Run
The code below depends on the X-ray finger run 189 HDF5 file as well as the binary
.datfile for an X-ray finger simulation.One thing we could learn from the below is that the size of the simulation (especially if the 1 arc minute figure error is used) is larger than the real signal we observed. The likely culprit, I imagine, is the size of the X-ray finger we use in the simulation. Potentially there are other reasons, but the uncertainty on the real X-ray finger makes that seems likely. Possibly only because of some kind of collimator for example.
NOTE: Given how tricky it is to match the simulation to our real data, for the time being I will not continue working on it. The general idea seems sound, but the real data is too low statistics and too noisy to yield a good result. To fix it we would likely need to:
- use an interpolation similar to the background interpolation in the limit calculation (without energy) to get a smoother image
- do some other data preprocessing to get a clearer signal.
import nimhdf5, ggplotnim, options import ingrid / tos_helpers import std / [strutils, tables] proc inRadius(x, y: float, radius: float): bool = let xC = x - 7.0 yC = y - 7.0 result = xC*xC + yC*yC <= radius*radius proc readData(file: string): DataFrame = const run = 189 withH5(file, "r"): # compute counts based on number of each pixel hit proc toIdx(x: float): int = (x / 14.0 * 256.0).round.int.clamp(0, 255) var ctab = initCountTable[(int, int)]() var df = readRunDsets(h5f, run = run, chipDsets = some(( chip: 3, dsets: @["centerX", "centerY"]))) .filter(f{float -> bool: inRadius(`centerX`, `centerY`, 4.5)}) .mutate(f{"xs" ~ toIdx(idx("centerX"))}, f{"ys" ~ toIdx(idx("centerY"))}) let xidx = df["xs", int] let yidx = df["ys", int] forEach x in xidx, y in yidx: inc cTab, (x, y) df = df.mutate(f{int -> float: "zs" ~ cTab[(`xs`, `ys`)].float }, f{float: "xs" ~ `xs` / 255.0 * 14.0}, f{float: "ys" ~ `ys` / 255.0 * 14.0}) .filter(f{float: `zs` > 2.5}) result = df import numericalnim type Grid = object #data: Tensor[float] ## Tensor storing the actual data width: int height: int cX: float = 127.5 cY: float = 127.5 interp: Interpolator2DType[float] shift: (float, float) ## shift in (x, y) rot: float ## Angle to rotate *around the center* scale: float = 1.0 ## Scale entire image by this factor proc toGrid(hmap: DataFrame, w, h: int): Grid = ## Regrids the data to the same gridding used in the simulation, i.e. ## 400x400 pixels. The input data is *not* gridded, but already has count ## numbers associated for each `xidx`, `yidx`. But duplicates remain (I think). # check the following line doesn't change anything #let df = df.unique(["xidx", "yidx"]) var t = zeros[float]([w, h]) echo hmap let zMax = if w > 300: hmap["zs", float].max #percentile(95) else: hmap["zs", float].percentile(95) for idx in 0 ..< hmap.len: let x = (hmap["xs", idx, float] / 14.0 * (w.float - 1.0)).round.int let y = (hmap["ys", idx, float] / 14.0 * (h.float - 1.0)).round.int let z = hmap["zs", idx, float] t[x, y] = clamp(z / zMax.float, 0.0, 1.0) # / zSum * pixPerArea).float #zMax / 784.597 # / zSum # TODO: add telescope efficiency abs. * 0.98 echo "Constructing grid" result = Grid(width: w, height: h, interp: newBilinearSpline(t, (0.0, 255.0), (0.0, 255.0)), # w.float - 1), (0.0, h.float - 1)), shift: (0.0, 0.0), rot: 0.0) echo "Done" #proc normalize(df: DataFrame): DataFrame = # doAssert "zs" in df # result = df.mutate(f{"zs" ~ `zs` / col("zs").max}) proc translate(g: var Grid, to: (float, float)) = g.shift = to proc rotate(g: var Grid, angle: float) = g.rot = angle.degToRad proc `[]`(g: Grid, x, y: int): float = ## Return position `(x, y)` at current translation and rotation ## 1. apply rotation var xR = (cos(g.rot) * (x.float - g.cX) - sin(g.rot) * (y.float - g.cY)) yR = (sin(g.rot) * (x.float - g.cX) + cos(g.rot) * (y.float - g.cY)) ## 2. apply scaling xR = xR / g.scale # Note: dividing by scale effectively decreases size yR = yR / g.scale # for scale < 1, as we change the arguments to interp ## 1b. re-add center xR = xR + g.cX yR = yR + g.cY ## 3. apply translation xR = xR + g.shift[0].float yR = yR + g.shift[1].float if xR < 0.0 or xR > 255.0 or yR < 0.0 or yR > 255.0: result = 0.0 else: result = g.interp.eval(xR, yR) proc score(g1, g2: Grid): float = ## Computes the test score on a fixed grid of 256x256 points for both grids. for y in 0 ..< 256: for x in 0 ..< 256: ## χ² test with weight 1 or Mean Squared Error: result += (g1[x, y] - g2[x, y])^2 proc setParam(g: var Grid, idx: int, val: float) = ## Index: ## - 0: X shift ## - 1: Y shift ## - 2: Rotation ## - 3: Scale case idx of 0: g.shift = (val , g.shift[1]) of 1: g.shift = (g.shift[0], val) of 2: g.rot = clamp(val.degToRad, -180.0, 180.0) of 3: g.scale = clamp(val, 0.1, 2.0) else: doAssert false, "Invalid branch." proc setParams(g: var Grid, val: array[4, float]) = for i in 0 ..< 4: g.setParam(i, val[i]) proc getParam(g: var Grid, idx: int): float = case idx of 0: result = g.shift[0] of 1: result = g.shift[1] of 2: result = g.rot of 3: result = g.scale else: doAssert false, "Invalid branch." template genGrad(name, paramIdx: untyped): untyped = proc `name`(g1: var Grid, g2: Grid, params: array[4, float]): float = ## Computes the numerical gradient along X/Y/rotation for the *current* shift and rotation var h = 1e-8 # some suitable small h let fx = score(g1, g2) # value at current shift g1.setParam(paramIdx, params[paramIdx] + h) # update parameter let fxh = score(g1, g2) # value at shifted by `h` g1.setParam(paramIdx, params[paramIdx]) # update parameter back result = (fxh - fx) / h echo "f(x) = ", fx, " vs f(x+h) = ", fxh, " grad = ", result genGrad(gradX, 0) genGrad(gradY, 1) genGrad(gradRot, 2) genGrad(gradScale, 3) proc gridToDf(g: Grid): DataFrame = # now compute the grid var xs = newSeqOfCap[float](g.width * g.height) var ys = newSeqOfCap[float](g.width * g.height) var zs = newSeqOfCap[float](g.width * g.height) for y in 0 ..< 256: for x in 0 ..< 256: xs.add x.float ys.add y.float zs.add g[x, y] result = toDf(xs, ys, zs) import std/strformat proc plotGrids(g1, g2: Grid, suffix = "") = echo "Plotting the two grids" let df1 = gridToDf(g1) let df2 = gridToDf(g2) var customInferno = inferno() customInferno.colors[0] = 0 # transparent ggplot(df1, aes("xs", "ys", fill = "zs")) + geom_raster(alpha = 0.3) + geom_raster(data = df2, alpha = 0.3) + minorGridLines() + scale_fill_gradient(customInferno) + xlim(0, 256) + ylim(0, 256) + ggsave(&"/tmp/grid_overlay{suffix}.pdf") template genIt(op: untyped): untyped = proc `op`[N: static int](a, b: array[N, float]): array[N, float] = for i in 0 ..< N: result[i] = `op`(a[i], b[i]) genIt(`+`) genIt(`-`) proc `*`[N: static int](val: float, a: array[N, float]): array[N, float] = for i in 0 ..< N: result[i] = val * a[i] proc main(data, sim: string, lr = 0.01, gradTol = 1e-6, absTol = 1.0, maxIter = 1000) = let gData = readData(data).toGrid(256,256) #.normalize() ## XXX: rotate and translate the simulated data, then apply gradient descent to recover the correct orientation. ## That must work, otherwise something is broken. #let gData = readCsv(sim).toGrid(400,400) #normalize() var gSimu = readCsv(sim).toGrid(400,400) #normalize() plotGrids(gData, gSimu) var params = [0'f64, 0, 0, 1.0] var score = score(gSimu, gData) var grad = [gradX(gSimu, gData, params), gradY(gSimu, gData, params), gradRot(gSimu, gData, params), gradScale(gSimu, gData, params)] echo "Starting gradient: ", grad, " score: ", score var i = 0 while abs(lr * abs(grad).max) > gradTol and score > absTol: # gradient descent until next iteration would change # less than some epsilon params = params - lr * grad # update the gradient gSimu.setParams(params) score = score(gSimu, gData) grad = [gradX(gSimu, gData, params), gradY(gSimu, gData, params), gradRot(gSimu, gData, params), gradScale(gSimu, gData, params)] echo "i = ", i, "\n\tθ = ", params, " ε = ", grad, " score: ", score inc i if i >= maxIter: break echo "Final result: ", params, " at gradient: ", grad, " score: ", score plotGrids(gData, gSimu, "_final") when isMainModule: import cligen dispatch main
- No spacer, no window
X-ray finger without spacer & window and with perfect mirrors and thickness 0.21 mm.
./raytracer \ --width 1200 --maxDepth 10 --speed 10.0 \ --nJobs 32 --vfov 30 --llnl --focalPoint \ --sourceKind skXrayFinger \ --rayAt 1.00 \ --ignoreWindow --ignoreSpacer --sensorKind sSum \ --energyMin 2.0 --energyMax 5.0
produced ./../CastData/ExternCode/RayTracing/out/image_sensor_0_2023-09-05T16:16:21+02:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_160000_width_400_height_400.dat from which we produce 4 different plots:
import shell, std/strutils const args = ["", "--switchAxes", "--invertY", "--switchAxes --invertY"] proc toName(arg: string): string = arg.replace(" ", "_").replace("--", "") let infile = "out/image_sensor_0_2023-09-05T16:16:21+02:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_160000_width_400_height_400.dat" for arg in args: let outfile = "/tmp/image_sensor_XrayFinger_sSum_no_spacer_no_window_" & toName(arg) & ".pdf" shell: one: cd "~/CastData/ExternCode/RayTracing/" "./plotBinary" -f ($infile) --dtype float --outfile ($outfile) ($arg)
- Spacer, but no window
./raytracer \ --width 1200 --maxDepth 10 --speed 10.0 \ --nJobs 32 --vfov 30 --llnl --focalPoint \ --sourceKind skXrayFinger \ --rayAt 1.00 \ --ignoreWindow --sensorKind sSum \ --energyMin 2.0 --energyMax 5.0
produced ./../CastData/ExternCode/RayTracing/out/image_sensor_0_2023-09-05T13:30:20+02:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_160000_width_400_height_400.dat from which we produce 4 different plots:
import shell, std/strutils const args = ["", "--switchAxes", "--invertY", "--switchAxes --invertY"] proc toName(arg: string): string = arg.replace(" ", "_").replace("--", "") let infile = "out/image_sensor_0_2023-09-05T13:30:20+02:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_160000_width_400_height_400.dat" for arg in args: let outfile = "/tmp/image_sensor_XrayFinger_sSum_spacer_no_window_" & toName(arg) & ".pdf" shell: one: cd "~/CastData/ExternCode/RayTracing/" "./plotBinary" -f ($infile) --dtype float --outfile ($outfile) ($arg)
- Spacer and window
./raytracer \ --width 1200 --maxDepth 10 --speed 10.0 \ --nJobs 32 --vfov 30 --llnl --focalPoint \ --sourceKind skXrayFinger \ --rayAt 1.00 \ --sensorKind sSum \ --energyMin 2.0 --energyMax 5.0
import shell, std/strutils const args = ["", "--switchAxes", "--invertY", "--switchAxes --invertY"] proc toName(arg: string): string = arg.replace(" ", "_").replace("--", "") let infile = "out/image_sensor_0_2023-09-05T16:32:12+02:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_160000_width_400_height_400.dat" for arg in args: let outfile = "/tmp/image_sensor_XrayFinger_sSum_spacer_window_" & toName(arg) & ".pdf" shell: one: cd "~/CastData/ExternCode/RayTracing/" "./plotBinary" -f ($infile) --dtype float --outfile ($outfile) ($arg)
And now with an imperfect mirror:
./raytracer \ --width 1200 --maxDepth 10 --speed 10.0 \ --nJobs 32 --vfov 30 --llnl --focalPoint \ --sourceKind skXrayFinger \ --rayAt 1.00 \ --sensorKind sSum \ --energyMin 2.0 --energyMax 5.0 \ --usePerfectMirror=false
import shell, std/strutils const args = ["", "--switchAxes", "--invertY", "--switchAxes --invertY"] proc toName(arg: string): string = arg.replace(" ", "_").replace("--", "") let infile = "out/image_sensor_0_2023-09-05T18:49:35+02:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_160000_width_400_height_400.dat" for arg in args: let outfile = "/tmp/image_sensor_XrayFinger_sSum_spacer_window_imperfectMirror_" & toName(arg) & ".pdf" shell: one: cd "~/CastData/ExternCode/RayTracing/" "./plotBinary" -f ($infile) --dtype float --outfile ($outfile) ($arg)
1.102.
This section was initially written as part of the thesis when we realized that using 6000 background events for each background run would end up using a large fraction (O(1/3)) of all background clusters.
As a result the idea came up to only use the outer chip data for background clusters. I implemented this and the sections below are the first MLP trained that way. This was not the final network we ended up using though. There were more issues with the synthetic data generation that – after being fixed – drastically decreased the performance of this model. See the next entries in this file.
1.102.1. Train an MLP on outer chip background data only extended
Started the training run . Note that we leave
out --backgroundChip 3 explicitly, to train on all outer chips and
not on the center chip at all!
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh300_mse.pt \ --plotPath ~/Sync/30_10_23_sgd_tanh300_mse_outer_chips/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6
Training of the first 100k epochs finished .
In order for this to work as intended, the following changes were made:
subsetPerRunnow is taken literal. No more hardcoded factors for background data or anything like thatbackgroundChipsadded to read background data for each of these chips, allowing us to explicitly exclude the center chip. We will have to see whether that causes us to interpret too many center chip clusters as X-rays because the diffusion and gas gain of the training data better matches the center chip for X-raysshufflenow uses the MLPDesc random number seed- added more fields to the MLPDesc HDF5 file about how many elements were used for training and for validation as well as how many elements were read for background in total.
Continue for another 200k now :
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh300_mse.pt \ --plotPath ~/Sync/30_10_23_sgd_tanh300_mse_outer_chips/ \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 200000 \ --continueAfterEpoch 100000
It finished some time around 19:00 to 20:00.
I've now modified the train_ingrid code a bit more to:
- allow handing an input model file via
--modelinstead of--modelOutpath - the user should not include the final checkpoint name anymore in the
initial training call
--modelOutpath. That will be generated based on the model layout - continuing training does not require the
--continueAfterEpochargument anymore. If further training is to be skipped use--skipTraining - paths can be changed
- learning rates can be changed. The old learning rates will be stored
in
--pastLearningRates
Continue further from 300k now, around :
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh300_mse.pt \ --plotPath ~/Sync/30_10_23_sgd_tanh300_mse_outer_chips/ \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 100000
Finished .
Continue for another 100k (slightly different command, because we now want to use the auto generated model. In the future the same command will work of course) :
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --plotPath ~/Sync/30_10_23_sgd_tanh300_mse_outer_chips/ \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 100000
Finished around .
Won't start another run for now, due to meeting at 2 pm. -> Meeting canceled, starting now . -> Finished .
Start another 100k at . Finished sometime while I was out.
Start another 100k, because there is an intriguing looking dip at the very end with a steeper slope!
And it finished with 800k epochs in total around .
The plots are:
That's a mighty fine result!
Last output of training and test validation:
Train set: Average loss: 0.0046 | Accuracy: 0.994 Test set: Average loss: 0.0047 | Accuracy: 0.9943 Test loss after training: 0.004683235776610672 with accuracy 0.994271375464684
A good chunk better than even the 485k epoch for 100523SGD, our previous best model which included center chip data. The question will be (tomorrow) as to how well this model generalizes to the center chip. But I'm not worried really.
See the HDF5 file ./resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_desc_v2.h5 for the real parameters as they were used to verify they are the same as in the command above.
[ ]Create effective efficiency plot[ ]Create background cluster plot for Run-2 and Run-3 data.
- Effective efficiency of newly trained MLP
Let's create the effective efficiency plot using
TimepixAnalysis/Tools/NN_playground/effective_eff_55_fe.nimfor 95%:./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/run2_run3_30_10_23_tanh300_effective_eff
which yields
./../Sync/run2_run3_30_10_23_tanh300_effective_eff/efficiency_based_on_fake_data_per_run_cut_val.pdf
which looks very good! Actually even a little bit better than fig. [BROKEN LINK: fig:background:mlp:effective_efficiencies].
And for 98%:
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --ε 0.98 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/run2_run3_30_10_23_tanh300_effective_eff_98
which yields
And for 85%:
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --ε 0.85 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit \ --plotDatasets \ --plotPath ~/Sync/run2_run3_30_10_23_tanh300_effective_eff_85
which yields ./../Sync/run2_run3_30_10_23_tanh300_effective_eff_85/efficiency_based_on_fake_data_per_run_cut_val.pdf
If now the network also produces a number of clusters as good or better than the old network that'd be splendid.
- Regenerate the training plots for thesis
We'll regenerate the plots and place them in the thesis figure directory by essentially rerunning the same commands as above, but handing the
--skipTrainingargument:./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --plotPath ~/phd/Figs/neuralNetworks/30_10_23_sgd_tanh300_mse_outer_chips/ \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --skipTraining
- Apply MLP to data
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{fkMLP, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 4 \ --dryRun
- Background clusters of new MLP
85%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.85_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.85_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ --zMax 30 \ --title "X-ray like clusters of CAST data MLP@80%" \ --outpath /tmp/ \ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 \ --suffix "_mlp_85" \ --backgroundSuppression
98%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.98_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.98_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ --zMax 30 \ --title "X-ray like clusters of CAST data MLP@98%" \ --outpath /tmp/ \ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 \ --suffix "_mlp_98" \ --backgroundSuppression
85%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ --zMax 30 \ --title "X-ray like clusters of CAST data MLP@80%" \ --outpath /tmp/ \ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 \ --suffix "_mlp_85_vetoes" \ --backgroundSuppression
98%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.98_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.98_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ --zMax 30 \ --title "X-ray like clusters of CAST data MLP@98%" \ --outpath /tmp/ \ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 \ --suffix "_mlp_98_vetoes" \ --backgroundSuppression
- Background rates (alone & compare to old best MLP)
First background rates on its own, 85%:
plotBackgroundRate \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.85_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.85_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ --centerChip 3 \ --names "MLP@0.8" --names "MLP@0.8" \ --names "MLP@0.8+V" --names "MLP@0.8+V" \ --title "Background rate in center 5·5 mm², MLP@80%" \ --showNumClusters \ --region crGold \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.2 \ --outfile background_rate_gold_mlp_0.8_plus_vetoes.pdf \ --outpath /tmp \ --quiet
90%:
plotBackgroundRate \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ --centerChip 3 \ --names "MLP@0.9" --names "MLP@0.9" \ --names "MLP@0.9+V" --names "MLP@0.9+V" \ --title "Background rate in center 5·5 mm², MLP@90%" \ --showNumClusters \ --region crGold \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.2 \ --outfile background_rate_gold_mlp_0.9_plus_vetoes.pdf \ --outpath /tmp \ --quiet
And now comparisons to the previously best model, both at 95%:
plotBackgroundRate \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933.h5 \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ ~/org/resources/lhood_limits_10_05_23_mlp_sEff_0.99/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh300_msecheckpoint_epoch_485000_loss_0.0055_acc_0.9933_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ --names "MLP@91" --names "MLP@91" \ --names "MLP91+V" --names "MLP91+V" \ --names "MLP@91 NEW" --names "MLP@91 NEW" \ --names "MLP91NEW+V" --names "MLP91NEW+V" \ --centerChip 3 \ --title "Background rate from CAST data" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_crGold_scinti_fadc_septem_line.pdf \ --outpath /tmp \ --useTeX \ --region crGold \ --energyMin 0.2 \ --quiet # --hideErrors \ # --hidePoints \ # # --applyEfficiencyNormalization \
Comparison of the different efficiencies for the MLP without vetoes:
plotBackgroundRate \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.98_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.98_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.85_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.85_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2.h5 \ --names "MLP@98" --names "MLP@98" \ --names "MLP@95" --names "MLP@95" \ --names "MLP@90" --names "MLP@90" \ --names "MLP@85" --names "MLP@85" \ --centerChip 3 \ --title "Background rate from CAST data" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_crGold_mlp_eff_comparison.pdf \ --outpath /tmp \ --useTeX \ --region crGold \ --energyMin 0.2 \ --quiet # --hideErrors \ # --hidePoints \ # # --applyEfficiencyNormalization \
And with scinti+fadc+line veto:
plotBackgroundRate \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.98_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.98_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_tanh300_outer_chip_training_30_10_23/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_SGD_300_2_vQ_0.99.h5 \ --names "MLP@98" --names "MLP@98" \ --names "MLP@95" --names "MLP@95" \ --names "MLP@90" --names "MLP@90" \ --names "MLP@85" --names "MLP@85" \ --centerChip 3 \ --title "Background rate from CAST data with scinti+fadc+line veto" \ --showNumClusters \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile background_rate_crGold_mlp_eff_comparison_scinti_fadc_line.pdf \ --outpath /tmp \ --useTeX \ --region crGold \ --energyMin 0.2 \ --quiet # --hideErrors \ # --hidePoints \ # # --applyEfficiencyNormalization \
- Generate plot of the ROC curve extended
The ROC curve plot is generated by
train_ingridif the--predictargument is given, specifically via thetargetSpecificRocCurveproc.Generate the ROC curve for the MLP trained on the outer chips:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --plotPath ~/phd/Figs/neuralNetworks/30_10_23_sgd_tanh300_mse_outer_chips/ \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --predict
1.103. and Understanding MLP at low energies
[ ]Look at runs 329 and 351 in particular (Ag-Ag target/filter kind) in context of diffusion determination -> These seem to have some systematic difference in them still I think.[ ]Now it is a bit more sensible, but it seems like for the 0.9kV
Total global efficiency = 0.7999918480476074
Test set: Average loss: 0.0848 | Accuracy: 0.9078 INFO: The integer column `count` has been automatically determined to be continuous. To overwrite this behavior add a `+ scalex/ydiscrete()` call to the pl otting chain. Choose `x` or `y` depending on which axis this column refers to. Or apply a `factor` to the column name in the `aes` call, i.e. `aes(…, facto r("count"), …)`. Target local Ag-Ag-6kV cutValue = 0.6467242836952209 eff = 0.8000325679856701
Test set: Average loss: 0.1032 | Accuracy: 0.8872 INFO: The integer column `count` has been automatically determined to be continuous. To overwrite this behavior add a `+ scalex/ydiscrete()` call to the pl otting chain. Choose `x` or `y` depending on which axis this column refers to. Or apply a `factor` to the column name in the `aes` call, i.e. `aes(…, facto r("count"), …)`. Target local Al-Al-4kV cutValue = 0.732017719745636 eff = 0.7998960498960499
Test set: Average loss: 0.6717 | Accuracy: 0.0523 INFO: The integer column `count` has been automatically determined to be continuous. To overwrite this behavior add a `+ scalex/ydiscrete()` call to the pl otting chain. Choose `x` or `y` depending on which axis this column refers to. Or apply a `factor` to the column name in the `aes` call, i.e. `aes(…, facto r("count"), …)`. Target local C-EPIC-0.6kV cutValue = 0.05473856329917908 eff = 0.7999526795220632
Test set: Average loss: 0.7496 | Accuracy: 0.0420 INFO: The integer column `count` has been automatically determined to be continuous. To overwrite this behavior add a `+ scalex/ydiscrete()` call to the pl otting chain. Choose `x` or `y` depending on which axis this column refers to. Or apply a `factor` to the column name in the `aes` call, i.e. `aes(…, facto r("count"), …)`. Target local Cu-EPIC-0.9kV cutValue = 0.03405002132058144 eff = 0.7999139414802066
Test set: Average loss: 0.0946 | Accuracy: 0.8947 INFO: The integer column `count` has been automatically determined to be continuous. To overwrite this behavior add a `+ scalex/ydiscrete()` call to the pl otting chain. Choose `x` or `y` depending on which axis this column refers to. Or apply a `factor` to the column name in the `aes` call, i.e. `aes(…, facto r("count"), …)`. Target local Cu-EPIC-2kV cutValue = 0.7672248125076294 eff = 0.7998602050326188
Test set: Average loss: 0.0031 | Accuracy: 0.9966 INFO: The integer column `count` has been automatically determined to be continuous. To overwrite this behavior add a `+ scalex/ydiscrete()` call to the pl otting chain. Choose `x` or `y` depending on which axis this column refers to. Or apply a `factor` to the column name in the `aes` call, i.e. `aes(…, facto r("count"), …)`. Target local Cu-Ni-15kV cutValue = 0.9883408546447754 eff = 0.8000298908982215
Test set: Average loss: 0.0023 | Accuracy: 0.9978 INFO: The integer column `count` has been automatically determined to be discrete. To overwrite this behavior add a `+ scalex/ycontinuous()` call to the pl otting chain. Choose `x` or `y` depending on which axis this column refers to. Target local Mn-Cr-12kV cutValue = 0.9918201684951782 eff = 0.8
Test set: Average loss: 0.0010 | Accuracy: 0.9990 INFO: The integer column `count` has been automatically determined to be discrete. To overwrite this behavior add a `+ scalex/ycontinuous()` call to the pl otting chain. Choose `x` or `y` depending on which axis this column refers to. Target local Ti-Ti-9kV cutValue = 0.9963032007217407 eff = 0.8000294724432655
see the Cu-EPIC-0.9kV and C-EPIC-0.6kV values.
It seems like our choice of output neuron is switched? Or rather the network's prediction is wrong?
The ROC curve technically has become even worse now lol.
Let's revisit:
1.103.1. TODO Diffusion fix [0/1]
The diffusion values were wrong for most CDL runs, because if
determined via determineDiffusion like the below, we were using the
wrong target energy. We always used 5.9 keV, resulting in the wrong
properties.
We fixed it and:
./determineDiffusion \ ~/CastData/data/DataRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/DataRuns2018_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --plotPath ~/phd/Figs/determineDiffusion/ \ --histoPlotPath ~/phd/Figs/determineDiffusion/histograms
NOTE: The above command is what we ran, but we moved the files to ./Figs/statusAndProgress/determineDiffusion/fixedDiffusionCDL/ because after we finally fixed everything we might rerun it again, who knows.
now gives
./../CastData/ExternCode/TimepixAnalysis/resources/cacheTab_diffusion_runs.h5
which contains a single table dataset now, because we use a custom
toH5 / fromH5 procedure for tables.
The plot is ./Figs/statusAndProgress/determineDiffusion/fixedDiffusionCDL/σT_per_run.pdf showing that the uncertainty via CvM is now very small, with one exception (or 2). The yellow spots are the Ag-Ag runs, as mentioned above!
[ ]Investigate Ag-Ag runs.
1.103.2. Looking at MLP with correct diffusion values for CDL runs
n:PROPERTIES: :CUSTOMID: sec:journal:051123:fixeddiffusion
:END:
With the diffusion values fixed, we first looked at the effective efficiency to see if the values for the CDL runs now looked better:
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_30_10_23_tanh300_effective_eff_95_fixedDiffusion/
which, going by the plot, ./../Sync/run2_run3_30_10_23_tanh300_effective_eff_95_fixedDiffusion/efficiency_based_on_fake_data_per_run_cut_val.pdf is not really the case!
The worst runs here though are Cu-EPIC-2kV, which are not the ones with the 'inverted' cut values / prediction outputs from below / above. See below:
Then we ran train_ingrid with the --predict option:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 --back ~/CastData/data/DataRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --plotPath ~/phd/Figs/neuralNetworks/30_10_23_sgd_tanh300_mse_outer_chips_fixedDiffusion/ \ --numHidden 300 --numHidden 300 \ --activation tanh --outputActivation sigmoid --lossFunction MSE --optimizer SGD \ --learningRate 7e-4 --simulatedData --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 --backgroundChips 1 --backgroundChips 2 --backgroundChips 4 --backgroundChips 5 --backgroundChips 6 \ --predict
which mainly produced the quoted output already mentioned in the above section and the plots:
The Aluminum has a huge chunk of data that is fully in the background range! Hence the low effective efficiency!
-> This one is just completely inverted.
-> Cu-EPIC-0.9kV is very broken
-> Cu-EPIC-2kV also has a good chunk on the left, similar to Al-Al-4kV above!
So now let's look at the prediction of the network for the corresponding fake data.
1.103.3. Looking at the NN prediction for fake data
I extended
./../CastData/ExternCode/TimepixAnalysis/Tools/NN_playground/nn_predict.nim
to work as a standalone program to compute the run local cut values
for the given data files.
And I added an option in the determineCutValue procedure of
nn_cuts.nim to produce a histogram of the NN output for the data.
Running:
./nn_predict \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --signalEff 0.8
yielded the plots in: ./Figs/statusAndProgress/neuralNetworks/runLocalCutsCDLPrediction/
where we can see that some of the runs
- Target: C-EPIC-0.6kV, run: 342
- Target: C-EPIC-0.6kV, run: 343
- Target: Cu-EPIC-0.9kV, run: 339
- Target: Cu-EPIC-0.9kV, run: 340
have a completely inverted prediction!
UPDATE: Looking into the fake event generation for the MLP training, I noticed the likely culprit: These runs have mostly extremely low diffusion values below 600!
But in our fake event generation for the MLP training we have the following diffusion sampling logic
# 3. sample a diffusion let σT = rnd.gauss(mu = 660.0, sigma = (diffusion[1] - diffusion[0] / 4.0))
where diffusion is:
let diffusion = @[550.0, 700.0] # μm/√cm, will be converted to `mm/√cm` when converted to DF
which means that we sample in a normal distribution with a sigma of
37 around 660. So below 600 barely get any fake X-rays!!
UPDATE 2: I just noticed that THERE IS EVEN
A BUG IN THE SAMPLING LOGIC. There is a missing parenthesis for the
diffusion parameter! So we actually have a sigma of
700 - 550 / 4 = 562.5!!!
What the heck!!!
[X]Run the code as is, printing the
σTvalues and see what we get../train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --modelOutpath ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips_testing_additional/ \ --plotPath ~/phd/Figs/neuralNetworks/30_10_23_sgd_tanh300_mse_outer_chips_fixedDiffusion/ \ --numHidden 300 --numHidden 300 \ --activation tanh --outputActivation sigmoid --lossFunction MSE --optimizer SGD --learningRate 7e-4 \ --simulatedData --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 --backgroundChips 1 --backgroundChips 2 --backgroundChips 4 --backgroundChips 5 --backgroundChips 6 \ --epochs 100000
σT = -349.2786469330247 σT = 673.0827513404888 σT = 1339.81017470335 σT = 375.3202836179698 σT = 279.7901226013589 σT = -54.46352298513648 σT = 757.6270717741737 σT = 464.7151428501154 σT = 1075.299359327855 σT = 525.282292375453 σT = -452.3085611825393 σT = 186.9591401583243 σT = 684.1493145310708 σT = 607.5930633308599 σT = 875.8938252595046 σT = 1211.994490752969 σT = 769.4028326611971 σT = 582.1565741578706 σT = 986.4413834680025 σT = 435.8008999447983 σT = 980.8439488317562 σT = -193.6147744885794 σT = 1191.811868521617 Found more than 1 or 0 cluster! Skipping. Number of clusters: 2 σT = 840.8752668800632 σT = 1669.755678114918 Found more than 1 or 0 cluster! Skipping. Number of clusters: 2 σT = 1075.003354683646 Found more than 1 or 0 cluster! Skipping. Number of clusters: 2 σT = 572.7477916295879 σT = 398.4177966160603 σT = 719.6147823480718 σT = 1229.567030912002 σT = 1003.407372089173 σT = 932.923706910155 σT = -547.1796240141737 σT = -353.8365784807355 σT = 646.4456514540479 σT = 1361.725946770055 σT = 1456.445411753667 σT = 945.6512103826537 σT = 834.9911542146369 σT = 1105.710979574551 σT = 1219.779892777278 σT = 1626.74927640482 σT = 168.570702411871 σT = 573.573485141321 σT = 728.329373540124 σT = 1495.006890524006 Found more than 1 or 0 cluster! Skipping. Number of clusters: 2 σT = 1481.450602352936 σT = 654.340107917818 σT = 1166.199712139452 σT = 1358.478142822759 σT = 1505.960731274281 σT = 1587.535164666273 σT = 113.6167117839664
YUP this gives horrible values.
I guess this explains why we get so many "Found more than 1 cluster"!!!!!!!!!!!!!!!!!!!!!!!!!!!!
So:
[ ]Continue training the network using a flat distribution between 550 and 700![X]Check the
σTvalues after switching to a flat rangeσT = 641.4507468480646 σT = 630.7897513713178 σT = 626.1310549652765 σT = 577.1992649764234 σT = 592.0799459762638 σT = 613.9328992175666 σT = 614.9282075375172 σT = 668.1154808362544
-> Looking sensible
[X]Copied the model to ./resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips_testing_additional/[X]start training:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips_testing_additional/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --plotPath ~/phd/Figs/neuralNetworks/30_10_23_sgd_tanh300_mse_outer_chips_fixedDiffusion_testing_additional/ \ --numHidden 300 --numHidden 300 \ --activation tanh --outputActivation sigmoid --lossFunction MSE --optimizer SGD --learningRate 7e-4 \ --simulatedData --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 --backgroundChips 1 --backgroundChips 2 --backgroundChips 4 --backgroundChips 5 --backgroundChips 6 \ --epochs 100000
Training finished . Note that the loss did not really go down very much anymore, see
Anyway, let's see what this model now does to our CDL runs.
[X]Run
nn_predictwith the new model:./nn_predict \ --model ~/org/resources/nn_devel_mixing/30_10_23_sgd_tanh300_mse_loss_outer_chips_testing_additional/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --signalEff 0.8
yielded the plots in: ./Figs/statusAndProgress/neuralNetworks/runLocalCutsCDLPrediction/additionalTrainingFixedDiffusion/ where we can see that the behavior for low energy X-rays has improved quite a bit. It's still very far away from optimal however. I think the training from this model on is not the most likely idea to succeed. Hence we'll train a new network now.
[X]Train a completely new MLP with sensible ranges. Start training now. We'll go with 300k epochs directly. -> Stopped at after 100k for the time being. Seems that at low energies the same problem persists.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_ochip_diffusion/ \ --plotPath ~/Sync/05_11_23_sgd_tanh300_mse_outer_chips/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 300000
Quick check:
I ran
nn_predicton the network snapshot at epoch 25k and the low energy targets still look roughly similar. Why? Is there something we are forgetting in our data sampling? Compared to what we create when we sample for the CDL runs that is? Checking now at 75k again. -> Unchanged. I suppose at this point I really need to understand how / why the network doesn't predict these well. Either: train an MLP only on low energy X-rays (for signal?) And/Or: try to understand the difference between what we generate for the training and what we generate for thenn_predictcalls. -> Another point is that the gas gain is very low in CDL and our gain sampling islet G = rnd.gauss(mu = (gains[1] + gains[0]) / 2.0, sigma = (gains[1] - gains[0]) / 4.0)
with
let gains = @[2400.0, 4000.0]
i.e. a mean of 3200 and σ of 400. Might need more on the lower end.
We stop this network training at 100k now and first try a network with different parameters:
let G = rnd.gauss(mu = (gains[1] + gains[0]) / 2.0, sigma = (gains[1] - gains[0]) / 3.0) let gain = GainInfo(N: 900_000.0, G: G, theta: rnd.rand(1.4 .. 2.4))
so a tighter theta parameter (1.4-2.4 instead of 0.4-2.4), larger N
(900k instead 100k) and wider allowed gains
(divide by 3 instead of 4).
[X]Let's train a network with those gain values
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_different_gains/ \ --plotPath ~/Sync/05_11_23_sgd_tanh300_mse_different_gains/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 300000
-> Ran
nn_predictafter the first 5k epochs and the trend is still the same in the low energy X-rays. Likely we need to try running a training on low energy events only. -> Stopped at after 75k epochs![X]Let's try to run a low energy training in parallel to the other
one. We'll see how well that works. Running with
let energy = rnd.rand(0.1 .. 10.0).keV let lines = @[FluorescenceLine(name: "Fake", energy: energy, intensity: 1.0)] # 2. sample a gas gain let G = rnd.gauss(mu = (gains[1] + gains[0]) / 2.0, sigma = (gains[1] - gains[0]) / 3.0) let gain = GainInfo(N: 900_000.0, G: G, theta: rnd.rand(1.4 .. 2.4))
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_only_up_2keV/ \ --plotPath ~/Sync/05_11_23_sgd_tanh300_mse_only_up_2keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 300000
-> Let's test this on nn_predict for epoch 35k
-> This does help!!
-> Stopped at after 50k epochs.
I THINK I FOUND SOMETHING:
## Generate the sampler sampling the conversion point if line.name notin fakeDesc.sampleTab: if fakeDesc.gasMixture.gases.len == 0: fakeDesc.gasMixture = initCASTGasMixture() fakeDesc.sampleTab[line.name] = fakeDesc.generateSampler(result)
DOES NOT CHANGE THE SAMPLER EVER FOR CHANGING ENERGIES!!!!!!!!!
So: combined with our changes to the gas gain and diffusion changes, if we fix the sampling of the energy I'm quite optimistic things should start looking up!
We fixed to to
let energy = rnd.rand(0.1 .. 10.0).keV let lineName = &"{energy.float:.2f}" ## <-- This should yield 1000 different absorption length samplers! let lines = @[FluorescenceLine(name: lineName, energy: energy, intensity: 1.0)]
This should guarantee to give us 1000 different absorption length samplers!
Let's train a network with this. We use the new gasgain and diffusion logic, but train again energies from 0 to 10 keV:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_fixed_absLength/ \ --plotPath ~/Sync/05_11_23_sgd_tanh300_mse_fixed_absLength/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 300000
The training finished . Quite a long time.
-> The change seems to work in terms of the sampling logic, because
the code outputs many USING λ = messages, which get less and less
the more samplers have already been produced!
-> But the sampling is much slower than before. Hmm, why? I guess
our 'default' sampler was very "efficient"? Not sure why other
samplers would be more expensive from the top of my head though.
-> The actual sampling of pixels really shouldn't be the
reason. Maybe the cluster reconstruction is the cause?
It took more than half an hour for the simulation of the 500k events
now. Curious.
-> The training progress looks somewhat similar to the previous
trained networks now, so relatively slow after the initial 5k
epochs.
Let's look at nn_predict for snapshot 15k:
./nn_predict \ --model ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_fixed_absLength/mlp_tanh_sigmoid_MSE_SGD_300_2checkpoint_epoch_15000_loss_0.0302_acc_0.9620.pt \ --signalEff 0.8
-> Crap. For now it looks like it also struggles with low energy events. :(
-> It is actually visible in the validation and training outputs: ./../Sync/05_11_23_sgd_tanh300_mse_fixed_absLength/training_output_log10.pdf in the form of the peak on the very left.
-> Let's see how this develops with longer training.
-> After rerunning after 175k epochs it's starting to turn! The Cu-EPIC 0.9kV is already most correct and also the C-EPIC 0.6kV is starting to move right!!
[ ]Can we change the loss function such that this is penalized much more? -> Might not be needed. Also better to pretrain on low energy clusters?
Let's start training a network for 50k epochs only on <3 keV data using: ./../CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 generated via:
./simulate_xrays \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --nFake 250000 \ --energyMin 0.1 --energyMax 3.0 \ --yEnergyMin 1.0 --yEnergyMax 0.0 \ --outfile ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --note "Energies linear decrease in frequency from 0.1 to 3.0 keV"
I wrote simulate_xrays to separate the simulation parts from the
training to avoid waiting so long before the actual training
starts.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_first_train_less3keV/ \ --plotPath ~/Sync/05_11_23_sgd_tanh300_mse_first_train_less3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 50000
This finished sometime after 3 am.
Let's continue with the uniform 0 to 10 keV data:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_first_train_less3keV/mlp_tanh_sigmoid_MSE_SGD_300_2checkpoint_epoch_50000_loss_0.0494_acc_0.9338.pt \ --plotPath ~/Sync/05_11_23_sgd_tanh300_mse_first_train_less3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 300000
Effective efficiency of "first trained to less 3 keV" model at epoch 140k (90k on uniform data):
./effective_eff_55fe ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 --model ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_first_train_less3keV/mlp_tanh_sigmoid_MSE_SGD_300_2checkpoint_epoch_140000_loss_0.0449_acc_0.9413.pt --ε 0.95 --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 --evaluateFit --plotDatasets --plotPath ~/Sync/run2_run3_05_11_23_first_train_less3keV/
-> Efficiency generally better on 55Fe data and most CDL, but significantly worse for some CDL data :'(
Effective efficiency of the initial model from yesterday trained on uniform energies with the correct absorption length for 300k epochs:
./effective_eff_55fe ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 --model ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_fixed_absLength/mlp_tanh_sigmoid_MSE_SGD_300_2.pt --ε 0.95 --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 --evaluateFit --plotDatasets --plotPath ~/Sync/run2_run3_05_11_23_fixed_absLength
Run combinations:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{fkMLP}" \ --mlpPath ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_first_train_less3keV/mlp_tanh_sigmoid_MSE_SGD_300_2checkpoint_epoch_330000_loss_0.0429_acc_0.9442.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_tanh300_outer_chip_training_05_11_23_first_train_less3keV_330k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 4 \ --dryRun
1.103.4. Fixing the totalCharge for CDL runs
After fixing the diffusion and absorption length in the sections above, one problem remaining is the fact that the total charge is badly predicted for CDL runs.
Let's take for example run 336 of the Cu-EPIC-2.0 kV tfkind
or one of the Cu-Ni-15kV runs, 320:
Neither the location of the total charge peak, nor its width matches the real data.
The issue I had with this today is that of course if the total charge goes into the MLP, then it shouldn't be a surprise that the effective efficiency is wrong if we produce fake data that doesn't match the real data we want to cut on.
E.g. see the 0.93 keV runs here: ./../Sync/run2_run3_30_10_23_tanh300_effective_eff_95_fixedDiffusion/efficiency_based_on_fake_data_per_run_cut_val.pdf that have a much, much lower efficiency than target.
So I thought again about the total charge and how we sample. This lead me to our line:
let calibFactor = linearFunc(@[calibInfo.bL, calibInfo.mL], gain.G) * 1e-6
which after some thinking assumes the linear gas gain vs energy
calibration factor relation also holds for CDL data. But we know it
does not, otherwise we wouldn't get such bad energy values for the CDL
runs if we just look at energyFromCharge.
So I decided to implement an improvement whereby the calibFactor is
computed specifically for each CDL run based on the fit parameters to
the peak of each run.
The peak position is used as
calibration factor = energy of line / peak position in charge
which is then added to CalibInfo in databaseRead.nim.
Similarly for the width in sampleTargetCharge:
result = rnd.gauss(mu = targetEnergyCharge, sigma = charge_σ)
we now use charge_σ, which is either the same as before
targetCharge * 0.075
or for CDL runs the actual width from the fit to the peak!
This leads to a great agreement of the total charge runs.
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_fixed_absLength/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_05_11_23_fixed_absLength/
After the fix it looks like this: ./../Sync/run2_run3_05_11_23_fixed_absLength/0.93_336/dsetPlots/totalCharge_ridgeline_comparison.pdf
or the 15 kV run from above:
Perfect.
Unfortunately, this still does not entirely fix the effective efficiency found for some CDL runs. :(
1.103.5. Determine best fit of real energy resolution to use for fake gen
For non CDL runs we still use
let charge_σ = if calibInfo.isCdl: calibInfo.peak_σ else: targetEnergyCharge * 0.075 # for 55Fe data this is about 10% # -> For better training data should change this to be energy dependent too! let targetCharge = rnd.sampleTargetCharge(targetEnergyCharge, charge_σ)
1.103.6. train_ingrid --predict for MLP first trained on < 3 keV
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 --back ~/CastData/data/DataRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_first_train_less3keV/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --plotPath ~/phd/Figs/neuralNetworks/05_11_23_mlp_sgd_tanh_first_less3keV/ \ --numHidden 300 --numHidden 300 --activation tanh --outputActivation sigmoid --lossFunction MSE --optimizer SGD --learningRate 7e-4 \ --simulatedData --backgroundRegion crAll --nFake 250_000 \ --backgroundChips 0 --backgroundChips 1 --backgroundChips 2 --backgroundChips 4 --backgroundChips 5 --backgroundChips 6 \ --predict
yields the plots in:
./../phd/Figs/neuralNetworks/05_11_23_mlp_sgd_tanh_first_less3keV/
We can see that the target specific predictions look quite good with this model. However, the target specific ROC curve is still a bit of a disappointment.
Some of the targets (0.6 kV, 0.9 kV, 2 kV and 4 kV) still drop below the LnL line at a signal efficiency around the 93% mark.
This plot would at least indicate that an efficiency of 90% should be the most 'interesting'.
1.103.7. train_ingrid --predict on uniformly trained MLP after absorption length fix
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 --back ~/CastData/data/DataRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_fixed_absLength/mlp_tanh_sigmoid_MSE_SGD_300_2.pt \ --plotPath ~/phd/Figs/neuralNetworks/05_11_23_mlp_sgd_tanh_fixed_absLength/ \ --numHidden 300 --numHidden 300 --activation tanh --outputActivation sigmoid --lossFunction MSE --optimizer SGD --learningRate 7e-4 \ --simulatedData --backgroundRegion crAll --nFake 250_000 \ --backgroundChips 0 --backgroundChips 1 --backgroundChips 2 --backgroundChips 4 --backgroundChips 5 --backgroundChips 6 \ --predict
yields ./../phd/Figs/neuralNetworks/05_11_23_mlp_sgd_tanh_fixed_absLength
here in particular
and
still disqualify this network. It sucks at those energies.
The combined ROC curve
looks comparable to the one of the network trained on <3 keV data first.
1.103.8. Play around with other parameters again maybe
- Larger learning rate trained on < 3 keV data
Larger learning rate trained on < 3 keV data first. Otherwise identical to 1.103.3:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/06_11_23_sgd_tanh300_mse_less3keV_lr_2em4/ \ --plotPath ~/Sync/06_11_23_sgd_tanh300_mse_less3keV_lr_2em4/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 2e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 50000
Finished .
Train set: Average loss: 0.0577 | Accuracy: 0.922
Test set: Average loss: 0.0576 | Accuracy: 0.9223
How does this compare to the old 7e-4 variant of the same?
hdfview ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_first_train_less3keV/mlp_desc_v2.h5
-> loss at 10 = 50k: 0.0494 -> testAccuracies at 10 = 50k: 0.93377
So a bit worse actually!
I think we don't need to continue on this avenue.
The loss looks like:
it is slower than for the one from yesterday.
- Larger hidden layers (500 neurons)
500 neurons instead of 300. Otherwise identical to 1.103.3:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/06_11_23_sgd_tanh500_mse_less3keV/ \ --plotPath ~/Sync/06_11_23_sgd_tanh500_mse_less3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 500 \ --numHidden 500 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer SGD \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 50000
Finished .
Train set: Average loss: 0.0494 | Accuracy: 0.934
Test set: Average loss: 0.0495 | Accuracy: 0.9340
How does this compare to the old 300 hidden neuron variant?:
hdfview ~/org/resources/nn_devel_mixing/05_11_23_sgd_tanh300_mse_first_train_less3keV/mlp_desc_v2.h5
-> loss at 10 = 50k: 0.0494 -> testAccuracies at 10 = 50k: 0.93377
So pretty much on par!
The loss:
at least still seems to decay somewhat.
Probably comparable performance to the other one though.
- ReLU, Adam, 500, sigmoid cross entropy
Something very different to what we trained lately:
This seems to still produce very large loss values. Maybe sigmoid output activation with MSE is better after all?
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/06_11_23_adam_relu500_cross_entropy_less3keV/ \ --plotPath ~/Sync/06_11_23_adam_relu500_cross_entropy_less3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 500 \ --numHidden 500 \ --activation relu \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 50000
Probably stopping this after 15k epochs, because the overtraining is extreme: ./../Sync/06_11_23_adam_relu500_cross_entropy_less3keV/training_output_log10.pdf vs.
and the loss
- ReLU, Adam, 500, sigmoid cross entropy
Something very different to what we trained lately:
This seems to still produce very large loss values. Maybe sigmoid output activation with MSE is better after all?
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/06_11_23_adam_relu500_mse_sigmoid_less3keV/ \ --plotPath ~/Sync/06_11_23_adam_relu500_mse_sigmoid_less3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 500 \ --numHidden 500 \ --activation relu \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 50000
Stopped at after 20k epoch snapshot. The network achieved strong overtraining again after 10k epochs.
But the test data doesn't look too bad, so
- let's try to use the network to predict CDL data:
./nn_predict \ --model ~/org/resources/nn_devel_mixing/06_11_23_adam_relu500_mse_sigmoid_less3keV/mlp_relu_sigmoid_MSE_Adam_500_2checkpoint_epoch_20000_loss_0.0472_acc_0.9513.pt \ --signalEff 0.8
the plots look rather weird. In some runs the prediction is a perfect 1.0. In others (like a C-EPIC-0.6kV run) ~30% is at 0, 70% at 1. hardly anything in between.
Might be an artifact of ReLU?
Anyhow, let's look at continuing to train for another 50k with uniform data:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/06_11_23_adam_relu500_mse_sigmoid_less3keV/mlp_relu_sigmoid_MSE_Adam_500_2checkpoint_epoch_20000_loss_0.0472_acc_0.9513.pt \ --plotPath ~/Sync/06_11_23_adam_relu500_mse_sigmoid_less3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 500 \ --numHidden 500 \ --activation relu \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 50000
Stopped after 35k. The loss is constant.
Let's run
train_ingridfor the ROC curve on this one at 35k:./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 --back ~/CastData/data/DataRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/06_11_23_adam_relu500_mse_sigmoid_less3keV/mlp_relu_sigmoid_MSE_Adam_500_2checkpoint_epoch_35000_loss_0.0477_acc_0.9513.pt \ --plotPath ~/phd/Figs/neuralNetworks/06_11_23_adam_relu500_mse_sigmoid_less3keV/ \ --numHidden 500 --numHidden 500 --activation relu --outputActivation sigmoid --lossFunction MSE --optimizer Adam --learningRate 7e-4 \ --simulatedData --backgroundRegion crAll --nFake 250_000 \ --backgroundChips 0 --backgroundChips 1 --backgroundChips 2 --backgroundChips 4 --backgroundChips 5 --backgroundChips 6 \ --predict
The plots in ./../phd/Figs/neuralNetworks/06_11_23_adam_relu500_mse_sigmoid_less3keV/ are at least partially problematic. The prediction is so strict 0 or 1 that we don't get any precision, resulting in a ROC curve that's extremely sharp (and useless).
-> Maybe better with linear + cross entropy after all.
[ ]Try (G)eLU?[X]Try Adam with tanh? -> See below.[ ]Try Adam again cross entropy
1.103.9. Adam with tanh
Let's see.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/06_11_23_adam_tanh500_mse_sigmoid_less3keV/ \ --plotPath ~/Sync/06_11_23_adam_tanh500_mse_sigmoid_less3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 500 \ --numHidden 500 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 50000
-> After 5k epochs we're already in the strong overtraining regime.
Stopping the training here.
Let's try only 300 neurons:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/06_11_23_adam_tanh300_mse_sigmoid_less3keV/ \ --plotPath ~/Sync/06_11_23_adam_tanh300_mse_sigmoid_less3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 50000
Same problem really. Strong overtraining. Stopped after 10k epochs .
Let's try Adam with cross entropy but using tanh: This at least should give us a smooth ROC curve that we can investigate (to see how it performs regardless of overtraining).
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/06_11_23_adam_tanh300_linear_cross_entropy_less3keV/ \ --plotPath ~/Sync/06_11_23_adam_tanh300_linear_cross_entropy_less3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 50000
When looking at the sigmoidCrossEntropy predictions keep in mind that by default we clamp to 50.
We'll change that to 5000 or so (plenty large) by default.
Let's run the prediction for this network:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --model ~/org/resources/nn_devel_mixing/06_11_23_adam_tanh300_linear_cross_entropy_less3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4464_acc_0.9474.pt \ --plotPath ~/Sync/06_11_23_adam_tanh300_linear_cross_entropy_less3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --predict
The ROC curve here looks pretty promising!
Significantly better pretty much everywhere with the exception of Cu-EPIC-0.9kV from about 90% on. But the rest beats everything to a rather crazy level.
Continue with 10k epochs for uniform energy data.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/06_11_23_adam_tanh300_linear_cross_entropy_less3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4464_acc_0.9474.pt \ --plotPath ~/Sync/06_11_23_adam_tanh300_linear_cross_entropy_less3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --epochs 15000
This training run is done. The training accuracy is pretty much perfect now, which is kind of crazy. The test accuracy hasn't really changed at all though.
Let's check the effective efficiency we get for the model at 15k epochs.
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/06_11_23_adam_tanh300_linear_cross_entropy_less3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_15000_loss_0.7418_acc_0.9458.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_06_11_23_adam_tanh300_linear_cross_entropy_less3keV/
-> This looks actually surprisingly reasonable!
Maybe worth a try to run this one through likelihood for background
rates etc.
Also running train_ingrid --predict on the 20k resulting snapshot:
./train_ingrid ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 --back ~/CastData/data/DataRuns2017_Reco.h5 --back ~/CastData/data/DataRuns2018_Reco.h5 --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 --model ~/org/resources/nn_devel_mixing/06_11_23_adam_tanh300_linear_cross_entropy_less3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_20000_loss_0.7605_acc_0.9455.pt --plotPath ~/Sync/06_11_23_adam_tanh300_linear_cross_entropy_less3keV_20k/ --datasets eccentricity --datasets skewnessLongitudinal --datasets skewnessTransverse --datasets kurtosisLongitudinal --datasets kurtosisTransverse --datasets length --datasets width --datasets rmsLongitudinal --datasets rmsTransverse --datasets lengthDivRmsTrans --datasets rotationAngle --datasets fractionInTransverseRms --datasets totalCharge --datasets σT --numHidden 300 --numHidden 300 --activation tanh --outputActivation linear --lossFunction sigmoidCrossEntropy --optimizer Adam --learningRate 7e-4 --simulatedData --backgroundRegion crAll --nFake 250_000 --backgroundChips 0 --backgroundChips 1 --backgroundChips 2 --backgroundChips 4 --backgroundChips 5 --backgroundChips 6 --clamp 5000 --predict
./../Sync/06_11_23_adam_tanh300_linear_cross_entropy_less3keV_20k/ -> The 5k version might actually be better!
[X]CREATE effective eff for 5k model as well.
Let's check the effective efficiency we get for the model at 5k epochs,
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/06_11_23_adam_tanh300_linear_cross_entropy_less3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4413_acc_0.9476.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/
finished .
The plots are in
./../Sync/run2_run3_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/,
in particular
This looks pretty damn good actually, as long as we just accept that a few of these CDL runs are just rubbish. Fortunately it is not the lowest energies and not every run of a particular type is low, giving credence to the fact that data quality in some runs is just very bad! This could be a winner.
- Apply to
likelihoodwith 5k model
[X]Run through likelihood for background.
Run likelihood combinations for the 5k model first. Note: we try to use 6 jobs for now. Let's see if that works, : -> So far 6 jobs in parallel seems to run without a problem. Each process uses about 6.2 GB. Has been stable for a while now.
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{fkMLP}" \ --mlpPath ~/org/resources/nn_devel_mixing/06_11_23_adam_tanh300_linear_cross_entropy_less3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4413_acc_0.9476.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 4 \ --dryRun
It finished . Took
Running all likelihood combinations took 3105.762787818909 s
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4413_acc_0.9476.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4413_acc_0.9476.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_adam_tanh300_linear_cross_entropy_5k" \ --backgroundSuppression
Background rate, comparison:
plotBackgroundRate \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/lhood_c18_R2_crAll_sEff_0.98_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4413_acc_0.9476.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/lhood_c18_R3_crAll_sEff_0.98_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4413_acc_0.9476.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4413_acc_0.9476.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4413_acc_0.9476.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4413_acc_0.9476.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4413_acc_0.9476.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/lhood_c18_R2_crAll_sEff_0.85_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4413_acc_0.9476.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_5k/lhood_c18_R3_crAll_sEff_0.85_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_5000_loss_0.4413_acc_0.9476.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --names "85" --names "85" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_cross_entropy_5k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
Hmm, these look quite a bit worse than the SGD model trained first 50k on low energy and then 300k on uniform data. Maybe the 15k model is better?
- Apply to
likelihoodwith 15k model
Run likelihood combinations for the 15k model now. Directly use 6 jobs.
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{fkMLP}" \ --mlpPath ~/org/resources/nn_devel_mixing/06_11_23_adam_tanh300_linear_cross_entropy_less3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_15000_loss_0.7418_acc_0.9458.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_15k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 6 \ --dryRun
Finished at some point earlier in 3140 s.
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_15000_loss_0.7418_acc_0.9458.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_15000_loss_0.7418_acc_0.9458.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_adam_tanh300_linear_cross_entropy_5k" \ --backgroundSuppression
Background rate, comparison:
plotBackgroundRate \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.98_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_15000_loss_0.7418_acc_0.9458.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.98_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_15000_loss_0.7418_acc_0.9458.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_15000_loss_0.7418_acc_0.9458.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_15000_loss_0.7418_acc_0.9458.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_15000_loss_0.7418_acc_0.9458.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_15000_loss_0.7418_acc_0.9458.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.85_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_15000_loss_0.7418_acc_0.9458.h5 \ ~/org/resources/lhood_mlp_06_11_23_adam_tanh300_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.85_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_300_2checkpoint_epoch_15000_loss_0.7418_acc_0.9458.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --names "85" --names "85" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_cross_entropy_15k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
A bit better than the 5k version maybe, but not spectacularly so.
1.103.10. Train Adam model with snapshots every 100 epochs
To get a better image of when the overtraining starts, let's look at an Adam training with snapshots every 100 epochs.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/07_11_23_adam_tanh300_linear_cross_entropy_less3keV_every100/ \ --plotPath ~/Sync/07_11_23_adam_tanh300_linear_cross_entropy_less3keV_every100/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 100
After only 300 epochs significant overtraining becomes visible
Crazy!
But the accuracy was actually pretty good (96%) after those 300 epochs.
Also look at such training using an Adam model but directly starting on uniform energy data. Maybe the better gradient algorithm is able to separate the low energy data as well?
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/07_11_23_adam_tanh300_linear_cross_entropy_uniform_every100/ \ --plotPath ~/Sync/07_11_23_adam_tanh300_linear_cross_entropy_uniform_every100/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 300 \ --numHidden 300 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 100
97% after 100 epochs. 98% after 200. At epoch 400-500 overtraining becomes really visible.
- Training tiny nets
How small can we go?
Let's start with 10 neurons:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/07_11_23_adam_tanh10_linear_cross_entropy_uniform_every100/ \ --plotPath ~/Sync/07_11_23_adam_tanh10_linear_cross_entropy_uniform_every100/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 100
If this still overtrains I'd be very surprised.
Still manages almost 97% accuracy after only 200 epochs.
This does not overtrain anymore
The fun thing is this network is so small, we can in principle print it as a matrix in the thesis.
We'll stop the training after 10k epochs and then let it continue only plotting progress every 1k epochs.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/07_11_23_adam_tanh10_linear_cross_entropy_uniform_every100/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_10000_loss_0.0426_acc_0.9846.pt \ --plotPath ~/Sync/07_11_23_adam_tanh10_linear_cross_entropy_uniform_every100/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --epochs 50000 \ --plotEvery 1000
Stopping training after 31k epochs . The loss has flatlined even on a log plot.
nn_predict, Effective efficiency andtrain_ingrid --predictfor 10 neuron network
./nn_predict \ --model ~/org/resources/nn_devel_mixing/07_11_23_adam_tanh10_linear_cross_entropy_uniform_every100/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_4500_loss_0.0441_acc_0.9842.pt \ --signalEff 0.8
Looking interesting!
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/07_11_23_adam_tanh10_linear_cross_entropy_uniform_every100/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_4500_loss_0.0441_acc_0.9842.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_07_11_23_adam_tanh10_4.5k \
Finished .
Let's see: ./../Sync/run2_run3_07_11_23_adam_tanh10_4.5k
Generally looks quite good. But a bit larger variation than in the large networks it seems.
Note that the larger Adam trained network exhibits a much better match of target efficiency though.
Using 15k model for
train_ingrid --predict:./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/07_11_23_adam_tanh10_linear_cross_entropy_uniform_every100/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.pt \ --plotPath ~/Sync/07_11_23_adam_tanh10_linear_cross_entropy_uniform_every100/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --predict
Wow, we might have a winner here. The ROC curve in particular looks quite remarkable.
Let's run all likelihood combinations for this one.
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{fkMLP}" \ --mlpPath ~/org/resources/nn_devel_mixing/07_11_23_adam_tanh10_linear_cross_entropy_uniform_every100/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 6 \ --dryRun
Finished .
Now run the same 95% case for tracking data: (or so) Note : I just added the other signal efficiencies into the same command.
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkMLP, +fkFadc, +fkScinti, fkLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/07_11_23_adam_tanh10_linear_cross_entropy_uniform_every100/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k_tracking/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 6 \ --tracking \ --dryRun
Yay, got segfaults at . Why? See section below. -> Fixed, running again, . -> Output:
Running all likelihood combinations took 4056.30966758728 s
And run the other veto setups of interest for the tiny MLP (no tracking!): I'm attempting to run 8 in total in parallel. 2 tracking, 6 of the below non tracking… Let's see what happens. Gonna re-run all of them, including the MLP only, because I just deleted a couple of the wrong files, oops! Over night.
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{fkMLP, +fkFadc, +fkScinti, fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/07_11_23_adam_tanh10_linear_cross_entropy_uniform_every100/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 6 \ --dryRun
Final output:
Running all likelihood combinations took 11828.16075730324 s
- Background rate & clusters
- No vetoes
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_adam_tanh10_linear_cross_entropy_15k" \ --backgroundSuppression
plotBackgroundClusters \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.85_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.85_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@85%" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_85_adam_tanh10_linear_cross_entropy_15k" \ --backgroundSuppression
Background rate, comparison:
plotBackgroundRate \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.98_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.98_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.85_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.85_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --names "85" --names "85" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh10_cross_entropy_15k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- Scinti+FADC+Line
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_scinti_fadc_line_adam_tanh10_linear_cross_entropy_15k" \ --backgroundSuppression
85%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@85%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_85_scinti_fadc_line_adam_tanh10_linear_cross_entropy_15k" \ --backgroundSuppression
plotBackgroundRate \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.98_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.98_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --names "85" --names "85" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh10_scinti_fadc_line_cross_entropy_15k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- Scinti+FADC+Septem+Line
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%+Scinti+FADC+Septem+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_scinti_fadc_septem_line_adam_tanh10_linear_cross_entropy_15k" \ --backgroundSuppression
85%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@85%+Scinti+FADC+Septem+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_85_scinti_fadc_septem_line_adam_tanh10_linear_cross_entropy_15k" \ --backgroundSuppression
plotBackgroundRate \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.98_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.98_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --names "85" --names "85" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh10_scinti_fadc_septem_line_cross_entropy_15k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- No vetoes
- Debug segfaults
Let's run a single command and see:
likelihood \ -f /home/basti/CastData/data/DataRuns2017_Reco.h5 \ --h5out /home/basti/org/resources/lhood_mlp_07_11_23_adam_tanh10_linear_cross_entropy_less3keV_15k_tracking//lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847_vQ_0.99.h5 \ --region=crAll \ --cdlYear=2018 \ --scintiveto --fadcveto --lineveto \ --mlp /home/basti/org/resources/nn_devel_mixing/07_11_23_adam_tanh10_linear_cross_entropy_uniform_every100/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.0421_acc_0.9847.pt \ --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --calibFile=/home/basti/CastData/data/CalibrationRuns2017_Reco.h5 \ --vetoPercentile=0.99 \ --nnSignalEff=0.95 \ --tracking
Segfault immediately. Let's compile in debug mode.
This ended up breaking our ./../CastData/data/DataRuns2017_Reco.h5 file. Time to reconstruct it…
Started at .
./runAnalysisChain \ -i ~/CastData/data \ --outpath ~/CastData/data \ --years 2017 \ --back --reco --logL --tracking
I say it will be done around 21:40. Let's see! -> It more or less finished . But it crashed in
likelihoodto compute the logL values, likely for the same reason as in the other code, see below. -> Had a minor issue with the LogReader as well, now fixedLet's compile
likelihoodin debug mode in--out:/tmp/logLand run the 2018 file to debug the crash there. Hm, no crash for the first few runs on the 2018 data.Let's compile in danger mode and try that again.
Could it have been some weird interaction between the different processes after all?
This is also running fine so far. Nope: we did get a segfault! Turns out the reason is our calculation of the
logLvalue in the septem veto code. There we calculate logL in case we need it for the septem veto!Seems to be all working now, I think!
- Background rate & clusters
- Training tiny net first < 3 keV data
Given that the tiny net above works pretty well, but slightly struggles for 0.6kV and 0.9kV data in particular, let's see if we train on 3 keV data first for a few thousand epochs.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh10_cross_entropy_less_3keV/ \ --plotPath ~/Sync/14_11_23_adam_tanh10_cross_entropy_less_3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 500
Decent training progress, given that low energy data is hard to separate:
Stopping at 15k now.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh10_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_15000_loss_0.1005_acc_0.9566.pt \ --plotPath ~/Sync/14_11_23_adam_tanh10_cross_entropy_less_3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 2000
So far (up to 24k now) the training hasn't really moved the needle at all. A bit confusing honestly. But maybe all higher energy X-rays simply fit perfectly into the right lobe of the signal data? Still, 95.7% accuracy is not exactly amazing.
-> Stopping training at 88k epochs. Improvements are extremely slow.
[X]Maybe try to find largest network that does not yet overtrain. 20 neurons? -> See section below.
Let's run the
--predictcommand on the last snapshot while training continues:./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh10_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_32000_loss_0.0997_acc_0.9569.pt \ --plotPath ~/Sync/14_11_23_adam_tanh10_cross_entropy_less_3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --predict
Done, let's have a look: ~/Sync/141123adamtanh10crossentropyless3keV/
-> The ROC curve looks extremely good again! Even better than the one of the network trained uniform data from the start.
The predictions for CDL 0.6, 0.9 and 2 kV data look much better especially!
- ./../Sync/14_11_23_adam_tanh10_cross_entropy_less_3keV/cdl_prediction_Cu-EPIC-0.9kV.pdf
- ./../Sync/14_11_23_adam_tanh10_cross_entropy_less_3keV/cdl_prediction_Cu-EPIC-2kV.pdf
Stopping the training at 50k now-> It is still improving a bit, let's wait a bit longer. -> good I didn't. A small jump at 52k, more to come? Or a sign that information about low energy events is getting worse? The shape of the output starts getting more like the 10 neuron network trained directly on uniform data. -> Stopped at 88k in the end.Let's run predict on final model:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh10_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.pt \ --plotPath ~/Sync/14_11_23_adam_tanh10_cross_entropy_less_3keV_88k/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --predict
Finished .
Let's look at: ~/Sync/141123adamtanh10crossentropyless3keV88k/
effective_eff_55fe
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh10_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_32000_loss_0.0997_acc_0.9569.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_14_11_23_adam_tanh10_l3keV_32k/
Done . While it looks reasonable, there is a jump in efficiency between Run-2 and Run-3 ./../Sync/run2_run3_14_11_23_adam_tanh10_l3keV_32k/efficiency_based_on_fake_data_per_run_cut_val.pdf
That makes me a bit worried.
Rerun with 90 and 95, 98% using the 88k epoch model that we use below:
90%
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh10_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.pt \ --ε 0.90 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_14_11_23_adam_tanh10_l3keV_mlp90_88k/
Ouch,
In Run-2 the real efficiency for the photo peak is horrific.
createAllLikelihoodCombinations
Start applying the likelihood method. First let's run only 95% case (because 30 neuron network is also still running):
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{fkMLP, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh10_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.95 \ --out ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 2 \ --dryRun
Finished .
Running all likelihood combinations took 7660.591343402863 s
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{fkMLP, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh10_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.9 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 5 \ --dryRun
Rerun everything after fix to septem & line veto:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkMLP, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh10_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.90 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 4 \ --dryRun
Rerun Run-2, see sec 1.106.2.
plotBackgroundClustersandplotBackgroundRate
- No vetoes
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_adam_tanh10_linear_cross_entropy_88k" \ --backgroundSuppression
90%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@90%" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_90_adam_tanh10_linear_cross_entropy_88k" \ --backgroundSuppression
Background rate, comparison:
plotBackgroundRate \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.98_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.98_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh10_cross_entropy_88k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- Scinti+FADC+Line
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_scinti_fadc_line_adam_tanh10_linear_cross_entropy_88k" \ --backgroundSuppression
85%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@85%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_85_scinti_fadc_line_adam_tanh10_linear_cross_entropy_88k" \ --backgroundSuppression
plotBackgroundRate \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.98_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.98_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh10_scinti_fadc_line_cross_entropy_88k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- Scinti+FADC+Septem+Line
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%+Scinti+FADC+Septem+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_scinti_fadc_septem_line_adam_tanh10_linear_cross_entropy_88k" \ --backgroundSuppression
85%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@85%+Scinti+FADC+Septem+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_85_scinti_fadc_septem_line_adam_tanh10_linear_cross_entropy_88k" \ --backgroundSuppression
plotBackgroundRate \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.98_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.98_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh10_scinti_fadc_septem_line_cross_entropy_88k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- No vetoes
- Adam network with 30 neurons
Does this also overtrain?
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/ \ --plotPath ~/Sync/14_11_23_adam_tanh30_cross_entropy_less_3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 30 \ --numHidden 30 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 1000
Starts to show very minor overtraining at around 10k epochs. So more neurons definitely not.
Stopping after 12k epochs to continue with uniform data.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_12000_loss_0.0935_acc_0.9594.pt \ --plotPath ~/Sync/14_11_23_adam_tanh30_cross_entropy_less_3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 30 \ --numHidden 30 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 2000
-> Stopping training at 64k epochs. Pretty stagnant now.
Let's run the
--predictcommand on the last snapshot while training continues:./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_28000_loss_0.0919_acc_0.9603.pt \ --plotPath ~/Sync/14_11_23_adam_tanh30_cross_entropy_less_3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --predict
Done .
Running again with 50k model :
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_50000_loss_0.0898_acc_0.9616.pt \ --plotPath ~/Sync/14_11_23_adam_tanh30_cross_entropy_less_3keV_50k/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --predict
Finished .
Look at ~/Sync/141123adamtanh30crossentropyless3keV50k/ -> looks very comparable to above! As kind of expected. But good to see.
The higher energy targets look a bit better here than in 10 neuron case above, but the 0.6kV target looks a bit worse.
effective_eff_55fe
Start
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_28000_loss_0.0919_acc_0.9603.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_14_11_23_adam_tanh30_l3keV_28k/
Finished . Also shows a slight
difference in Run-2 and Run-3.
Rerun for 90%
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.pt \ --ε 0.9 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_14_11_23_adam_tanh30_l3keV_mlp90_38k/
createAllLikelihoodCombinations
Start applying the likelihood method: NOTE: later trained network might be better!
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{fkMLP, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.90 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 5 \ --dryRun
Finished .
Rerun after fixing the septem veto. For now only single efficiency and not standalone MLP (that remains unchanged):
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkMLP, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.95 \ --out ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 6 \ --dryRun
Finished
Running all likelihood combinations took 2472.750953912735 s
Rerunning everything now, due to the septem veto behavior, at least affecting everything that includes the septem or line veto.
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkMLP, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 4 \ --dryRun
[X]I STILL HAVE TO RECONSTRUCT RUN-2 AGAIN!!! -> See sec. 1.106.2
plotBackgroundClusters,plotBackgroundRate
- No vetoes
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_adam_tanh30_linear_cross_entropy_38k" \ --backgroundSuppression
90%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@90%" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_90_adam_tanh30_linear_cross_entropy_38k" \ --backgroundSuppression
Background rate, comparison:
plotBackgroundRate \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.98_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.98_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh30_cross_entropy_38k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- Scinti+FADC+Line
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_scinti_fadc_line_adam_tanh30_linear_cross_entropy_38k" \ --backgroundSuppression
85%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@85%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_85_scinti_fadc_line_adam_tanh30_linear_cross_entropy_38k" \ --backgroundSuppression
plotBackgroundRate \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.98_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.98_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh30_scinti_fadc_line_cross_entropy_38k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- Scinti+FADC+Septem+Line
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%+Scinti+FADC+Septem+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_scinti_fadc_septem_line_adam_tanh30_linear_cross_entropy_38k" \ --backgroundSuppression
85%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@85%+Scinti+FADC+Septem+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_85_scinti_fadc_septem_line_adam_tanh30_linear_cross_entropy_38k" \ --backgroundSuppression
plotBackgroundRate \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.98_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.98_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/lhood_c18_R3_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh30_scinti_fadc_septem_line_cross_entropy_38k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
Uhhh, something fishy is going on here. Nothing is left in the background rate!
See section below.
- No vetoes
- Debug septem veto behavior
[ ]Debug septem veto!!![ ]Does this also happen for septem veto alone? -> Just run on Run-3 alone and see what happens. Add some echos and run with plots
[X]Get likelihood command from
createAllLikelihoodCombinationsThis for example:likelihood \ -f /home/basti/CastData/data/DataRuns2018_Reco.h5 \ --h5out /home/basti/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k//lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5 \ --region=crAll --cdlYear=2018 \ --scintiveto --fadcveto --septemveto --lineveto \ --mlp /home/basti/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.pt \ --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --calibFile=/home/basti/CastData/data/CalibrationRuns2018_Reco.h5 \ --vetoPercentile=0.99 \ --nnSignalEff=0.95
[X]Look output file for this:
hdfview /home/basti/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k//lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611_vQ_0.99.h5
-> It says total passed events 1673 -> This has 25 clusters on chip 3 in run 240. compared to
hdfview /home/basti/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576_vQ_0.99.h5
which says 5006. Not such a big difference, so that's very weird. -> This file has 72 clusters on chip 3 in run 240.
A factor of 3 difference is quite bizarre, but I would have expected even more given the background rate plot.
[X]Run
likelihoodcommand and see. For now only run in run 240.likelihood \ -f /home/basti/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/test_R3_all_vetoes.h5 \ --region=crAll --cdlYear=2018 \ --scintiveto --fadcveto --septemveto --lineveto \ --mlp /home/basti/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.pt \ --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --calibFile=/home/basti/CastData/data/CalibrationRuns2018_Reco.h5 \ --vetoPercentile=0.99 \ --nnSignalEff=0.95 \ --run 240 \ --plotSeptem
-> The plots look a bit funny in some aspects.
I get the feeling that in the Septemboard data we do indeed NOT 'invert' the pixel data. We don't call any function that performs such an inversion. And unless I'm missing something, given that we work with the raw pixel data as input (and that is never converted), the data "would need" to be inverted as well? Also: when we see the septemboard in event displays, I'm pretty sure our chip 2 is on the left of the center row and chip 4 on the right. If we inverted they would be switched!
-> WE DO USE toXPix. IN LIKELIHOOD! That would explain some things!
-> AT THE SAME TIME the argument to these procedures is the
cl.centerXfield! That has already been inverted due to how reconstruction works? Or not?From
geometry.nim:when T is Pix or T is PixTpx3: (result.centerX, result.centerY) = applyPitchConversion(pos_x, pos_y, NPIX) elif T is PixInt or T is PixIntTpx3: if useRealLayout: (result.centerX, result.centerY) = (toRealXPos(pos_x), toRealYPos(pos_y)) else: (result.centerX, result.centerY) = applyPitchConversion(pos_x, pos_y, NPIX * 3) else: error("Invalid type: " & $T)
this means we use
toRealXPosinstead ofapplyPitchConversionor anything that injects the inversion foruseRealLayout.toRealXPosis justproc toRealXPos*(x: int|float): float = (float(x) + 0.5) * (XSize / XSizePix.float)
So clearly NO inversion.
So I think we need to use
toXPixNormaleverywhere inlikelihood.nim?The only code branch affecting us (using real septem layout) is:
let cX = toXPix(clusterTup[1].centerX) let cY = toYPix(clusterTup[1].centerY)
inside
applySeptemVeto.Let's make that change there and rerun the code.
Ok, that should not change much, because this is only used to determine the chip the cluster is on. But inverting x does not change the result. The center chip is always going to be in the center!
[X]ChecklvRegularin use when no septem veto -> Seems to be correct in output HDF5 files
For example plot: Septem event of event 10172 and run 240. Center cluster energy: is at least extremely bizarre. At the very least there is a center dot where it does not belong.
Let's debug the center clusters points.
For some reason
xCenter/yCenteris(0,0)in this event. That's the red dot for some reason.??? Because the red dot is exactly in the center -> Ahh! septem geometryx/yCenterare 0 here, because they are only set when the line veto kicks in (a cluster to be checked for the line veto. Not the case in this event due tolvRegularNoHLC. the only cluster is part of the big cluster)Ohh, do we trigger a ggplotnim bug here? That the data is considered discrete (single entry) and thus it's placed at the center of the plot instead where it belongs? -> Yes. The issue was that we set the
dcKindX/YinfilledIdentityGeombased on the individual X and y scales. This seems nonsensical to me. So now we use thefilledScales.discreteX/Yvalue instead. If either x or y scales are discrete, how could we have a non discrete geom there?But the current logic assigns
discreteX/Yfields such that they are set totrueongeoms.anyIt(it.kind == dcDiscrete)essentially. That also seems very bad.-> Ok, this works now.
Back to where we were. Why do some of these look so weird? In particular above 2 keV there's barely anything. Let's look at that.
Ok some events are just utterly wrong: Septem event of event 1498 and run 240 for example. There is no large cluster, and yet the
septemVetoedis true. It is clearly line vetoed, but that does not mean the septem veto should be true. -> Ok, I introduced that changing fromseptemVetoPassedtoseptemVetoed. There are cases when theifcondition is never met so the veto is never set to false.I think the issue is that the reconstructed septem cluster ends up having different properties slightly (likely the charge?) and thus does not get predicted correctly. -> The cut value is the same in both branches (
buildSeptemEventand body ofapplySeptemVeto) -> Output fromnn_predict.nim predictproc:Data index 436: DataFrame with 22 columns and 1 rows: Idx centerX centerY hits eccentricity skewnessLongitudinal skewn essTransverse kurtosisLongitudinal kurtosisTransverse length width rmsLongitudinal rmsTransverse lengthDivRmsTrans rotationAngle energyFromCharge fractionInTransverseRms totalCharge gasGain eventNumber Type Target σT dtype: float float float float float float float float float float float float float float float float float float int string string float 0 3.288 1.371 7 2.557 -0.7306 -0.7589 -1.214 -1.063 2.94 1.262 1.181 0.4619 6.365 0.1241 0.167 0 2.115e+04 3.004 1498 back C-EPIC-0.6kV 0.62 for path: /reconstruction/run_240/chip_3for index 436 (which is the data index for chip 3 for event 1498). Note the total charge 2.115e+04. Let's compare that to the charge in the septem cluster.
UHHHHHHHHH, there are many clusters in
evaluateClusterfor whomtotChargeis 0!! -> Yeah, there really seems to be a mismatch between the pixels as we access it later and how we insert data.Phew, what a trip.
The fundamental issue was the following:
- the charge tensor storing the charge values of all pixels was always put into a real layout septemboard tensor
- The pixels added into the tensor were only using tight layout pixel values (which by itself is not a problem, the real layout tensor simply has some additional empty padding)
- in the lookup of the pixels in
evaluateClusterthe pixel information however WAS in the real coordinates, so the pixel lookup looked at the wrong pixels! Thus charge remained empty. - In addition to that, we were always performing the cluster finding on the real layout, instead on the tight layout. This means the septem veto was much less efficient for any event at the bottom of the chip (in particular)!
See for example: Septem event of event 19506 and run 240. Center cluster energy: which
All events we looked at (after fixes!) are found in ./Figs/statusAndProgress/debugSeptemVeto/septem_events_debug_missing_charge_run240.pdf
Page 252 for event 19506 to see that the clustering algo now correctly identifies events over the bottom chip boundary.
The angles are still correct though, because we convert to the real coordinates after clustering, but before cluster reconstruction! See for example page 257 of event 19642.
Now try rerunning the likelihood application. -> I looked at it yesterday evening and it still is broken.
Two possible reasons:
- the
nnPredvalue is still different for each individual cluster in the septem events compared to the original cluster - our change to
passed -> septemVetoedbroke the veto
Let's investigate, starting again with the same run 240, command from above.
I think it may be (2). Example event: Septem event of event 21860 and run 240. Center cluster energy: That one has a single cluster on the center chip that is not connected. While it is line vetoed (fair enough) it is also septem vetoed, which does not make sense, indicating that our logic for
septemVetoedis probably wrong!Ok, the chip on the center cluster is simply, once again, not kept by the MLP itself:
Index was: 6463 Chip center cluster: 3 nnVeto? true == center? true for center: 3 nnVeto : true for -1.699134230613708 < -1.243452227115631 for energy: 0.120909825335327
so it seems the cause is actually rather point (1).
Event number: 21860 has nnPred: 0.6899389028549194
Has a charge:
1.65e+04
and 5 hits.
In the septem veto code:
Total charge: 16500.64730257308 num pixels: 5
so the charge seems to match!
What else is it then?
In
evaluateCluster:Total charge: 16500.64730257308 num pixels: 5 CLUSTER DF IN `evaluateCluster` yielded: -1.699134230613708 centerX = 17.55 centerY = 31.92 eccentricity = 1.705 energyFromCharge = 0.1209 fractionInTransverseRms = 0 hits = 5 kurtosisLongitudinal = -1.546 kurtosisTransverse = -1.755 length = 1.506 lengthDivRmsTrans = 4.276 likelihood = inf rmsLongitudinal = 0.6006 rmsTransverse = 0.3522 rotationAngle = 1.803 skewnessLongitudinal = -0.3915 skewnessTransverse = -0.1545 totalCharge = 1.65e+04 width = 0.8081 σT = 0.62 Chip center cluster: 3 nnVeto? true == center? true for center: 3 nnVeto : true for -1.699134230613708 < -1.243452227115631 for energy: 0.120909825335327 Index was: 6463
vs the original prediction:
Pred data for group: /reconstruction/run_240/chip_3 Target = C-EPIC-0.6kV Type = back centerX = 11.02 centerY = 12.7 eccentricity = 3.122 energyFromCharge = 0.1295 eventNumber = 21860 fractionInTransverseRms = 0 gasGain = 3.016 hits = 5 kurtosisLongitudinal = -1.486 kurtosisTransverse = 0.1102 length = 1.707 lengthDivRmsTrans = 8.044 rmsLongitudinal = 0.6625 rmsTransverse = 0.2122 rotationAngle = 1.803 skewnessLongitudinal = -0.2226 skewnessTransverse = -1.393 totalCharge = 1.65e+04 width = 0.5799 σT = 0.62
YEAH, that is clearly busted. How can the numbers be so different? My only explanation is that it must be related to our septem event reconstruction.
The cluster data being reconstructed is the following pixels:
Cluster data: @[(x: 319, y: 570, ch: 53), (x: 315, y: 562, ch: 85), (x: 311, y: 582, ch: 66), (x: 323, y: 592, ch: 42), (x: 323, y: 591, ch: 50)]
Let's compare with the raw data in the H5 file.
import ingrid / [ingrid_types, tos_helpers] import os, stats import nimhdf5 const p = "~/CastData/data/DataRuns2018_Reco.h5" const run = 240 const chip = 3 const dataIndex = 6463 let tp = special_type(uint8) var pixs = newSeq[Pix]() withH5(p, "r"): let xs = h5f[recoPath(run, chip).string / "x", tp, uint8] let ys = h5f[recoPath(run, chip).string / "y", tp, uint8] echo "Total: ", xs.len, " and ", ys.len let xi = xs[dataIndex] let yi = ys[dataIndex] for i in 0 ..< xi.len: pixs.add (x: xi[i], y: yi[i], ch: 0'u16) echo pixs # convert them to real pixels echo pixs.chpPixToSeptemPix(3) echo pixs.chpPixToSeptemPix(3).septemPixToRealPix echo "And real layout:" var pxs = newSeq[float]() var pys = newSeq[float]() for p in pixs.chpPixToSeptemPix(3).septemPixToRealPix: pxs.add toRealXPos(p.x) pys.add toRealYPos(p.y) echo pxs echo pys echo pxs.mean echo pys.mean
UPDATE : I'm a dummy. I was reading x data also for y here. Ignore the next debugging sessions below. Jump to 1.103.10.4.4.
Check, we get 5 pixels. The x coordinates are correct, but in y we get too large spacing. Do we accidentally scale along the y axis? Hmm, converting manually from chip pixels to septemboard pixels and then to the real layout keeps the correct spacing. Do we accidentally convert too much (i.e. again?)? It is not just applying
septemToRealPixtwice as we can see. Guess we need to dig into the code.In
getPixels(inprivate/veto_utils.nim), we already see:PIX raw: (57, 221) pix : (x: 313, y: 477, ch: 53) pC : (x: 319, y: 570, ch: 53) PIX raw: (53, 213) pix : (x: 309, y: 469, ch: 85) pC : (x: 315, y: 562, ch: 85) PIX raw: (49, 233) pix : (x: 305, y: 489, ch: 66) pC : (x: 311, y: 582, ch: 66) PIX raw: (61, 243) pix : (x: 317, y: 499, ch: 42) pC : (x: 323, y: 592, ch: 42) PIX raw: (61, 242) pix : (x: 317, y: 498, ch: 50) pC : (x: 323, y: 591, ch: 50)
so the y component of the raw data is already off?
allChipDatato blame? In all chip data we use the data we read inreadAllChipData, which is defined inlikelihoodand just doesresult = AllChipData(x: newSeq[seq[seq[uint8]]](numChips), y: newSeq[seq[seq[uint8]]](numChips), ToT: newSeq[seq[seq[uint16]]](numChips), charge: newSeq[seq[seq[float]]](numChips)) for i in 0 ..< numChips: result.x[i] = h5f[group.name / "chip_" & $i / "x", vlenXY, uint8] result.y[i] = h5f[group.name / "chip_" & $i / "y", vlenXY, uint8] result.ToT[i] = h5f[group.name / "chip_" & $i / "ToT", vlenCh, uint16] result.charge[i] = h5f[group.name / "chip_" & $i / "charge", vlenCh, float]
there's not much that could go wrong here?
Outputting the data in
getPixelsviaallChipData.y[chip][6463]directly yields the same, wrong, numbers. huh?So, given that the raw data looks to be the same, the differences must come from the geometry calculation, no?
Maybe it's an issue of the center of the cluster. Well, the rotation angle is exactly the same, so that also seems unlikely.
The center position in
geometryis (posx, posy argument to calcGeometry):for center: (17.54686299615877, 31.92122350230415)
same as calculated in snippet above!
Let's try to call
calcGeometryfrom here with the original and real layout data and see what we get. We will hardcode the rotation angle from above.import ingrid / [ingrid_types, tos_helpers] import os, stats, sequtils import nimhdf5 const p = "~/CastData/data/DataRuns2018_Reco.h5" const run = 240 const chip = 3 const dataIndex = 6463 let tp = special_type(uint8) var pixs = newSeq[Pix]() withH5(p, "r"): let xs = h5f[recoPath(run, chip).string / "x", tp, uint8] let ys = h5f[recoPath(run, chip).string / "y", tp, uint8] echo "Total: ", xs.len, " and ", ys.len let xi = xs[dataIndex] let yi = ys[dataIndex] for i in 0 ..< xi.len: pixs.add (x: xi[i], y: yi[i], ch: 0'u16) #var pxs = newSeq[PixInt]() #for p in pixs.chpPixToSeptemPix(3).septemPixToRealPix: # pxs.add (x: toRealXPos(p.x), y: toRealYPos(p.y), ch: 0) # now call calcGeometry proc cX[T](p: seq[T]): float = p.mapIt(it.x.float).mean proc cY[T](p: seq[T]): float = p.mapIt(it.y.float).mean let (pcx, pcy) = applyPitchConversion(cX(pixs), cY(pixs), 256) let pxs = pixs.chpPixToSeptemPix(3).septemPixToRealPix let (pcxR, pcyR) = (toRealXPos(cX(pxs)), toRealYPos(cY(pxs))) echo (pcx, pcy), " with ", pixs echo calcGeometry(pixs, pcx, pcy, 1.803, useRealLayout = false) echo (pcxR, pcyR), " with ", pxs echo calcGeometry(pxs, pcxR, pcyR, 1.803, useRealLayout = true)
The numbers are identical.The 'identicalness' was only because I was reading the doubly printed lines… I was echoing twice. But this does not explain why the reco240.h5 code snippet below printed the 'new' numbers?!This must imply that the numbers we compare to are wrong, no?
Let's read the raw numbers for, e.g. the rms values for the same index.
import ingrid / [ingrid_types, tos_helpers] import os, stats, sequtils import nimhdf5 const p = "~/CastData/data/DataRuns2018_Reco.h5" const run = 240 const chip = 3 const dataIndex = 6463 let tp = special_type(uint8) var pixs = newSeq[Pix]() withH5(p, "r"): let rmsT = h5f[recoPath(run, chip).string / "rmsTransverse", float] echo rmsT.len echo rmsT[dataIndex]
which yields the same number as in the output above. What the hell?
Let's reconstruct run 240 and see what happens?
Run 240 already exists as a raw H5 file in ./../CastData/data/2018_2/raw_240.h5
so let's go from there:
reconstruction -i ~/CastData/data/2018_2/raw_240.h5 --out /tmp/reco_240.h5
and let's read from that:
import ingrid / [ingrid_types, tos_helpers] import os, stats, sequtils import nimhdf5 const p = "/tmp/reco_240.h5" const run = 240 const chip = 3 const dataIndex = 6463 let tp = special_type(uint8) var pixs = newSeq[Pix]() withH5(p, "r"): let rmsT = h5f[recoPath(run, chip).string / "rmsTransverse", float] let evs = h5f[recoPath(run, chip).string / "eventNumber", int] echo rmsT.len echo rmsT[dataIndex] echo "event: ", evs[dataIndex]
UPDATE: : I'm exceptionally confused. I swear the above snippet initially reproduced the same
0.3 number for rmsT instead of the now (old / correct) 0.21 number. What is going on. Ok, for the snippet where I call ~calcGeometrymanually above, I simply misread my own output. But why did the code here produce what I expected, causing me to get even more confused? Honestly, I don't know what to believe anymore. I guess my must have imagined it?!?!… is… our… reconstructed data broken? What? This can't be a Nim regression in
stats, can it? Seems unlikely, there haven't been any meaningful changes there in over 2 years. Is it a regression? A fix? The most related thing I can think of is that we introduced always using the long axis based on the max / min differences. But that should if anything imply that the rmsLongitudinal value should then be the correct one. Bizarre.Huh, running reconstruction in single threaded mode and printing the pixels + the geometry calc output:
Cluster data: @[(x: 57, y: 221, ch: 53), (x: 53, y: 213, ch: 85), (x: 49, y: 233, ch: 66), (x: 61, y: 243, ch: 42), (x: 61, y: 242, ch: 50)] for center: (11. 0165, 12.6995) (rmsLongitudinal: 0.662548681986465, rmsTransverse: 0.2122009519252882, eccentricity: 3.122270074545826, rotationAngle: 1.802620123362616, skewnessLongitudin al: -0.2226153757621063, skewnessTransverse: -1.393148992657465, kurtosisLongitudinal: -1.485727185875586, kurtosisTransverse: 0.1102275455215613, length: 1. 706952117272804, width: 0.5798664786334701, fractionInTransverseRms: 0.0, lengthDivRmsTrans: 8.044036097791816)
this matches the numbers for the original data after all???
What? The code snippet above now also produces the correct result???
Anyway, we need to understand why
calcGeometrygives such different numbers for the real and single chip coordinates.Let's repeat the code here to leave the above untouched (if we change more and more):
import ingrid / [ingrid_types, tos_helpers] import os, stats, sequtils import nimhdf5 const p = "~/CastData/data/DataRuns2018_Reco.h5" const run = 240 const chip = 3 const dataIndex = 6463 let tp = special_type(uint8) var pixs = newSeq[Pix]() withH5(p, "r"): let xs = h5f[recoPath(run, chip).string / "x", tp, uint8] let ys = h5f[recoPath(run, chip).string / "y", tp, uint8] echo "Total: ", xs.len, " and ", ys.len let xi = xs[dataIndex] let yi = ys[dataIndex] for i in 0 ..< xi.len: pixs.add (x: xi[i], y: yi[i], ch: 0'u16) #var pxs = newSeq[PixInt]() #for p in pixs.chpPixToSeptemPix(3).septemPixToRealPix: # pxs.add (x: toRealXPos(p.x), y: toRealYPos(p.y), ch: 0) # now call calcGeometry proc cX[T](p: seq[T]): float = p.mapIt(it.x.float).mean proc cY[T](p: seq[T]): float = p.mapIt(it.y.float).mean let (pcx, pcy) = applyPitchConversion(cX(pixs), cY(pixs), 256) let pxs = pixs.chpPixToSeptemPix(3).septemPixToRealPix let (pcxR, pcyR) = (toRealXPos(cX(pxs)), toRealYPos(cY(pxs))) echo (pcx, pcy), " with ", pixs echo calcGeometry(pixs, pcx, pcy, 1.803, useRealLayout = false) echo (pcxR, pcyR), " with ", pxs echo calcGeometry(pxs, pcxR, pcyR, 1.803, useRealLayout = true)
The rotated coordinates are clearly similar but different.
I think I found the issue. While there is a slight difference in our
PITCHvariable and the equivalent intoRealX/YPos, the actual cause is…. our damn inversion of the X coordinates inapplyPitchConversion!Due to that inversion, the geometric properties change for some reason that I don't fully understand.
This also likely explains my previous 'confusion' above! I had compiled the
reconstructionprogram without the inversion the other day to test it. By doing that the produced rmsTransverse will indeed have been the ~0.3 value instead of the 0.21.So question:
- What do we want to do with this knowledge?
- Why is there a difference due to the rotation?
Output of
reconstructionincluding the 256 - x pitch conversion:Cluster data: @[(x: 57, y: 221, ch: 53), (x: 53, y: 213, ch: 85), (x: 49, y: 233, ch: 66), (x: 61, y: 243, ch: 42), (x: 61, y: 242, ch: 50)] for center: (11. 0165, 12.6995) (rmsLongitudinal: 0.662548681986465, rmsTransverse: 0.2122009519252882, eccentricity: 3.122270074545826, rotationAngle: 1.802620123362616, skewnessLongitudin al: -0.2226153757621063, skewnessTransverse: -1.393148992657465, kurtosisLongitudinal: -1.485727185875586, kurtosisTransverse: 0.1102275455215613, length: 1. 706952117272804, width: 0.5798664786334701, fractionInTransverseRms: 0.0, lengthDivRmsTrans: 8.044036097791816)
Now without:
Cluster data: @[(x: 57, y: 221, ch: 53), (x: 53, y: 213, ch: 85), (x: 49, y: 233, ch: 66), (x: 61, y: 243, ch: 42), (x: 61, y: 242, ch: 50)] for center: (3.1 185, 12.6995) (rmsLongitudinal: 0.6001520258271371, rmsTransverse: 0.3518771744452651, eccentricity: 1.705572482140331, rotationAngle: 1.802620129698567, skewnessLongitudi nal: -0.391515795064999, skewnessTransverse: -0.1545464167312244, kurtosisLongitudinal: -1.546467541782199, kurtosisTransverse: -1.755435457911516, length: 1 .504769548869993, width: 0.8073218718662321, fractionInTransverseRms: 0.0, lengthDivRmsTrans: 4.276405684006827)
Yeah. That is the reason. It even makes sense. By inverting the x coordinate our rotation angle is not aligned with the coordinates anymore. Computing the rotation angle of a cluster and then inverting the coordinates, requires the angle to be inverted too. E.g. an angle of 30° of a track in (x, y) becomes a track of -30° in (-x, y).
This is why in
calcGeometrywe actually write-rot_angleas arguments to sin/cos.Also, one reason this has not been super obvious so far is that for very spherical clusters (those likely to be found as X-ray like and thus part of septem & line veto) the difference is between long and small axis is small. Therefore rotating to a wrong coordinate system that is not really aligned with long and short axis is not so important. And for any method not using the MLP the difference is even less pronounced, because only the 3 variables entering LnL are relevant.
Questions:
[X]Did we use subtract from 768 in the past for septem / line veto? -> I don't think so. Now for sure we don't anymore at all.[ ]Did we reconstruct the 2017 data file two days (?) ago using a wrongreconstructionfile, i.e. one that did not contain theNPIX - xpart? -> Better just recreate that file again.
With the rotation angle taken into account based on the
useRealLayout, our cluster info fromevaluateClusternow looks like:CLUSTER DF IN `evaluateCluster` yielded: 0.1691101491451263 centerX = 17.55 centerY = 31.92 eccentricity = 3.122 energyFromCharge = 0.1209 fractionInTransverseRms = 0 hits = 5 kurtosisLongitudinal = -1.486 kurtosisTransverse = 0.1102 length = 1.708 lengthDivRmsTrans = 8.043 likelihood = inf rmsLongitudinal = 0.6631 rmsTransverse = 0.2124 rotationAngle = 1.803 skewnessLongitudinal = 0.2226 skewnessTransverse = -1.393 totalCharge = 1.65e+04 width = 0.5804 σT = 0.62 Chip center cluster: 3 nnVeto? false == center? true for center: 3 nnVeto : false for 0.1691101491451263 < -1.243452227115631 for energy: 0.120909825335327
which now is ~identical to the previous info from direct prediction:
Pred data for group: /reconstruction/run_240/chip_3 Target = C-EPIC-0.6kV Type = back centerX = 11.02 centerY = 12.7 eccentricity = 3.122 energyFromCharge = 0.1295 eventNumber = 21860 fractionInTransverseRms = 0 gasGain = 3.016 hits = 5 kurtosisLongitudinal = -1.486 kurtosisTransverse = 0.1102 length = 1.707 lengthDivRmsTrans = 8.044 rmsLongitudinal = 0.6625 rmsTransverse = 0.2122 rotationAngle = 1.803 skewnessLongitudinal = -0.2226 skewnessTransverse = -1.393 totalCharge = 1.65e+04 width = 0.5799 σT = 0.62
The minor differences are due to:
applyPitchConversionstill usingnpix - x, where it should benpix - 1 - xapplyPitchConversionstill using aPITCH = 0.055value, when the real one (e.g. used in real layout) is slightly larger.
With this the event: Septem event of event 21860 and run 240 is now not vetoed by the septem veto, as it should be.
Let's run the
likelihoodonly on Run 3 data (need to reconstruct Run-2 again) and see what we get:likelihood \ -f /home/basti/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/R3_mlp_all_vetoes.h5 \ --region=crAll \ --cdlYear=2018 \ --scintiveto --fadcveto --septemveto --lineveto \ --mlp /home/basti/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.pt \ --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --calibFile=/home/basti/CastData/data/CalibrationRuns2018_Reco.h5 \ --vetoPercentile=0.99 \ --nnSignalEff=0.95
Finished .
It still produces completely bizarre results.
Compare with the
*_before_any_fixes.pdfplots in the same directory for what it looked like before this section.I'm now just running likelihood on a single run and then plotting the clusters etc. to try to learn something.
Ok, for example this event: Septem event of event 98659 and run 276 should absolutely be part of the resulting output file and be damn near in the center. -> But IT IS NOT.
ALSO: in the HDF5 file the datasets have 7430 cluster entries. But in the cluster plot the top only says 4320??? -> This could be due to our energyMin 0.2 filter & general noisy pixel filter!
Why is the event number above not there anymore though?
-> The reason was:
if (useSeptemVeto and not septemVetoed) or (useLineVeto and lineVetoed): ## If `passed` is still false, it means *no* reconstructed cluster passed the logL now. Given that ## the original cluster which *did* pass logL is part of the septem event, the conclusion is that ## it was now part of a bigger cluster that did *not* pass anymore. passedInds.excl septemFrame.centerEvIdx passedEvs.excl evNum
which I had forgotten to adjust when going from
passedtoseptemVetoed. Thenotin front ofseptemVetoedneeded to be removed!This fixed it!
- Adam network with 50 neurons
Let's look at a network with 50 neurons in parallel.
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/07_11_23_adam_tanh50_linear_cross_entropy_uniform_every100/ \ --plotPath ~/Sync/07_11_23_adam_tanh50_linear_cross_entropy_uniform_every100/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 50 \ --numHidden 50 \ --activation tanh \ --outputActivation linear \ --lossFunction sigmoidCrossEntropy \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 100
This one starts to see a tiny amount of overtraining near epoch 1200 and onwards.
Gets more and more significant (still minor though), up to 3.5k now.
Stopping at 4.2k .
1.103.11. TODO notes
[X]note about addition of data incalibInfo[X]plots and commands for before and after in/Sync/
[ ]effective efficiency plots of firstless3keV and fixedabsLength[ ]background rate & clusters and commands of firstless3keV -> in terms of background & efficiency it actually seems very promising. But we'd have to argue around the behavior for some CDL runs. -> Maybe add in conclusion about MLP that better data for validation would be extremely useful. TAKE MORE XRAY TUBE DATA!!!!!
1.103.12. TODO
[ ]Check background diffusion is used correctly with its -40 offset -> I believe so, but check[X]Check what the prediction of the MLP looks like for e.g. run 335, 336, 337 for the fake data (Cu-EPIC 2kV). As we saw in the quoted section above
Target local Cu-EPIC-0.9kV cutValue = 0.03405002132058144 eff = 0.7999139414802066
this seems to imply even the fake data ends up on the 'left' side of the prediction plot? -> See above.
[X]Check the fake event generation logic (parameter range) to see how it compares to the data produced for training. -> See the UPDATE in sec. 1.103.3[ ]Implement writing the simulated data to HDF5 files of the/reconstructionform!!! -> Ideally we would also store the raw data though :/ -> Still would be useful to have general idea of what data looks like and more importantly to start training quicker![ ]Pre-train e.g. 50k epochs with a dataset heavily biased towards low energies!
[ ]Maybe the total charge really is a problem in our datasets for CDL? -> Maybe train another network without total charge, i.e. without any charge information at all? Or can we improve the total charge we recover? -> We fixed the total charge usingCalibInfo, by attaching the correct calibration factor to use for the CDL runs based on the fit peak position. For the width we now use the width of the fit to the peak.[ ]What would the effective efficiency of low energy CDL runs look like if we disable the neighboring pixel logic?
1.104.
Today working on applying the correct coordinates to the limit calculation.
See ./../phd/thesis.html for our explanation as to why exchanging X and Y axes is enough for the cluster data to rotate the clusters to the 90° coordinate system that describes the system used by TrAXer.
[X]Implement switching axes intomcmc_limit_calculation[X]Verify that switching the coordinates works as expected by producing a plot of the clusters as read. -> It seems to work as expected. Run sanity checks first without switching axes to some test place:
./mcmc_limit_calculation sanity --limitKind lkMCMC --rombergIntegrationDepth 3 --backgroundInterp --sanityPath /tmp/testSanity
And now run with the switching to some other place:
./mcmc_limit_calculation sanity --limitKind lkMCMC --rombergIntegrationDepth 3 --backgroundInterp \ --sanityPath /tmp/testSanitySwitched \ --switchAxes
Comparing the
background_clusters.pdfplot shows the axes are clearly switched. Also thinking about the non switched data by imagining it from the opposite side (mirrored "into" the plot) and rotating then by 90° clockwise gives the same result as the switched data indeed. The plots stored here:[X]Implement TrAXer output to common CSV format we read in
mcmc_limit_calculationfor the raytracing heatmap -> Should be done inplotBinary-> Implemented. Example:./plotBinary \ --dtype float \ -f out/image_sensor_0_2023-11-11T21:16:32+01:00__dx_14.0_dy_14.0_dz_0.1_type_float_len_1000000_width_1000_height_1000.dat \ --invertY \ --out /t/axion_image_figure_errors.pdf \ --inPixels=false \ --title "Axion image, axion-electron" \ --xrange 7.0 \ --gridpixOutfile ~/org/resources/limitClusterCorrectRotationDevelop/test_gridpix_file.csv
Note that most arguments are irrelevant for the CSV file!
invertYis really the only parameter that affects the output file.[X]Implement switching axes inplotBackgroundClusters-> Done.[X]Create background cluster plot of the old tracking candidates with correct rotation and the new axion image, to get a rough idea where we end up in terms of candidates (I have a bad feeling about this!) First test without switching using MLP95%:
plotBackgroundClusters \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --outpath ~/org/Figs/statusAndProgress/limitClusterCorrectRotation/ \ --suffix "mlp_candidates_run2_run3_no_switching" \ --energyMin 0.2 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0
-> Looking good.
Now with switching:
plotBackgroundClusters \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --outpath ~/org/Figs/statusAndProgress/limitClusterCorrectRotation/ \ --suffix "mlp_candidates_run2_run3_switching" \ --energyMin 0.2 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0 \ --switchAxes
-> Still looking fine! (and it seems even this way we are lucky with the candidates!)
Now also with new axion image:
plotBackgroundClusters \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2017.h5 \ /home/basti/Sync/lhood_tracking_scinti_line_mlp_0.95_2018.h5 \ --outpath ~/org/Figs/statusAndProgress/limitClusterCorrectRotation/ \ --suffix "mlp_candidates_run2_run3_switching_new_axion_image" \ --energyMin 0.2 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage /home/basti/org/resources/limitClusterCorrectRotationDevelop/test_gridpix_file.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0 \ --switchAxes
-> I guess we will still be lucky! (Unless the new MLP brings us a bunch of new clusters in the center)
[ ]Create likelihood combinations for the new tiny MLP
- 85%, 90%, 95%, 98% with scinti+fadc+line, all vetoes
- Start with 95% + scinti+fadc+line & with and without tracking
-> Will add these to sec. 1.103.10.2!
[ ]Calculate limit for previous best case scenario (95% tiny MLP + scinti + fadc + line)[ ]Run limits on different setups (with new axion image & switched axes):- LnL @ 80%, 90%, without vetoes, scinti+fadc+line, all vetoes
- Old MLP @ 95% + scinti + fadc + line (as reference)
- tiny MLP @ 85%, 90%, 95%, 98% without vetoes, scinti+fadc+line, all vetoes
1.105.
See sec. 1.103.10.4.4 for the notes on debugging the broken septem veto for (at least) the Adam 30 neuron MLP. Continue work today .
[ ]WE need to rerun the random coincidence calculations for the vetoes after our septemboard and line veto related fixes inlikelihood![ ]NOTE this might also explain why the lnL method + septem veto got worse at some point! From < 10k clusters total over the entire center chip to > 10k after changes! -> In other words we need to rerun all likelihood files that contain septem or line vetoes! (line veto alone should be safe, but who knows)[ ]I STILL HAVE TO RECONSTRUCT RUN-2 AGAIN!!! -> see below
1.106.
Let's reconstruct one run from Run-2, then compare some cluster
properties for a few clusters to the existing DataRuns2017Reco.h5
file to see if we actually reconstructed it using the not-inverted
reconstruction or not.
Raw data:
raw_data_manipulation -p /mnt/4TB/CAST/Data/2017/DataRuns/Run_100_171128-06-46.tar.gz -r back -o /tmp/run_100.h5
Reconstruction:
reconstruction -i /tmp/run_100.h5 --out /tmp/reco_100.h5
Now compare:
import ingrid / [ingrid_types, tos_helpers] import os, stats, sequtils import nimhdf5, datamancer const run = 100 const chip = 3 const DsetSet = XrayReferenceDsets - { igNumClusters, igFractionInHalfRadius, igRadiusDivRmsTrans, igRadius, igBalance, igLengthDivRadius } + {igEventNumber} let Dsets = DsetSet.toSeq.mapIt(it.toDset()) proc read(f: string): DataFrame = withH5(f, "r"): result = readRunDsets(h5f, run = run, chipDsets = some(( chip: chip, dsets: Dsets) )) let dfN = read("/tmp/reco_100.h5") let dfE = read("~/CastData/data/DataRuns2017_Reco.h5") proc printRow(df: DataFrame) = doAssert df.len == 1, "not 1 element" for k in getKeys(df).sorted: echo k, " = ", df[k, Value][0] for i in 0 ..< 3: dfN[i].printRow dfE[i].printRow echo "\n\n==============================\n\n"
- The DataRuns file does seem to be different. So we need to re-reconstruct.
- But more importantly: why are there different events in each
file???
-> The contents of the new file are not sorted by event
number. Ughhhh
-> For one: we reconstructed in the single threaded branch of
reconstruction-> In theraw_data_manipulationoutput the data is already not sorted!
While debugging the event numbering / file ordering issue in raw, I
noticed the usage of the batchFileReading procedure. What the hell
was the point of that? All the data is kept in memory for the duration
of the proc anyways?
-> It was a regression of this commit > c366538275c66c7ff6c5e86330899130b389d76b > AuthorDate: Sun Jan 29 02:52:21 2023 +0100 > [raw] improve handling of list of raw files & name parsing I forgot to extract the sorted filenames when making that change. I > took the unsorted files.
I just spent some time trying to improve the multithreading logic in
raw_data_manipulation, but everything I tried pretty much ended up
being either worse or also crashed. :( A cligen solution I wrote is
really quite nice, but suffers from severe performance issues, due to
the forking. The HDF5 library goes crazy in that for some reason
(based on perf).
[X]wrote a refresher for the CacheTab HDF5 files, ./../CastData/ExternCode/TimepixAnalysis/Tools/refresh_serialized_file.nim.
I tried to parallelize the septem veto logic.
Let's try to run it on run 276:
likelihood -f /home/basti/CastData/data/DataRuns2018_Reco.h5 --h5out /tmp/test_R3_septem_276_perf_fix.h5 --region=crAll --cdlYear=2018 --septemVeto --mlp /home/basti/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.pt --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 --calibFile=/home/basti/CastData/data/CalibrationRuns2018_Reco.h5 --vetoPercentile=0.99 --nnSignalEff=0.95 --run 276
Well, I think it's faster, but it eats a huge amount of memory. It topped out at 50.5 GB, likely due to the memory requirements of all the septem board data?
I implemented batching for this. On my first attempt it seems like the code was running in ~55 s for run 276 as up there. With 1000 per batch it was suddenly almost 80s, and 5000 was 83 s? -> Those times don't make any sense. The code has been running close to 4 minutes or so now.
For 500 batched events:
likelihood -f /home/basti/CastData/data/DataRuns2018_Reco.h5 --h5out --ml 6040.76s user 71.74s system 1684% cpu 6:02.93 total
-> Ahh, it was 6 minutes in total then, I think.
For 500 batched, running with 28 threads:
likelihood -f /home/basti/CastData/data/DataRuns2018_Reco.h5 --h5out --ml 10266.94s user 88.74s system 2397% cpu 7:12.02 total
This doesn't make a whole lot of sense to me.
Running with 100k:
likelihood -f /home/basti/CastData/data/DataRuns2018_Reco.h5 --h5out --ml 1457.86s user 182.48s system 1476% cpu 1:51.08 total
Oh my goodness. I forgot to filter to those events that are part of
the passedEvs!!!
Well, not quite. That's part of buildSeptemEvents!
But now we filter before computing the batch slices. So that
septemDf only contains O(5k) events in the first place.
No batching, 28 threads:
likelihood -f /home/basti/CastData/data/DataRuns2018_Reco.h5 --h5out --ml 1089.03s user 201.38s system 1327% cpu 1:37.18 total
Batching of 1k events, 28 threads:
likelihood -f /home/basti/CastData/data/DataRuns2018_Reco.h5 --h5out --ml 3152.81s user 134.29s system 2196% cpu 2:29.64 total
This peaks around 12 GB. But quite a lot slower!
Single threaded:
likelihood -f /home/basti/CastData/data/DataRuns2018_Reco.h5 --h5out --ml 380.71s user 9.13s system 99% cpu 6:30.16 total
So we gain some, but not huge amounts. A good factor of 3.
Still, I'll take it.
With 20 threads and 500 events per batch:
likelihood -f /home/basti/CastData/data/DataRuns2018_Reco.h5 --h5out --ml 2930.59s user 81.10s system 1654% cpu 3:02.08 total
this doesn't cause excessive RAM usage at least.
With 20 threads and no batching:
likelihood -f /home/basti/CastData/data/DataRuns2018_Reco.h5 --h5out --ml 931.76s user 127.05s system 1007% cpu 1:45.04 total
Crazy difference still.
I honestly don't fully understand why the scaling is the way it is when changing batch sizes. Surely it should scale better with more threads and also better with changed batch sizes? Do we accidentally do too much maybe?
It's pretty brutal how slow DBSCAN actually is. It's 8416 clusters in the run that pass the MLP cut at 95%.
Counting events, batched 500:
EVENT COUNTER : 8416
Unbatched:
EVENT COUNTER : 8416
-> So simple overcounting or the like is not the cause.
[X]Check performance for single threaded version- At 5000 events per batch it topped out around the 8GB mark
- At 500 events it uses ~6.3GB at most
[X]Check if Weave multithreading works here with more than 20 threads -> It seems to work without problems. Bizarre. (tested 28) Anything related to uint8 / uint16 types?[X]Check the output of the septem number of before / after events. -> The ./../../../t/septem_veto_before_after.txt shows the exact same numbers regardless of the approach we use. Good to know.[ ]Rerun the likelihoods for Run-2.
1.106.1. Recreate DataRuns2017_Reco.h5
Time to rerun it again.
Started at :
./runAnalysisChain \ -i ~/CastData/data \ --outpath ~/CastData/data \ --years 2017 \ --back --reco --logL --tracking
It finished in about 29 min:
The entire analysis chain took: 28.74584188858668 min
Time to rerun the MLP data classification again for vetoes + Run-2.
1.106.2. Create all likelihood combinations
Rerun all Run-2 combinations and Run-2+Run-3 for lnL cut method.
-> Hmm, I'm not sure if this was actually faster in any way! I think it was actually slower and more or less ran out of RAM at points (maybe it had to swap and was so slow for that reason?) Anyway, before we start another run, we should rethink our approach / parameters.
- Tanh30 Adam MLP
-> Rerun Run-2:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --regions crAll \ --vetoSets "{+fkMLP, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh30_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_30_2checkpoint_epoch_38000_loss_0.0906_acc_0.9611.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_14_11_23_adam_tanh30_cross_entropy_less3keV_38k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 2 \ --dryRun
Running all likelihood combinations took 31379.83979129791 s
- Tanh10 Adam MLP
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --regions crAll \ --vetoSets "{+fkMLP, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/14_11_23_adam_tanh10_cross_entropy_less_3keV/mlp_tanh_linear_sigmoidCrossEntropy_Adam_10_2checkpoint_epoch_88000_loss_0.0974_acc_0.9576.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.90 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_14_11_23_adam_tanh10_cross_entropy_less3keV_88k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 2 \ --dryRun
Running all likelihood combinations took 26816.16909456253 s
- LnL
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkLogL, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.7 --signalEfficiency 0.8 --signalEfficiency 0.9 \ --out ~/org/resources/lhood_lnL_17_11_23_septem_fixed \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 2 \ --dryRun
Running all likelihood combinations took 17967.36839723587 s
- Scinti+FADC+Line
Background clusters, 90%:
plotBackgroundClusters \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R2_crAll_sEff_0.9_lnL_scinti_fadc_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R3_crAll_sEff_0.9_lnL_scinti_fadc_line_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data LnL90%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_lnL_90_scinti_fadc_line" \ --backgroundSuppression
80%
plotBackgroundClusters \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data LnL80%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_lnL_80_scinti_fadc_line" \ --backgroundSuppression
plotBackgroundRate \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R2_crAll_sEff_0.9_lnL_scinti_fadc_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R3_crAll_sEff_0.9_lnL_scinti_fadc_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R2_crAll_sEff_0.7_lnL_scinti_fadc_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R3_crAll_sEff_0.7_lnL_scinti_fadc_line_vQ_0.99.h5 \ --names "90" --names "90" \ --names "80" --names "80" \ --names "70" --names "70" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_lnL_scinti_fadc_line.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- Scinti+FADC+Septem+Line
Background clusters, 90%:
plotBackgroundClusters \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R2_crAll_sEff_0.9_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R3_crAll_sEff_0.9_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data LnL90%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_lnL_90_scinti_fadc_septem_line" \ --backgroundSuppression
80%
plotBackgroundClusters \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data LnL80%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_lnL_80_scinti_fadc_septem_line" \ --backgroundSuppression
plotBackgroundRate \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R2_crAll_sEff_0.9_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R3_crAll_sEff_0.9_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R2_crAll_sEff_0.7_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ ~/org/resources/lhood_lnL_17_11_23_septem_fixed/lhood_c18_R3_crAll_sEff_0.7_lnL_scinti_fadc_septem_line_vQ_0.99.h5 \ --names "90" --names "90" \ --names "80" --names "80" \ --names "70" --names "70" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_lnL_scinti_fadc_septem_line.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- Scinti+FADC+Line
1.107.
Having rerun the likelihood classification over night now, one thing
I noticed that worries me is the effective efficiency.
Based on the nnEffectiveEff fields in the HDF5 files, e.g.
-> These three files show effective efficiencies much higher than the target ~/org/resources/lhoodmlp141123adamtanh30crossentropyless3keV38k/lhoodc18R2crAllsEff0.9scintifadcseptemlinemlpmlptanhlinearsigmoidCrossEntropyAdam302checkpointepoch38000loss0.0906acc0.9611vQ0.99.h5 ~/org/resources/lhoodmlp141123adamtanh30crossentropyless3keV38k/lhoodc18R2crAllsEff0.95scintifadcseptemlinemlpmlptanhlinearsigmoidCrossEntropyAdam302checkpointepoch38000loss0.0906acc0.9611vQ0.99.h5 ~/org/resources/lhoodmlp141123adamtanh30crossentropyless3keV38k/lhoodc18R2crAllsEff0.98scintifadcseptemlinemlpmlptanhlinearsigmoidCrossEntropyAdam302checkpointepoch38000loss0.0906acc0.9611vQ0.99.h5
-> and these show efficiencies much lower than the target. What is going on? ~/org/resources/lhoodmlp141123adamtanh10crossentropyless3keV88k/lhoodc18R2crAllsEff0.95scintifadcseptemlinemlpmlptanhlinearsigmoidCrossEntropyAdam102checkpointepoch88000loss0.0974acc0.9576vQ0.99.h5 ~/org/resources/lhoodmlp141123adamtanh10crossentropyless3keV88k/lhoodc18R2crAllsEff0.98scintifadcseptemlinemlpmlptanhlinearsigmoidCrossEntropyAdam102checkpointepoch88000loss0.0974acc0.9576vQ0.99.h5 ~/org/resources/lhoodmlp141123adamtanh10crossentropyless3keV88k/lhoodc18R2crAllsEff0.9scintifadcseptemlinemlpmlptanhlinearsigmoidCrossEntropyAdam102checkpointepoch88000loss0.0974acc0.9576vQ0.99.h5
The values in the HDF5 file are so different from the plots like:
(not that this looks good!!)
because when calling
meanEffectiveEff we pass readEscapeData =
false to the actual proc. That means the escape data, which is
predicted well, is excluded, causing the prediction to be way too low.
The reason for this is probably that the output prediction developed 3 distinct peaks for the efficiency. Depending on the data they may land in different peaks. If simulated data is more in one than the other, the effective cut values needed may be completely off, I imagine.
FUCKING HELL
I think the fake training data contained the inverted rotation angles
due to missing npix - x in applyPitchConversion. If I change the
code in geometry to use
let rotAngle = if not useRealLayout: rot_angle # for single chip we invert `x ↦ -x`. Thus need to invert angle too. else: rotAngle
instead of the correct
let rotAngle = if not useRealLayout: -rot_angle # for single chip we invert `x ↦ -x`. Thus need to invert angle too. else: rotAngle
the predictions for the MLP for CDL data (nn_predict) all come out
close to 1.
But if I do the same with the correct version, the network predicts every single cluster to be background.
So that means:
[X]Regenerate the fake data HDF5 files[X]Train another network (and another…)[ ]Check it works[ ]…profit?
At least now there is a chance that all this will look better in the end than it did before.
[X]I will delete the existingcacheTabsfor effective efficiency and run local cut values, because they are 'tainted'.
More fucking hell???
I noticed that my fix for the above (the let rotAngle lines), did
not take into account that the rotAngle variable then would be
inverted completely, implying that the
result.rotationAngle = rotAngle
line would receive the inverted rotation angle! What. A. Pain. In. The. Butt.
Let's check the rotation angles of the new DataRuns2017/8_Reco.h5
files (and for sanity calibration & CDL), as well as FakeData files.
Are they all positive / negative etc? And more importantly, is it fine
if we just apply abs(rotAngle) to the rotation angle field of the
cluster? An angle between 0-180° is all the information available
anyway, no? We don't perform a fit of the track "direction" after all,
so we are rotationally symmetric around mod 180° values.
Run-2:
hdfview ~/CastData/data/DataRuns2017_Reco.h5
-> all negative
Run-3:
hdfview ~/CastData/data/DataRuns2018_Reco.h5
-> all positive
Calibration Run-2:
hdfview ~/CastData/data/CalibrationRuns2017_Reco.h5
-> all positive
Calibration Run-3:
hdfview ~/CastData/data/DataRuns2018_Reco.h5
-> all positive
CDL
hdfview ~/CastData/data/CDL_2019/CDL_2019_Reco.h5
-> all positive
calibration-cdl
hdfview ~/CastData/data/CDL_2019/calibration-cdl-2018.h5
-> all positive
Fake 0-3 keV
hdfview ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5
-> all negative
Fake 0-10 keV
hdfview ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5
-> all negative
So the negative ones are:
- Run-2 background, fake 0-3 and fake 0-10.
We can either invert the angles for all datasets with a script or regenerate everything.
Let's write the script.
import ingrid / [tos_helpers, ingrid_types] import std / [sequtils, math, os] import nimhdf5 const files = ["~/CastData/data/DataRuns2017_Reco.h5", "~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5", "~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5"] for f in files: withH5(f, "rw"): let fileInfo = h5f.getFileInfo() for run in fileInfo.runs: for chip in fileInfo.chips: # check if chip group in file (may not be, for fake) let path = recoPath(run, chip) if path.string in h5f: echo "Run: ", run, " chip: ", chip # read rotationAngle var dset = h5f[(path.string / "rotationAngle").dset_str] let fixed = dset[float].mapIt(it.abs) # write abs values back dset[dset.all] = fixed
Run the code:
ntangle ~/org/journal.org && nim c -r fixup_rotation_angles_abs.nim
Check the files by hand again:
- Run-2 background: positive
- Fake 0-3 keV: positive
- Fake 0-10 keV: positive
[X]Also: make sure to recompile every tool usinggeometry! After our change toabs(rotAngle)(for reference full args):[X]reconstructionnim c -d:danger reconstruction
[X]likelihoodnim cpp -d:cuda -d:blas=openblas -d:useMalloc -d:danger likelihood[X]nn_predictnim cpp -d:cuda -d:blas=openblas -d:useMalloc -d:danger nn_predict[X]effective_eff_55fenim cpp -d:cuda -d:blas=openblas -d:useMalloc -d:danger effective_eff_55fe[X]train_ingridnim cpp -d:cuda -d:blas=openblas -d:useMalloc -d:danger train_ingrid[X]simulate_xraysnim cpp -d:cuda -d:blas=openblas -d:useMalloc -d:danger simulate_xrays
1.107.1. Regenerate fake data
[X]Just noticed a bug where I forgot to adjust the random number seed for each process insimulate_xrays… Fixed and rerunning
First the decreasing 0.1 to 3 keV data:
MT=true ./simulate_xrays \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --nFake 250000 \ --energyMin 0.1 \ --energyMax 3.0 \ --yEnergyMin 1.0 \ --yEnergyMax 0.0 \ --outfile ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --note "Energies linear decrease in frequency from 0.1 to 3.0 keV"
Now the uniform data:
MT=true ./simulate_xrays \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --nFake 250000 \ --energyMin 0.1 \ --energyMax 10.0 \ --yEnergyMin 1.0 \ --yEnergyMax 1.0 \ --outfile ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --note "Uniform energy distribution 0.1 to 10 keV"
Both done .
1.107.2. Train another Adam MLP with linear output
Now let's retrain again. And again after not only fixing the first rotation angle bug, but also fixing the actual rotation angle bug!
Here I will overwrite the trained networks (delete the data). There is no value in them really.
Let's retrain two more MLPs similar to the previous best, but using sigmoid + MSE. That way the prediction is more restricted to a very small range, allowing more sane cut values.
- Adam tanh 10 sigmoid MSE
Train with the now correct fake data:Train with now not only correct fake data (really, I promise!), but also correct Run-2 data!
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh10_sigmoid_mse_3keV/ \ --plotPath ~/Sync/17_11_23_adam_tanh10_sigmoid_mse_3keV \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 1000
Stopped after 15k .Stopped after 25k .Start uniform training:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh10_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_25000_loss_0.0258_acc_0.9650.pt \ --plotPath ~/Sync/17_11_23_adam_tanh10_sigmoid_mse_3keV \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 1000
Stopped after 84k .
Not that much improvement over the epochs.
nn_predict
Prediction:
./nn_predict \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh10_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.pt \ --signalEff 0.9 \ --plotPath ~/Sync/17_11_23_adam_tanh10_sigmoid_mse_3keV/
Now the values are all positive at least! Some of them are pegged to 1 though, which is a bit of a shame? Do we need more bins? I mean it 'just' means all the fake data falls exactly into the last bin. Ah -> no binning. We're asking for the 90th percentile in this case. This just implies the upper 90 percent are clearly at 1.0 exactly.
From this point of view a linear + cross entropy output layer is better (despite the inverse problem becoming more relevant!)
effective_eff_55fe
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh10_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_17_11_23_adam_tanh10_sigmoid_mse_84k/
Finished .
This has the same Run-2, Run-3 discrepancy. But otherwise it looks pretty nice!
90%
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh10_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.pt \ --ε 0.90 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_17_11_23_adam_tanh10_sigmoid_mse_mlp90_84k/
train_ingrid --predict
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh10_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.pt \ --plotPath ~/Sync/17_11_23_adam_tanh10_sigmoid_mse_3keV \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --predict
The ROC curve
looks very good! All the CDL predictions also look as expected.
createAllLikelihoodCombinations
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{fkMLP, +fkFadc, +fkScinti, +fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh10_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.90 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 4 \ --dryRun
Running all likelihood combinations took 11732.32791733742 s
The multithreaded version yesterday with 2 jobs:
Running all likelihood combinations took 26816.16909456253 s
plotBackgroundClusters,plotBackgroundRate
- No vetoes
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_adam_tanh10_sigmoid_mse_84k" \ --backgroundSuppression
90%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@90%" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_90_adam_tanh10_sigmoid_mse_84k" \ --backgroundSuppression
Background rate, comparison:
plotBackgroundRate \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.98_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.98_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.85_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.85_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --names "85" --names "85" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh10_mse_sigmoid_84k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- Scinti+FADC+Line
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_scinti_fadc_line_adam_tanh10_sigmoid_mse_84k" \ --backgroundSuppression
85%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@85%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_85_scinti_fadc_line_adam_tanh10_sigmoid_mse_84k" \ --backgroundSuppression
plotBackgroundRate \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.98_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.98_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --names "85" --names "85" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh10_scinti_fadc_line_mse_sigmoid_84k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- Scinti+FADC+Septem+Line
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%+Scinti+FADC+Septem+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_scinti_fadc_septem_line_adam_tanh10_sigmoid_mse_84k" \ --backgroundSuppression
85%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@85%+Scinti+FADC+Septem+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_85_scinti_fadc_septem_line_adam_tanh10_sigmoid_mse_84k" \ --backgroundSuppression
plotBackgroundRate \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.98_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.98_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh10_sigmoid_mse_84k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_84000_loss_0.0254_acc_0.9655_vQ_0.99.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --names "85" --names "85" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh10_scinti_fadc_septem_line_mse_sigmoid_84k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- No vetoes
- Adam tanh 30 sigmoid MSE
And a 30 neuron network:Start at
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh30_sigmoid_mse_3keV/ \ --plotPath ~/Sync/17_11_23_adam_tanh30_sigmoid_mse_3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 30 \ --numHidden 30 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 1000
Stopped at 15k .Stopped after 25kNow continue with uniform data:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh30_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_25000_loss_0.0253_acc_0.9657.pt \ --plotPath ~/Sync/17_11_23_adam_tanh30_sigmoid_mse_3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 30 \ --numHidden 30 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 1000
Stopped after 55k .Stopped after 82k epochs .nn_predict
Prediction:
./nn_predict \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh30_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.pt \ --signalEff 0.9 \ --plotPath ~/Sync/17_11_23_adam_tanh30_sigmoid_mse_3keV
All positive fortunately.
effective_eff_55fe
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh30_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_17_11_23_adam_tanh30_sigmoid_mse_82k/
Finished .
-> Somewhat similar to the 10 neuron network BUT the spread is much smaller and the CDL data is better predicted! This might our winner.
90%
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh30_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.pt \ --ε 0.90 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_17_11_23_adam_tanh30_sigmoid_mse_mlp90_82k/
train_ingrid --predict
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh30_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.pt \ --plotPath ~/Sync/17_11_23_adam_tanh30_sigmoid_mse_3keV/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 30 \ --numHidden 30 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --predict
Finished
Compared to the ROC curve of tanh10, this ROC curve looks a little bit worse. But the effective efficiency predictions are quite a bit better.
So we will choose based on what happens with the background and other efficiencies.
createAllLikelihoodCombinations
Starting :
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{fkMLP, +fkFadc, +fkScinti, fkSeptem, fkLineVeto, fkExclusiveLineVeto}" \ --mlpPath ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh30_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 4 \ --dryRun
Running all likelihood combinations took 11730.39050412178 s
Despite only 2 jobs in the multithreaded case below (but excluding fkMLP alone!!) it was still more than twice slower. So single threaded is the better way.
Multithreaded took:
Running all likelihood combinations took 31379.83979129791 s
The above was initially run without the Septem veto alone! (and the numbers reflect that)
Running now only the case of base model + septem veto. Starting .
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkMLP, +fkFadc, +fkScinti, fkSeptem}" \ --mlpPath ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh30_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.pt \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.85 --signalEfficiency 0.9 --signalEfficiency 0.95 --signalEfficiency 0.98 \ --out ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/ \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 8 \ --dryRun
Finished .
Running all likelihood combinations took 3432.131967782974 s
plotBackgroundClusters,plotBackgroundRate
- No vetoes
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5 ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_adam_tanh30_sigmoid_mse_82k" \ --backgroundSuppression
90%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@90%" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_90_adam_tanh30_sigmoid_mse_82k" \ --backgroundSuppression
Background rate, comparison:
plotBackgroundRate \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.98_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.98_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.95_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.95_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.85_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.85_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --names "85" --names "85" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh30_mse_sigmoid_82k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- Scinti+FADC+Line
Background clusters, 95%:
plotBackgroundClusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_scinti_fadc_line_adam_tanh30_sigmoid_mse_82k" \ --backgroundSuppression
85%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@85%+Scinti+FADC+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_85_scinti_fadc_line_adam_tanh30_sigmoid_mse_82k" \ --backgroundSuppression
plotBackgroundRate \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.98_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.98_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.9_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --names "85" --names "85" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh30_scinti_fadc_line_mse_sigmoid_82k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- Scinti+FADC+Septem+Line
Background clusters, 95%:
plotbackgroundclusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@95%+Scinti+FADC+Septem+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_95_scinti_fadc_septem_line_adam_tanh30_sigmoid_mse_82k" \ --backgroundSuppression
85%
plotBackgroundClusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ --zMax 30 --title "X-ray like clusters of CAST data MLP@85%+Scinti+FADC+Septem+Line" \ --outpath /tmp/ --filterNoisyPixels \ --energyMin 0.2 --energyMax 12.0 --suffix "_mlp_85_scinti_fadc_septem_line_adam_tanh30_sigmoid_mse_82k" \ --backgroundSuppression
plotBackgroundRate \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.98_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.98_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.9_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_septem_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ --names "98" --names "98" \ --names "95" --names "95" \ --names "90" --names "90" \ --names "85" --names "85" \ --centerChip 3 --title "Background rate from CAST" \ --showNumClusters --showTotalTime --topMargin 1.5 --energyDset energyFromCharge \ --outfile background_rate_adam_tanh30_scinti_fadc_septem_line_mse_sigmoid_82k.pdf \ --outpath /tmp \ --region crGold \ --energyMin 0.2
- No vetoes
1.107.3. OUTDATED Train another Adam MLP with linear output
This section is outdated, due to bad synthetic data!
I have updated the output directories to include a _bad_reco suffix!
Let's retrain two more MLPs similar to the previous best, but using sigmoid + MSE. That way the prediction is more restricted to a very small range, allowing more sane cut values.
- Adam tanh 10 sigmoid MSE
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh10_sigmoid_mse_3keV_bad_reco/ \ --plotPath ~/Sync/17_11_23_adam_tanh10_sigmoid_mse_3keV_bad_reco \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 1000
Stopped after 26k .
Start uniform training:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh10_sigmoid_mse_3keV_bad_reco/mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_26000_loss_0.0144_acc_0.9808.pt \ --plotPath ~/Sync/17_11_23_adam_tanh10_sigmoid_mse_3keV_bad_reco \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 10 \ --numHidden 10 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 1000
It finished sometime before .
Let's run our usual bunch of checks, for the 125k snapshot
nn_predict
./nn_predict \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh10_sigmoid_mse_3keV_bad_reco/mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_125000_loss_0.0140_acc_0.9812.pt \ --signalEff 0.9 \ --plotPath ~/Sync/17_11_23_adam_tanh10_sigmoid_mse_3keV/
Looking interesting!
effective_eff_55fe
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh10_sigmoid_mse_3keV_bad_reco/mlp_tanh_sigmoid_MSE_Adam_10_2checkpoint_epoch_125000_loss_0.0140_acc_0.9812.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --plotPath ~/Sync/run2_run3_17_11_23_adam_tanh10_sigmoid_mse_125k_bad_reco
Finished .
- Adam tanh 30 sigmoid MSE
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_0_to_3keV_decrease.h5 \ --modelOutpath ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh30_sigmoid_mse_3keV_bad_reco/ \ --plotPath ~/Sync/17_11_23_adam_tanh30_sigmoid_mse_3keV_bad_reco/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 30 \ --numHidden 30 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 1000
Stopped at 20k .
Now continue with uniform data:
./train_ingrid \ ~/CastData/data/CalibrationRuns2017_Reco.h5 \ ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --back ~/CastData/data/DataRuns2017_Reco.h5 \ --back ~/CastData/data/DataRuns2018_Reco.h5 \ --simFiles ~/CastData/data/FakeData/fakeData_500k_uniform_energy_0_10_keV.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh30_sigmoid_mse_3keV_bad_reco/mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_20000_loss_0.0141_acc_0.9812.pt \ --plotPath ~/Sync/17_11_23_adam_tanh30_sigmoid_mse_3keV_bad_reco/ \ --datasets eccentricity \ --datasets skewnessLongitudinal \ --datasets skewnessTransverse \ --datasets kurtosisLongitudinal \ --datasets kurtosisTransverse \ --datasets length \ --datasets width \ --datasets rmsLongitudinal \ --datasets rmsTransverse \ --datasets lengthDivRmsTrans \ --datasets rotationAngle \ --datasets fractionInTransverseRms \ --datasets totalCharge \ --datasets σT \ --numHidden 30 \ --numHidden 30 \ --activation tanh \ --outputActivation sigmoid \ --lossFunction MSE \ --optimizer Adam \ --learningRate 7e-4 \ --simulatedData \ --backgroundRegion crAll \ --nFake 250_000 \ --backgroundChips 0 \ --backgroundChips 1 \ --backgroundChips 2 \ --backgroundChips 4 \ --backgroundChips 5 \ --backgroundChips 6 \ --clamp 5000 \ --plotEvery 1000
Stopped after 45k .
1.108.
[ ]Train one more network with linear + cross entropy[ ]Train one more SGD + 300 neuron network
1.108.1. Limit calculation
It's finally time to test and then run the limit calculation.
Things to test:
[X]Run a limit with known input file (lnL 80 + line) and old raytracing as reference! (we didn't break anything!), sec. [BROKEN LINK: sec:journal:18_11_23:old][X]Run a limit with known input file (lnL 80 + line) and old raytracing as reference with new solar axion flux! (we didn't break anything!), sec. [BROKEN LINK: sec:journal:18_11_23:old_limit_new_flux][X]Run a limit with known input file (lnL 80 + line) and new raytracing image, 1.108.1.2[X]Run a limit with known input file (lnL 80 + line) and new raytracing image + axes rotated, 1.108.1.4[X]Run a limit with known input file (lnL 80 + line) and new raytracing image + axes rotated, new random coincidence / veto efficiency, 1.108.1.5[X]Run a limit with known input file (lnL 80 + line) and new raytracing image + axes rotated, new telescope efficiency, 1.108.1.6[X]Run a limit with new input file (new mlp 95 + line) and everything else, 1.108.1.7
Once we have run all these and verified it all looks good, we'll start running all limits using ./../CastData/ExternCode/TimepixAnalysis/Analysis/runLimits.nim.
Command from runLimits:
mcmc_limit_calculation \ limit \ -f ($fRun2) -f ($fRun3) \ --years 2017 --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --nmc ($nmc) ($energyStr) ($axionStr) ($suffix) ($path) --outpath ($outpath)
[ ]ModifyrunLimitsto switch axes!
- Old files limit
Note: axion model & image don't need to be handed, because we want to reuse the old values:
axionModel = "/home/basti/CastData/ExternCode/AxionElectronLimit/axion_diff_flux_gae_1e-13_gagamma_1e-12.csv", axionImage = "/home/basti/org/resources/axion_images/axion_image_2018_1487_93_0.989AU.csv", ## Default corresponds to mean Sun-Earth distance during 20
Running:
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ --path "" \ --years 2017 \ --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --energyMin 0.2 \ --energyMax 12.0 \ --outpath ~/org/resources/lhood_limits_phd_tests \ --suffix "nmc_1k_lnL_80_scinti_fadc_line_test" \ --path "" \ --nmc 1000
-> There was a segfault producing the output H5 file for the limit context. Debugging now … Fixed, was due to addition of
trackingDf, but us not checking if given DF isnilindatamancer/serialize.-> Finished :
Expected limit: 6.966468310440489e-21 Generating group /ctx/trackingDf Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 10.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_limits_phd_tests/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0323_σb_0.0028_posUncertain_puUncertain_σp_0.0500nmc_1k_lnL_80_scinti_fadc_line_test.h5
This comes out to 8.34e-23 GeV⁻¹.
While the below is running now , let's check the result and see if it is compatible. The table I have in the phd thesis lists:
0.8 1000 LnL false true 0.6744 6.3147e-23 8.0226e-23 which unfortunately is quite a bit better. Why? Is the input file a different one? This limit was produced in the sec. ./Doc/StatusAndProgress.html with the command mentioned there. This references notes about it in this file. Sec. 1.64.1 produces the HDF5 logL files. Sec. 1.64.3 runs all the limit calculations for the LnL cases and then 1.66 contains the initial command to produce the table from all HDF5 outputs. The output files were written to ./resources/lhood_lnL_04_07_23/limits.
Let's compare the HDF5 output files:
- ./resources/lhood_limits_phd_tests/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0323_σb_0.0028_posUncertain_puUncertain_σp_0.0500nmc_1k_lnL_80_scinti_fadc_line_test.h5 -> The new file
- ./resources/lhood_lnL_04_07_23/limits/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0323_σb_0.0028_posUncertain_puUncertain_σp_0.0500_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5
Ahh! The 'big' difference is that the tracking time is 159.93 h in the new file and still 161.11 h in the old file.
The 159 h is correct though, no? -> Yes.
There are less events in the new HDF5 background data (12549 vs 16014 in the old). Ahh! But this is probably because we use
--energyMin 0.2here now.[X]Find the output file producing the value in the phd thesis
- Old files, new axion flux
[/]
Need to recreate the new axion flux :
./readOpacityFile \ --suffix "_0.989AU" \ --distanceSunEarth 0.9891144450781392.AU \ --fluxKind fkAxionElectronPhoton \ --plotPath ~/org/Figs/statusAndProgress/axionProduction/axionElectronRealDistance \ --outpath ~/org/resources/axionProduction/axionElectronRealDistance
with
outpathset in theconfig.tomlfile. Done.The file of interest for the limit calculation is ./resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv
Now run the limit using this axion flux model. Running :
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --path "" \ --years 2017 \ --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --energyMin 0.2 \ --energyMax 12.0 \ --outpath ~/org/resources/lhood_limits_phd_tests \ --suffix "nmc_1k_lnL_80_sfl_new_axion_flux" \ --path "" \ --nmc 1000
Finished
Expected limit: 6.725575061268316e-21 Generating group /ctx/trackingDf Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_limits_phd_tests/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0323_σb_0.0028_posUncertain_puUncertain_σp_0.0500nmc_1k_lnL_80_sfl_new_axion_flux.h5
which yields 8.20096034698e-23 GeV⁻¹.
Now whether this is down to statistical uncertainty or just improvements due to changes in the flux, I do not know.
- Old files, new axion flux, new raytracing image
Time to include the new axion image! The axion image is produced here ./../phd/thesis.html. This will be a very interesting one, because it is almost the most important difference.
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --path "" \ --years 2017 \ --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --energyMin 0.2 \ --energyMax 12.0 \ --outpath ~/org/resources/lhood_limits_phd_tests \ --suffix "nmc_1k_lnL_80_sfl_new_axion_flux_new_image" \ --path "" \ --nmc 1000
Done
Expected limit: 6.52489827514329e-21 Generating group /ctx/trackingDf Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_limits_phd_tests/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0323_σb_0.0028_posUncertain_puUncertain_σp_0.0500nmc_1k_lnL_80_sfl_new_axion_flux_new_image.h5
Yields 8.07768424435e-23 GeV⁻¹
Huh. Our new axion image actually improves things? Interesting.
- Old files, new axion flux, new raytracing image, rotated
Now run including the switch of axes.
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --path "" \ --years 2017 \ --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --energyMin 0.2 \ --energyMax 12.0 \ --outpath ~/org/resources/lhood_limits_phd_tests \ --suffix "nmc_1k_lnL_80_sfl_new_axion_flux_new_image_rotated" \ --path "" \ --switchAxes \ --nmc 1000
Results after fix of
switchAxesinContext:Expected limit: 6.212885795040392e-21 Generating group /ctx/trackingDf Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_limits_phd_tests/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0323_σb_0.0028_posUncertain_puUncertain_σp_0.0500nmc_1k_lnL_80_sfl_new_axion_flux_new_image_rotated.h5
Result before:
Expected limit: 6.212885795040392e-21 Generating group /ctx/trackingDf Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_limits_phd_tests/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0323_σb_0.0028_posUncertain_puUncertain_σp_0.0500nmc_1k_lnL_80_sfl_new_axion_flux_new_image_rotated.h5
yields 7.88218611493e-23 GeV⁻¹.
why is everything I do improving the limit? The expected limit should be identical in these cases, no? This implies for sure that
- something is wrong
- or the variance is just very large.
The variance must be very large, because the no candidate limit is larger than for one of the cases above!
Huh,
switchAxesisfalsein the output file! -> The reason was that while I handedswitchAxescorrectly to the output files, I did not hand it correctly to the construction of theContextobject! So the value was kept false, despite technically being true. I'll recompile and rerun. Rerun both to update the HDF5 file, but also to test my theory on determinism. The result should be identical. -> Exactly the same.[X]Check theswitchAxesfield in the output H5 file. Great, it was alsofalsein the files above. So we're not seeing any real effect in the change.[X]Q: Why is there a difference anyway? Isn't our result deterministic? We use fixed random seeds, no? Ah, maybe different seeds on different processes being differently fast (due to other threads / activity), thus resulting in effectively number of samples? But that doesn't quite make sense, each process runs a fixed number ofnmc! -> See above!
- Old files, new axion flux, new raytracing image, rotated, new veto eff
The new random coincidence values are:
septemVetoRandomCoinc = 0.8311, # only septem veto random coinc based on bootstrapped fake data lineVetoRandomCoinc = 0.8539, # lvRegular based on bootstrapped fake data septemLineVetoRandomCoinc = 0.7863, # lvRegularNoHLC based on bootstrapped fake data
These values will become the new default.
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --septemVetoRandomCoinc 0.8311 \ --lineVetoRandomCoinc 0.8539 \ --septemLineVetoRandomCoinc 0.7863 \ --path "" \ --years 2017 \ --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --energyMin 0.2 \ --energyMax 12.0 \ --outpath ~/org/resources/lhood_limits_phd_tests \ --suffix "nmc_1k_lnL_80_sfl_new_axion_flux_new_image_rotated_new_veto_eff" \ --path "" \ --switchAxes \ --nmc 1000
Done
Expected limit: 6.289161368274779e-21 Generating group /ctx/trackingDf Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_limits_phd_tests/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0323_σb_0.0028_posUncertain_puUncertain_σp_0.0500nmc_1k_lnL_80_sfl_new_axion_flux_new_image_rotated_new_veto_eff.h5
Note that the difference in the limit is not very large, because the input files did not use the septem veto at all (which shows larger differences in the new numbers)
- Old files, new axion flux, new raytracing image, rotated, new telescope eff
In order to use the new telescope effective area, we need to recreate the
let combEffDf = readCsv("/home/basti/org/resources/combined_detector_efficiencies.csv")
CSV file. This file is produced in ./Doc/StatusAndProgress.html.
The new script is ./../CastData/ExternCode/TimepixAnalysis/Tools/septemboardDetectionEff/septemboardDetectionEff.nim using
xrayAttenuationand allowing to read different effective area files for the LLNL telescope.To compute the old:
./septemboardDetectionEff --outpath /tmp/
and new:
./septemboardDetectionEff \ --outpath ~/org/resources/ \ --llnlEff ~/org/resources/llnl_cast_nature_jaime_data/2016_DEC_Final_CAST_XRT/EffectiveArea.txt \ --sep ' '
which we can now use in the limit:
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv \ --path "" \ --years 2017 \ --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --energyMin 0.2 \ --energyMax 12.0 \ --outpath ~/org/resources/lhood_limits_phd_tests \ --suffix "nmc_1k_lnL_80_sfl_new_axion_flux_new_image_rotated_new_veto_eff_new_tel_eff" \ --path "" \ --switchAxes \ --nmc 1000
Expected limit: 7.108243578231789e-21 Generating group /ctx/trackingDf Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_limits_phd_tests/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0323_σb_0.0028_posUncertain_puUncertain_σp_0.0500nmc_1k_lnL_80_sfl_new_axion_flux_new_image_rotated_new_veto_eff_new_tel_eff.h5
Ouch, still pretty bad :( yields 8.43089556334e-23 GeV⁻¹ as an expected limit :'(
Anyway, this needs to be compared to the 8.34e-23 GeV⁻¹ from the initial section here (which was on the high side, mind you), or down to about 8.02e-23 if we believe our initial table above. So with the MLP we should gain quite a bit and now we might even be able to use the Septem veto as well. So let's hope for the best.
[X]Make the combined efficiency CSV file an argument in the limit code -> Implemented ascombinedEfficiencyFile
- Mini detour
Got this result first due to forgetting that in
mcmc_limitwe again multiply with LLNL, which is not needed for the new CSV file!Expected limit: 1.603021310208126e-20 Generating group /ctx/trackingDf Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_limits_phd_tests/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0323_σb_0.0028_posUncertain_puUncertain_σp_0.0500nmc_1k_lnL_80_sfl_new_axion_flux_new_image_rotated_new_veto_eff_new_tel_eff.h5
Uhh, WHAT. -> Ah. It's because
Efficiencynow includes the LLNL efficiency, while in the old CSV it did not.Let's run with the old file using the new input file mechanism to see if everything works…
mcmc_limit_calculation \ limit \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R2_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ -f /home/basti/org/resources/lhood_lnL_04_07_23/lhood_c18_R3_crAll_sEff_0.8_lnL_scinti_fadc_line_vQ_0.99.h5 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies_LLNL_eff_2013JCAP.csv \ --path "" \ --years 2017 \ --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --energyMin 0.2 \ --energyMax 12.0 \ --outpath ~/org/resources/lhood_limits_phd_tests \ --suffix "nmc_1k_lnL_80_sfl_new_axion_flux_new_image_rotated_new_veto_eff_old_tel_eff_new_file" \ --path "" \ --switchAxes \ --nmc 1000
If this produces the same result as the last section above, we'll run with the CSV file using the 2013 JCAP, but produced with the new script. Maybe we mess up our efficiency somewhere. -> Stopped this when I realized my mistake.
- New files, new axion flux, new raytracing image, rotated
For the new flux, we use the new Adam Tanh30 MLP with the same sets of vetoes.
The directory is ./resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/.
mcmc_limit_calculation \ limit \ -f ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ -f ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv \ --path "" \ --years 2017 \ --years 2018 \ --σ_p 0.05 \ --limitKind lkMCMC \ --energyMin 0.2 \ --energyMax 12.0 \ --outpath ~/org/resources/lhood_limits_phd_tests \ --suffix "nmc_1k_mlp_95_sfl_new_axion_flux_new_image_rotated_new_veto_eff_new_tel_eff" \ --path "" \ --switchAxes \ --nmc 1000
Expected limit: 6.049116214356978e-21 Generating group /ctx/trackingDf Generating group /ctx/axionModel Serializing Interpolator by evaluating 0.001 to 15.0 of name: axionSpl Serializing Interpolator by evaluating 0.0 to 10.0 of name: efficiencySpl Serializing Interpolator by evaluating 0.2 to 12.0 of name: backgroundSpl Generating group /ctx/backgroundDf Wrote outfile /home/basti/org/resources/lhood_limits_phd_tests/mc_limit_lkMCMC_skInterpBackground_nmc_1000_uncertainty_ukUncertain_σs_0.0276_σb_0.0028_posUncertain_puUncertain_σp_0.0500nmc_1k_mlp_95_sfl_new_axion_flux_new_image_rotated_new_veto_eff_new_tel_eff.h5
This looks decent.
7.77760645338e-23 GeV⁻¹
Still worse though than the best expected limit for the old expected limits.
1.108.2. Test limits conclusion
I suppose this means everything seems to be working reasonably well.
Each of these limits above took on the order of 10 minutes. More statistics would be better in theory.
We have how many cases to run?
In ./resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/
- we have 12 R2 HDF5 files
In ./resources/lhood_lnL_17_11_23_septem_fixed/
- we have 6 R2 files
We should regenerate the likelihood cut method files without vetoes as reference.
1.108.3. Rerun the estimation of random coincidence estimation
This is important due to our changes to the septem veto & geometry stuff!
See sec. ./../phd/thesis.html for the code.
cd ~/phd
ntangle thesis.org && nim c code/analyze_random_coinc_and_efficiency_vetoes.nim
Run. Running now .
code/analyze_random_coinc_and_efficiency_vetoes \ --septem --line --septemLine --eccs \ --outpath ~/phd/resources/estimateRandomCoinc/ \ --jobs 16 \ --dryRun
Let's see if 16 jobs is being too greedy!
It finished some time between and . Pretty fast actually when running with 16 jobs! Each job max peaked at like 2.5GB. So running this many was not a problem at all.
The files in ./../phd/resources/estimateRandomCoinc/ generally produce good results. But surprisingly, the line veto is less efficient than before and the scaling for the eccentricity line veto cut is less strong.
The plot (produced by the above mentioned thesis section):
shows this.
[X]Can this be related to our rotation angle? Maybe the angle is inverted / needs to be and the slope is actually not correct?
Let's run
likelihoodfor the N-th time withplotSeptem. For the case of no septem veto:likelihood \ -f /home/basti/CastData/data/DataRuns2018_Reco.h5 \ --h5out /tmp/test_R3_line_ecc.h5 \ --region=crAll --cdlYear=2018 \ --scintiveto --fadcveto --lineveto \ --cdlFile=/home/basti/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --calibFile=/home/basti/CastData/data/CalibrationRuns2018_Reco.h5 \ --lnL \ --vetoPercentile=0.99 \ --signalEfficiency=0.8 \ --run 240 \ --plotSeptem
-> The plots all look fine to me. The main reason seem to be two fold (correlated):
- the fact that we don't include the spacing for the chips anymore for cluster finding, means more clusters are found together
- the line veto does not include the 'large clusters' as a pure veto. I.e. a large cluster that the original cluster is part of nevertheless has to point at the original cluster. In many, many cases the events that the original one is part of however is massive and the resulting track through its center goes anywhere but the original cluster
Together this seems to be sensible.
Given the increase in efficiency for both combined there is a good chance that the best limit will anyhow come from the combined application.
[X]Update the table in the thesis[ ]Update the text about the thesis etc[X]Update the numbers we use for the limit calculation.
1.108.4. Regenerate likelihood cut method without vetoes
We'll just produce them quickly:
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{fkLogL}" \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.7 --signalEfficiency 0.8 --signalEfficiency 0.9 \ --out ~/org/resources/lhood_lnL_17_11_23_septem_fixed \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 8 \ --dryRun
This should be very quick. Done.
Running all likelihood combinations took 173.1257138252258 s
Only 3 minutes? Wow!
1.108.5. Regenerate likelihood cut method with septem veto only
Start :
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkLogL,fkFadc,fkScinti,fkSeptem}" \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.8 \ --out ~/org/resources/lhood_lnL_17_11_23_septem_fixed \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 8 \ --dryRun
shell> Writing of all chips done Running all likelihood combinations took 2382.861197710037 s
Done!
-> Note, I initially only ran {+fkLogL,+fkFadc,+fkScinti,fkSeptem}
before I realized I need the other combinations (only FADC and FADC +
Scinti), too!
Starting this now :
./createAllLikelihoodCombinations \ --f2017 ~/CastData/data/DataRuns2017_Reco.h5 \ --f2018 ~/CastData/data/DataRuns2018_Reco.h5 \ --c2017 ~/CastData/data/CalibrationRuns2017_Reco.h5 \ --c2018 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --regions crAll \ --vetoSets "{+fkLogL,fkFadc,fkScinti}" \ --fadcVetoPercentile 0.99 \ --signalEfficiency 0.8 \ --out ~/org/resources/lhood_lnL_17_11_23_septem_fixed \ --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --multiprocessing \ --jobs 8 \ --dryRun
Done!
Running all likelihood combinations took 321.9797019958496 s
1.108.6. Run all limits
MLP limits in: In ./resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/
- we have 12 R2 HDF5 files
LnL limits in: In ./resources/lhood_lnL_17_11_23_septem_fixed/
- we now have 9 R2 files
We will place the resulting limit files all into the same folder. Both LnL and MLP, ./resources/lhood_limits_21_11_23/
We will use 2500 toys for now. Unless this ends up being too slow for the lnL cases without any vetoes. -> it takes 30s for one 150k chain (lnL 70). We build three, so 90s per toy limit. This means it should take about 30*3*2500/32=7031.25 s, so a bit less than 2 hours. Hmm. Definitely on the long side for this. And this is for lnL at 70% right now. LnL 90 might take way too long.
LnL limits running:
./runLimits \ --path ~/org/resources/lhood_lnL_17_11_23_septem_fixed/ \ --prefix "lhood_c18_R2_crAll_*" \ --outpath ~/org/resources/lhood_limits_21_11_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv \ --switchAxes \ --nmc 2500 \ --dryRun
Finished
Computing single limit took 864.9960966110229 s Computing all limits took 43548.35760474205 s
MLP limits running:
./runLimits \ --path ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/ \ --prefix "lhood_c18_R2_crAll_*" \ --outpath ~/org/resources/lhood_limits_21_11_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv \ --switchAxes \ --nmc 2500 \ --dryRun
It crashed at some point during the night:
shell> Expected limit: 6.509787030565791e-21 shell> syncio.nim(868) writeFile shell> Error: unhandled exception: cannot open: /home/basti/org/resources/lhood_limits_21_11_23//mc_limit_lkMCMC_skInterpBackground_nmc_2500_uncertainty_ukUncertain_σs_0.0288_σb_0.0028_posUncertain_puUncertain_σp_0.0500lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.csv [IOError] Error when executing: mcmc_limit_calculation limit -f /home/basti/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 -f /home/basti/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R3_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 --years 2017 --years 2018 --σ_p 0.05 --limitKind lkMCMC --nmc 2500 --energyMin 0.2 --energyMax 12.0 --axionModel /home/basti/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv --suffix=lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99 --path "" --outpath /home/basti/org/resources/lhood_limits_21_11_23/ --axionImage /home/basti/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv --switchAxes=true --combinedEfficiencyFile /home/basti/org/resources/combined_detector_efficiencies.csv err> ERROR: The previous command (see log file: /home/basti/org/resources/lhood_limits_21_11_23/mcmc_limit_output_nmc_2500_suffixArg.log) did not finish successfully. Aborting.
Uhhh, running ls or lc on the CSV filename for which it says
"cannot open", throws the error "file name too long"!
Damn, what is the file name limit on ext4?
A file name (not path length!) on ext4 is limited to 255 bytes, but
mc_limit_lkMCMC_skInterpBackground_nmc_2500_uncertainty_ukUncertain_σs_0.0288_σb_0.0028_posUncertain_puUncertain_σp_0.0500lhood_c18_R2_crAll_sEff_0.85_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.csv
is 259 bytes!
-> I now use the input filename without the given suffix as another
directory. Then we do not pass any suffixes to
mcmc_limit_calculation.
Rerun now :
./runLimits \ --path ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/ \ --prefix "lhood_c18_R2_crAll_*" \ --outpath ~/org/resources/lhood_limits_21_11_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv \ --switchAxes \ --nmc 2500 \ --dryRun
-> I had to restart it again at around 10:30 or so, because I forgot
to make sure to create the output directory in
mcmc_limit_calculation.
Now it's running correctly though.
Computing single limit took 440.6358246803284 s Computing all limits took 48734.33882236481 s
It finished some time during the night of .
Now running for MLP+Septem . This should be quick:
./runLimits \ --path ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/ \ --prefix "lhood_c18_R2_crAll_*" \ --outpath ~/org/resources/lhood_limits_21_11_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv \ --switchAxes \ --nmc 2500 \ --dryRun
Finished .
Computing single limit took 809.5403854846954 s Computing all limits took 4373.218421459198 s
1.109.
Time to look at the limit calculations of the last days!
./generateExpectedLimitsTable --path ~/org/resources/lhood_limits_21_11_23/ --prefix "mc_limit_lkMCMC"
We need to turn that into the form we use in the thesis, but we'll get to that.
[X]We should run the likelihood stuff + limits also for pure classifier + septem veto cases. This might be good, given that the septem veto is now actually more efficient than the line veto alone. -> We'll add those to thecreateAllLikelihoodCombinationcalls in sec.[X]Run limits for MLP + septem only![ ]Run limits for LnL + septem only! -> Not done yet, only because currentlyrunLimitswould also run all the lnL + FADC, lnL + FADC + scinti cases, which we don't really want to run, as that would be very slow.
1.109.1. DONE Tracking time
In our limit code now we end up having a tracking time of 159 hours, more or less. But in the thesis we still write 161 hours. And this number also appears in the summary table of the CAST chapter. That chapter computes it based on the data itself, so what gives?
See sec. 1.86.1.
-> So yeah, the 159.8 h is the correct number, not the 161 h.
-> Actually, it turned out to be 160.37 h now. That is however now
compatible with writeRunList as well. So that value it is.
[X]Do we need to update the table in the CAST summary chapter? -> Yes. -> Done.[X]Why is there suddenly more time? -> The reason was that we had accidentally commented out the- deadTimepart ingetExtendedRunInfo! Probably to test. -> The ~ half an hour more in tracking, I'm not entirely sure though.
1.110.
[ ]ExtendrunLimitsto offer more choice what to run? -> So that we can filter to not run e.g. specific veto setups? ->[ ]Run limits with more statistics for best cases. Problem: the MLP setups without vetoes contain way too many candidates to be evaluated quickly. So we will only rerun the best cases that use some vetoes, I think. In the thesis we can justify our hesitance to use the no veto cases by the extremely non uniform background, which increases the risk of issues due to underestimating position uncertainty or other systematic effects.[X]Change theprocessed.txtto include the number of toy candidates to compute. This way rerunning the code with a different--nmcdoes not count the files as 'done'. -> Manually renamed file to ./resources/lhood_limits_21_11_23/processed_2500.txt
Let's see what using ~*scinti* in the prefix does to the files we would calculate limits for:
./runLimits \ --path ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/ \ --prefix "lhood_c18_R2_crAll_*scinti*" \ --outpath ~/org/resources/lhood_limits_21_11_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv \ --switchAxes \ --nmc 15000 \ --dryRun
-> This would essentially run all MLP with at least some vetoes (meaning septem, line or both, plus FADC, scinti). It would also run the 85% cases, which are the worst in the table.
Still, these are very fast, so I think it should be fine.
Before we waste time implementing some time saver, let's just run it.
Starting .
Still, this will probably take a while… A good 1 1/2 h or so if one chain takes O(3.5 s).
I stopped it in the middle-ish of one limit calc at around . I'll continue now . Finally finished .
Computing single limit took 4963.147039413452 s Computing all limits took 45108.01824736595 s
Those numbers of course don't include the ~12 hours yesterday! So all in all it ran for about a full day.
1.111.
Yesterday I realized that the limit code still did not use the correct effective efficiency from the input files. It still used the last efficiency, which for the latest MLP meant that it overestimates the efficiency a bit.
For the 50k nmc run from last night I fixed it.
So we need to rerun all MLP cases.
For the LnL we do not need to rerun anything!
For now at first though we will only rerun the MLP + any veto cases with 2500 samples. This should be pretty quick and give us the majority of important stuff for the table. Then we can rerun these with 15k samples and at the end the no veto cases with maybe only 1k samples.
./runLimits \ --path ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/ \ --prefix "lhood_c18_R2_crAll_*scinti*" \ --outpath ~/org/resources/lhood_limits_21_11_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv \ --switchAxes \ --nmc 2500 \ --dryRun
Done.
Computing single limit took 818.3639938831329 s Computing all limits took 13074.90723443031 s
Now let's run the no MLP case with 1k:
/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k//home/basti/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/lhood_c18_R2_crAll_sEff_0.9_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.h5
We use the ~exclude argument to make sure we don't run any veto
files.
Starting :
./runLimits \ --path ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/ \ --prefix "lhood_c18_R2_crAll_*" \ --exclude "_scinti_" \ --outpath ~/org/resources/lhood_limits_21_11_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv \ --switchAxes \ --nmc 1000 \ --dryRun
I stopped it around or so Continuing now . The first 3 were done at that point, leaving the last one. This last one finished in less than an hour:
Computing all limits took 3058.317391633987 s
Time to compute the expected limit table.
Finally, let's run the MLP + line veto (no septem) with 15k. We might run one septem + line veto case later, too. Starting .
./runLimits \ --path ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/ \ --prefix "lhood_c18_R2_crAll_*line*" \ --exclude "_septem_" \ --outpath ~/org/resources/lhood_limits_21_11_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv \ --switchAxes \ --nmc 15000 \ --dryRun
Finished
Computing all limits took 37715.85584521294 s
Finally:
./runLimits \ --path ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/ \ --prefix "lhood_c18_R2_crAll_*98*septem_line*" \ --outpath ~/org/resources/lhood_limits_21_11_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv \ --switchAxes \ --nmc 15000 \ --dryRun
and
./runLimits \ --path ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k/ \ --prefix "lhood_c18_R2_crAll_*95*septem*" \ --exclude "line" \ --outpath ~/org/resources/lhood_limits_21_11_23/ \ --energyMin 0.2 --energyMax 12.0 \ --axionModel ~/org/resources/axionProduction/axionElectronRealDistance/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_0.989AU.csv \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv \ --switchAxes \ --nmc 15000 \ --dryRun
1.112.
Questions/comments for Klaus:
- Did you ask Igor whether he wants to be 2nd corrector?
- I'll go ahead and upload the data one of these days.
- Is there anything that makes it not allowed to upload the repository of my thesis before publication?
General things to do once draft is done:
[X]Write Chuck- Proofread people:
[X]Johanna: raytracing appendix[X]Markus / Tobi for Septemboard chapter[X]Lucian for detector and general stuff[X]Cristian for theory chapter[X]Stephan for theory chapter[X]Maybe Chuck for limit chapter[X]Roberto for the general stuff[X]Cristina for general stuff
- Upload data to Zenodo
- Push
phdrepository to Github -> Do not push Org file yet - Push all other notes, log files, etc. to backblaze / somewhere
- Fix HTML export of thesis
- make extended sections appear as folded by default
- Create a 'title page', maybe similar to the style common for ML
releases of papers? A picture, an abstract, some short and simple
overview / results?
Like this https://nihalsid.github.io/mesh-gpt/
based on https://nerfies.github.io/
Or https://qtransformer.github.io/
from https://jonbarron.info/
Or https://voyager.minedojo.org/
- CAST, detector, reconstruction, background, MLP, training synthetic, vetoes, limit calculation, results
- https://phd.vindaar.de: Should not contain the thesis until public, if I understood Klaus correctly now.
- Update all links from relative links in thesis to ones based on some fixed prefix
- Write mail to CS guy
[X]Write mail to Prüfungsbüro about whether my application was handed in back in the day -> Ask about application -> Ask about whether Igor is allowed to be 2nd corrector -> Put Klaus in CC- Write mail to Igor AFTER mail to Prüfungsbüro
Still to do:
[X]Include image of LLNL telescope[X]Fix images in CAST appendix[X]Check for any big TODO parts in the thesis document[X]Rewrite Polya section in calibration[ ]Finish appendices (after sending to people)[X]Fix ∫, ∂ unicode characters used in plot caption -> That was painful. We finally got it to work using thecombofontpackage! https://tex.stackexchange.com/a/514984[X]Fix introduction of chapter 8[X]Fix introduction of CAST chapter 9[X]Fix first sec of chapter 10[X]Fix introduction of background chapter 11
1.113.
Promotionsordnung:
- Apparently this is version 2019 https://www.uni-bonn.de/de/forschung-lehre/forschung-und-lehre-medien/promovierende-und-postdocs-medien/2019_mnf_promotionsordnung_lesefassung_final.pdf
- Version 2022 (latest?): https://www.mnf.uni-bonn.de/dokumente-1/promotionsordnung_mnf_2022.pdf
Website
- https://www.mnf.uni-bonn.de/de/promotion/leitfaden/anmeldungpromverfahren
- https://www.mnf.uni-bonn.de/de/promotion/leitfaden/promverfahren
Führungszeugnis:
- Bonn website: https://www.bonn.de/vv/produkte/Fuehrungszeugnis.php
- Online application: http://www.fuehrungszeugnis.bund.de/
[X]Beantragt and created an account with them.[X]Sent a contact request to Zenodo asking about Github accounts.
Things left to do until thesis really done
[ ]Finish appendices of thesis[ ]Compute axion-photon limit[ ]Compute chameleon limit[ ]Increase font size of all plots[ ]Possibly move some FADC stuff to appendix[ ]Compute a χ² plot of our likelihood. What does it look like?[ ]Reference / talk about conversion probability changing with axion mass, how limit changes, bla blub[ ]Whatever else appears on a reread
1.114.
Recreate all the reconstruction files on my laptop:
./runAnalysisChain \ -i ~/CastData/data \ --outpath ~/CastData/data \ --years 2017 --years 2018 \ --back --calib --reco --logL --tracking
1.115.
Note about totalNum total number of clusters printed in
plotBackgroundClusters and written to attributes of each chip in
likelihood as the Total number of clusters attribute:
That is the total number of clusters.
However, in the table ./../phd/thesis.html produced from the code in ./../phd/thesis.html we count the total number of events on the center chip.
Hence, the numbers differ quite a bit! 1.454508e6 for the table and about 1.6e6 for the likelihood attribute of the number of clusters!
1.116.
While doing revision on the thesis, I noticed that the comparison of the fake generated data with the real data for run 241 did not match my expectation anymore. Not only were energy and hits quite wrong, but also the total charge was very wrong.
I debugged the fake_event_generator just now to see if I understand
why / how to fix it and I came to the conclusion, that our crutch of
multiplying the gain by 0.9 to get the correct match is not needed
anymore? Using the real gain both the energy and total charge match
very well now.
I wonder about 3 things:
[X]How does that affect the effective efficiencies we compute? Maybe they are better now?[ ]Did we train the MLP with that behavior in the data? -> Probably?[X]Does the algorithm break if used for low energy runs in the CDL data? -> It does not seem to be the case!
We should really investigate this in theory.
The plot from the thesis is here:
~/org/Figs/statusAndProgress/fakeEventSimulation/run241_thesis_totalCharge_weird/ingrid_properties_run_241_ridgeline_kde_by_run.pdf
Generate new fake data without using gain * 0.9:
./fake_event_generator \ like \ -p ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --run 241 \ --outpath /tmp/test_fakegen_run241_noGainMod.h5 \ --outRun 241 \ --tfKind Mn-Cr-12kV \ --nmc 50000n
and plot
./plotDatasetGgplot \ -f ~/CastData/data/CalibrationRuns2018_Reco.h5 \ -f /t/test_fakegen_run241_noGainMod.h5 \ --run 241 \ --plotPath /t/test_compare_v2/ \ --names Real \ --names Fake
which yields
1.116.1. Effective efficiencies
What do we need to evaluate the effective efficiencies?
Now we compute the effective efficiencies (from ./../phd/thesis.html):
WRITE_PLOT_CSV=true USE_TEX=true ./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh30_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --generatePlots --generateRunPlots \ --plotPath ~/phd/Figs/neuralNetworks/17_11_23_adam_tanh30_sigmoid_mse_3keV/effectiveEff0.95/
Adapt to our debugging needs:
./effective_eff_55fe \ ~/CastData/data/CalibrationRuns2017_Reco.h5 ~/CastData/data/CalibrationRuns2018_Reco.h5 \ --model ~/org/resources/nn_devel_mixing/17_11_23_adam_tanh30_sigmoid_mse_3keV/mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662.pt \ --ε 0.95 \ --cdlFile ~/CastData/data/CDL_2019/CDL_2019_Reco.h5 \ --evaluateFit --plotDatasets \ --generatePlots --generateRunPlots \ --plotPath ~/org/Figs/statusAndProgress/neuralNetworks/effectiveEff0.95_noGainMod/
The old effective efficiency plot from the thesis:
compared with the new one:
shows that there may even be a slight improvement in the efficiencies!
But fortunately the numbers are very similar, so all in all I think we
can just leave it as it is (i.e. keep the fix, but don't redo
everything after, i.e. background and limits).
1.116.2. Does the algorithm now break for low energy cases?
This does not seem to be the case either.
See for example
Compared with
If anything, the rmsTransverse is closer now! And the others are unchanged too. So we'll just keep it as it is, and that's all.
1.117.
Things still left to do until I can hand in:
[X]Write 'reading guide' for axion theory[X]Use custom IDs for export(setq org-latex-prefer-user-labels t)-> This fixes candidate with weights caption.[X]Reference tip of red giant branch stars when discussing gae[X]add 5/95, 1σ ranges for limits to expected limit table[X]started![X]done for gae limits[X]chameleon limit[X]axion-photon limit
[X]Add statistical uncertainty for observed limits, based on computing 100 · real limits, use σ of that[X]Write limits as bounds[X]ADD Likelihood sampled space for axion-photon & chameleon limit![X]Read the appendix[ ]Upload all data to zenodo[ ]Raw data (CAST + CDL)[X]in preparation[X]in progress
[ ]log files[ ]Reconstructed data files[ ]phd repository without thesis files (figures and resources)[ ]add my own notes[ ]As the upload is kind of impossible, send a mail to the Zenodo support and ask them about it.
[ ]Reference zenodo datasets in "about thesis" (?) and "Reconstruct all data etc"[ ]Reference "reconstruct all data" appendix? Maybe in "about" or in introduction?[X]Make https://phd.vindaar.de point somewhere with an explanation that everything is upcoming, thesis once published, links to zenodo etc[X]Put the abstract (I have to write anyway) on the page already.[X]Verify that the link works now. Before we needed to append/index.htmlmanually -> Works now. I had to disable "append URL to path" for it to directly link to theindex.htmlfile.[ ]Consider to move to github pages?[ ]Consider switching to sans serif font? In CSS file set
font-family: "DejaVu Sans", serif;
[X]For print version change code snippets to use white background, different color scheme! -> Implemented the change. Requires the choice of the correctSETUPFILEat the top and for the print version we need to run a code snippet that disables the background color being set to a dark color.[ ]Tag git repositories of code[ ]Tag phd repository with "hand in" tag
Outside of the document itself:
[X]Write (max 3000 character) summary of the thesis[ ]Finalize. The current version in ./../phd/website/index.html is about 500 characters too long.
[X]Write mail to Klaus[X]Write mail to Promotionsbüro, asking to hand in[X]Check what I need, CV etc. and prepare that.
Antrag zum Promotionsverfahren:
[-]Antrag (1x)[X]Filled in[X]Take photo[X]Added photo[ ]printed
[-]Igor extra information[X]ready to print[ ]printed
[ ]Promotion (6x)[ ]printed
[-]Gedruckt und digital:[ ]abstract (6x)[X]Shortened[ ]printed
[-]Lebenslauf (6x)[X]ready to print[ ]printed
[ ]Eidesstaatliche Erklärung (1x)[ ]printed
[-]Kopie Passport (1x)[X]Ready to print[ ]print
1.118.
Update plots on poster for group (corridor).
Poster we start with
The following sizes are in pixels as reported by Inkscape, i.e. using a DPI of 96.
Candidates + axion image: W = 779.909 H = 584.932
Differential solar flux: W = 896.599 H = 533.128
Background rate: W = 653.056 H = 395.094
Window transmission: W = 827.505 H = 460.388
The font used in the poster so far is Calibri at a text size of 36.
Let's try to make all plots look nice, use reasonable text sizes and if possible use the same actual sizes on the poster.
1.118.1. Understand required sizes
- the poster is in A0
- A0 size: W = 33.110 inch H = 46.811 inch
- Inkscape size in pixel at 96 DPI: W = 3178.583 px H = 4493.858 px
- LaTeX uses 72 DPI for
bp(which we use) - target width W'
- A0 in LaTeX pixels: W = 2383.92 H = 3370.392
- that means: fWidth = W' / A0inkscapepx textWidth = A0latexpx
1.118.2. ggplotnim support for theme via TOML file
I added support in ggplotnim to change the appearance of a plot using a TOML file read at runtime.
1.118.3. Generate axion image + candidates
Straight from ./../phd/thesis.html:
The required fWidth for this plot is:
779.909 / 3178.583 = 0.245363735979
Let's aim for 800 pixel width instead: 800 / 3178.583 = 0.251684477014
The command then is:
TEXT_PRECISION=3 plotBackgroundClusters \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k_tracking/lhood_c18_R2_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ ~/org/resources/lhood_mlp_17_11_23_adam_tanh30_sigmoid_mse_82k_tracking/lhood_c18_R3_crAll_sEff_0.95_scinti_fadc_line_mlp_mlp_tanh_sigmoid_MSE_Adam_30_2checkpoint_epoch_82000_loss_0.0249_acc_0.9662_vQ_0.99.h5 \ --outpath ~/org/Figs/IAXO_poster/ \ --suffix "mlp_0.95_scinti_fadc_line_tracking_candidates_axion_image_with_energy_radius_85" \ --energyMin 0.2 --energyMax 12.0 \ --filterNoisyPixels \ --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionElectronPhoton_0.989AU_1492.93mm.csv \ --energyText \ --colorBy energy \ --energyTextRadius 85.0 \ --switchAxes \ --useTikZ \ --singlePlot \ --textWidth 2383.92 \ --fWidth 0.251684477014
which of course uses the candidate files calculated for the thesis as
well as the axion image created for that. The TEXT_PRECISION
environment variable reduces the precision of the printed text of the
cluster energies to 3 places.
This uses the theme:
[Theme] titleFont = "font(18.0)" labelFont = "font(18.0)" tickLabelFont = "font(14.0)" tickLength = 10.0 tickWidth = 2.0 gridLineWidth = 2.0 legendFont = "font(14.0)" legendTitleFont = "font(18.0, bold = true)" facetHeaderFont = "font(18.0, alignKind = taCenter)" baseLabelMargin = 0.5 annotationFont = "font(9.0, family = \"monospace\")" continuousLegendHeight = 2.2 continuousLegendWidth = 0.5 discreteLegendHeight = 0.6 discreteLegendWidth = 0.6 plotMarginRight = 5.0 plotMarginLeft = 3.0 plotMarginTop = 1.0 plotMarginBottom = 2.5 canvasColor = "#7fa7ce" baseScale = 1.5
Which we'll likely use as well for the other plots (with changed margins, potentially).
The canvas color is the lighter blue (e.g. 7 chip detector bullet point) on the poster.
1.118.4. Generate differential solar axion flux
First we need to rerun readOpacityFile to generate a CSV file that
contains the different fluxes (including all contributions).
./readOpacityFile \ --suffix "_all" \ --distanceSunEarth 1.AU \ --fluxKind fkAll \ --plotPath ~/org/Figs/IAXO_poster/ \ --outpath ~/org/resources/IAXO_poster/ \ --g_aN 1e-9
import ggplotnim proc customTheme(): Theme = tomlTheme("~/org/resources/IAXO_poster/differential_flux_theme.toml") let fCol = r"Flux [$\SI{1e20}{keV⁻¹.m⁻².yr⁻¹}$]" let df = readCsv("~/org/resources/IAXO_poster/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-09_fluxKind_fkAll_all.csv") .rename(f{fCol <- "Flux / keV⁻¹ m⁻² yr⁻¹"}) .mutate(f{string -> string: "type" ~ (if `type` == "57Fe Flux": r"\ce{^{57}Fe} Flux" else: `type`)}, f{float: fCol ~ idx(fCol) / 1e20}) ggplot(df, aes("Energy [keV]", fCol, color = "type")) + geom_line(size = 1.5) + themeLatex(fWidth = 0.251684477014, textWidth = 2383.92, width = 600, baseTheme = customTheme) + xlim(0, 15) + ggsave("~/org/Figs/IAXO_poster/differentia_axion_flux_poster.pdf")
1.118.5. Generate background rate plot of Tpx3 data
For the background rate plot of the Timepix3 data, the most important part is of course finding the correct data file for the Tpx3 data.
Fortunately, in ./../CastData/data/Tpx3Data/lhood_tpx3_background_cast_cdl.h5 we already find the most likely candidate.
The background rate plot on the current poster contains data of 747
hours. The above H5 file also contains that many hours (2692789.0
seconds of totalDuration).
The construction of that data file is still contained in our zsh history. I'll extract the relevant pieces from the history now:
: 1656511048:0;parse_raw_tpx3 -p ~/CastData/data/Tpx3Data/Data --out /t/tpx3_background.h5 --runType rtBackground
: 1656509503:0;parse_raw_tpx3 -p ~/CastData/data/Tpx3Data/Data/Fe --out /t/tpx3_calibration.h5 --runType rtCalibration
: 1657213131:0;raw_data_manipulation -p tpx3_calibration.h5 -r calib -o /tmp/raw_tp3_calibration.h5 --tpx3
: 1657213241:0;raw_data_manipulation -p tpx3_background.h5 -r back -o /tmp/raw_tp3_background.h5 --tpx3
: 1657213610:0;reconstruction raw_tp3_calibration.h5 --out reco_tpx3_calibration.h5
: 1657215431:0;reconstruction raw_tp3_background.h5 --out reco_tpx3_background.h5
: 1657216099:0;reconstruction reco_tpx3_calibration.h5 --only_charge
: 1657216105:0;reconstruction reco_tpx3_calibration.h5 --only_fe_spec
: 1657216223:0;reconstruction reco_tpx3_calibration.h5 --only_gas_gain
: 1657216252:0;reconstruction reco_tpx3_calibration.h5 --only_energy_from_e
: 1657216258:0;reconstruction reco_tpx3_background.h5 --only_charge
: 1657216262:0;reconstruction reco_tpx3_background.h5 --only_gas_gain
: 1657216886:0;plotData --h5file reco_tpx3_background.h5 --runType rtBackground --chips 0 --chips 3 --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml --applyAllCuts --h5Compare ~/CastData/data/DataRuns2018_Reco.h5 --ingrid
: 1657274452:0;likelihood reco_tpx3_background.h5 --h5out lhood_tpx3_background_cast_cdl.h5 --altCdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 --altRefFile ~/CastData/data/CDL_2019/XrayReferenceFile2018.h5 --cdlYear=2018 --region=crGold
: 1657274468:0;likelihood reco_tpx3_background.h5 --altCdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 --altRefFile ~/CastData/data/CDL_2019/XrayReferenceFile2018.h5 --cdlYear=2018 --region=crGold --computeLogL
: 1657274472:0;likelihood reco_tpx3_background.h5 --h5out lhood_tpx3_background_cast_cdl.h5 --altCdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 --altRefFile ~/CastData/data/CDL_2019/XrayReferenceFile2018.h5 --cdlYear=2018 --region=crGold
: 1657275328:0;plotData --h5file lhood_tpx3_background_cast_cdl.h5 --runType rtBackground --chips 0 --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml --applyAllCuts --cuts '("toaRms", 0.0, 0.55)' --eventDisplay -1
Note: the entire CDL stretching logic is in ./../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/private/cdl_stretching.nim and some in ./../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/private/likelihood_utils.nim.
And also see the notes ./tpx3_effective_efficiency.html and
.
So then we just need to redo the background rate plot of the Tpx3 data file mentioned above!
plotBackgroundRate \ ~/CastData/data/Tpx3Data/lhood_tpx3_background_cast_cdl.h5 \ --centerChip 0 \ --combName "Tpx3" --combYear 2023 \ --title "GridPix3 IAXO prototype background rate" \ --showNumClusters \ --region crGold \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.0 \ --outfile background_rate_tpx3_cast_cdl.pdf \ --outpath ~/org/Figs/IAXO_poster/ \ --useTeX \ --textWidth 2383.92 \ --fWidth 0.245363735979 \ --quiet
And now the real background rate plot of the data reconstructed (and again understood) in sec. 1.118.5.2.
CUSTOM_THEME=~/org/Figs/IAXO_poster/background_theme.toml plotBackgroundRate \ /mnt/1TB/Uni/Tpx3Data/lhood_crAll_tpx3_lnL80_tot_cut.h5 \ --centerChip 0 \ --combName "Tpx3" --combYear 2023 \ --title "GridPix3 IAXO prototype background rate, " \ --showNumClusters \ --region crGold \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.0 \ --toaCutoff 20.0 --readToA \ --filterNoisyPixels \ --outfile background_rate_tpx3_cast_cdl.pdf \ --outpath ~/org/Figs/IAXO_poster/ \ --useTeX \ --textWidth 2383.92 \ --fWidth 0.251684477014 \ --quiet
The used theme is
[Theme] titleFont = "font(18.0)" labelFont = "font(18.0)" tickLabelFont = "font(14.0)" tickLength = 10.0 tickWidth = 2.0 gridLineWidth = 2.0 legendFont = "font(14.0)" legendTitleFont = "font(18.0, bold = true)" facetHeaderFont = "font(18.0, alignKind = taCenter)" baseLabelMargin = 0.5 annotationFont = "font(9.0, family = \"monospace\")" continuousLegendHeight = 2.2 continuousLegendWidth = 0.5 discreteLegendHeight = 0.6 discreteLegendWidth = 0.6 plotMarginRight = 1.0 plotMarginLeft = 2.5 plotMarginTop = 1.0 plotMarginBottom = 2.5 canvasColor = "#7fa7ce" baseScale = 1.5
which is identical to the one used in sec. 1.118.3 for the axion candidate plot, just with a smaller margin on the right side.
- List of Tpx3 runs for background / calibration in 2022
Run file as contained in ./../CastData/data/Tpx3Data/Data (or ./../../../mnt/1TB/Uni/Tpx3Data/Data/ on laptop)
basti at voidRipper in ~/CastData/data/Tpx3Data/Data λ tree . -— DataTake2022-05-1816-54-23.h5 -— DataTake2022-05-1820-21-53.h5 -— DataTake2022-05-1909-34-07.h5 -— DataTake2022-05-1917-36-56.h5 -— DataTake2022-05-2005-18-43.h5 -— DataTake2022-05-2008-01-16.h5 -— DataTake2022-05-2015-58-53.h5 -— DataTake2022-05-2209-41-19.h5 -— DataTake2022-05-2316-19-43.h5 -— DataTake2022-05-2316-21-10.h5 -— DataTake2022-05-2316-26-19.h5 -— DataTake2022-05-2316-30-19.h5 -— DataTake2022-05-2316-31-24.h5 -— DataTake2022-05-2317-13-48.h5 -— DataTake2022-05-2407-45-08.h5 -— DataTake2022-05-2415-54-18.h5 -— DataTake2022-05-2418-52-50.h5 -— DataTake2022-05-2421-04-38.h5 -— DataTake2022-05-2514-22-28.h5 -— DataTake2022-05-2516-35-06.h5 -— DataTake2022-05-2610-23-11.h5 -— DataTake2022-05-2707-33-39.h5 -— DataTake2022-05-2714-05-01.h5 -— DataTake2022-05-2800-01-52.h5 -— DataTake2022-05-2817-27-49.h5 -— DataTake2022-05-3016-08-56.h5 -— DataTake2022-05-3114-23-33.h5 -— DataTake2022-05-3117-08-36.h5 -— DataTake2022-06-0112-27-41.h5 -— DataTake2022-06-0115-57-41.h5 -— DataTake2022-06-0216-48-09.h5 -— DataTake2022-06-0223-45-56.h5 -— DataTake2022-06-0311-18-21.h5 -— DataTake2022-06-0314-14-13.h5 -— DataTake2022-06-0319-36-53.h5 -— DataTake2022-06-0500-21-18.h5 -— DataTake2022-06-0716-02-23.h5 -— DataTake2022-06-0816-57-34.h5 -— DataTake2022-06-0916-00-26.h5 -— DataTake2022-06-0917-07-15.h5 -— DataTake2022-06-1015-27-58.h5 -— DataTake2022-06-1316-07-22.h5 -— DataTake2022-06-1408-08-36.h5 -— DataTake2022-06-1416-45-02.h5 -— DataTake2022-06-1516-43-52.h5 -— DataTake2022-06-1712-04-35.h5 -— DataTake2022-06-1810-06-42.h5
-— DataTake2022-05-1917-00-39.h5 -— DataTake2022-05-2015-17-08.h5 -— DataTake2022-05-2316-32-21.h5 -— DataTake2022-05-2415-11-21.h5 -— DataTake2022-05-2515-55-02.h5 -— DataTake2022-05-2713-22-28.h5 -— DataTake2022-05-3015-29-08.h5 -— DataTake2022-05-3116-21-53.h5 -— DataTake2022-06-0115-17-15.h5 -— DataTake2022-06-0215-58-30.h5 -— DataTake2022-06-0313-26-09.h5 -— DataTake2022-06-0715-22-08.h5 -— DataTake2022-06-0816-17-02.h5 -— DataTake2022-06-0915-12-44.h5 -— DataTake2022-06-1014-46-37.h5 -— DataTake2022-06-1315-21-10.h5 -— DataTake2022-06-1415-58-15.h5 -— DataTake2022-06-1516-04-34.h5 -— DataTake2022-06-1711-00-41.h5 2 directories, 66 files
- Reconstruct Tpx3 data again
Raw data parsing (adjust path accordingly on your computer):
parse_raw_tpx3 -p Data --out tpx3_background.h5 --runType rtBackground parse_raw_tpx3 -p Data/Fe --out tpx3_calibration.h5 --runType rtCalibration
Raw data parsing:
raw_data_manipulation -p tpx3_calibration.h5 -r calib -o raw_tpx3_calibration.h5 --tpx3 raw_data_manipulation -p tpx3_background.h5 -r back -o raw_tpx3_background.h5 --tpx3
[-]NOTE: Before running the reconstruction related things, update the ToT calibration with the file given by Markus! -> No, not needed, because the ToT calibration Markus sent us is for 'his' detector and not the one we are analyzing here (W15 A7 is this one, Markus is G6 W15).
Reconstruction:
reconstruction -i raw_tpx3_calibration.h5 --out reco_tpx3_calibration.h5 && reconstruction -i raw_tpx3_background.h5 --out reco_tpx3_background.h5 && reconstruction -i reco_tpx3_calibration.h5 --only_charge && reconstruction -i reco_tpx3_calibration.h5 --only_fe_spec && reconstruction -i reco_tpx3_calibration.h5 --only_gas_gain && reconstruction -i reco_tpx3_calibration.h5 --only_energy_from_e && reconstruction -i reco_tpx3_background.h5 --only_charge && reconstruction -i reco_tpx3_background.h5 --only_gas_gain && reconstruction -i reco_tpx3_background.h5 --only_energy_from_e
Calculate likelihood values:
likelihood -f reco_tpx3_background.h5 \ --cdlYear 2018 --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --Fe55 reco_tpx3_calibration.h5 \ --computeLogL
Likelihood cut:
likelihood -f reco_tpx3_background.h5 \ --cdlYear 2018 --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --lnL \ --signalEfficiency 0.8 \ --Fe55 reco_tpx3_calibration.h5 \ --region crAll \ --h5out lhood_crAll_tpx3_lnL80.h5
And plots:
ESCAPE_LATEX=true plotBackgroundRate lhood_crAll_tpx3_lnL80.h5 \ --centerChip 0 \ --combName "Tpx3" --combYear 2022 \ --showNumClusters --region crGold \ --showTotalTime --topMargin 1.5 \ --energyDset energyFromCharge \ --outfile tpx3_bckg_lnL80_test.pdf \ --outpath ~/org/Figs/statusAndProgress/Timepix3/Testing \ --useTeX --quiet
The resulting background rate plot
exhibits the same extremely high background at low energies as the one produced from the file that is still on my desktop at home.
Thinking about the reasons, 2 things occurred to me:
the file we used last to create plot contained the suffix
*_tot_cut. In the notes here I don't find a reference, but searching on Discord in the GasDet channel yields the information when searching for "tot cut":Vindaar — 08/19/2022 12:51 PM the ToT cut is implemented now (here 4 < x < 250) Attachment file type: acrobat totperpixelrun0chip0regioncrAlltoaLength-0.020.0applyAlltrue.pdf 12.55 KB (for a single 55Fe run in this case) 4 is apparently too low as the big peak is still there tobi — 08/19/2022 12:52 PM yes 5? das zählen der bins ergibt das es 5 sein müsste Vindaar — 08/19/2022 4:10 PM here's ~all plots for a single 55Fe run without ToT < 5 & > 250 Attachment file type: acrobat all.pdf 1.03 MB Vindaar — 08/19/2022 6:07 PM plots (with ToA < 20 & masked regions) of the clusters left after likelihood when cutting of ToT < 5 and > 250 before. Attachment file type: acrobat allbackgroundtotcut.pdf 977.30 KB Attachment file type: acrobat backgroundrate0show2014falseseparatefalse.pdf 16.88 KB
So it seems to be a ToT cut in the range 5 to 250. How do we filter these outside of doing it when running the raw data manipulation code (there via the config.toml file)? Can we maybe do it in a different way?
The other reason, which will be related to the first, is the occupancy map of the detector. We know that there are regions that are quite noisy. So we should ideally also filter out those regions for the background rate plot. From the config.toml file of
karaPlot./../CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.tomlregions = [1, 2] [MaskRegions.1] applyFile = ["reco_tpx3_background.h5", "lhood_tpx3_background_cast_cdl.h5"] applyDset = [] x = [150, 250] # mask x range from <-> to y = [130, 162] # mask y range from <-> to [MaskRegions.2] applyFile = ["reco_tpx3_background.h5", "lhood_tpx3_background_cast_cdl.h5"] applyDset = [] x = [125, 135] # mask x range from <-> to y = [110, 120] # mask y range from <-> to
we see the regions that should be masked for this detector.
UPDATE: Looking at the implementation of the
plotBackgroundRatescript to include the mask regions fromkaraPlotmade me realize that the background rate script already takes into account ONLY THE TPX3 noisy areas. I accidentally used the--filterNoisyPixelsin my thesis for the full chip background rates, meaning they are actually too high (because the noisy areas are not filtered, but non-noisy are).With the noisy pixels filtered the plot looks like:
ESCAPE_LATEX=true plotBackgroundRate lhood_crAll_tpx3_lnL80.h5 \ --centerChip 0 \ --combName "Tpx3" --combYear 2022 \ --showNumClusters --region crGold \ --showTotalTime --topMargin 1.5 \ --energyDset energyFromCharge \ --filterNoisyPixels \ --toaCutoff 20 --readToA \ --outfile tpx3_bckg_lnL80_noisy_filtered.pdf \ --outpath ~/org/Figs/statusAndProgress/Timepix3/Testing \ --useTeX --quiet
which results in
which pretty much looks like what we expect.
[X]Rerun the raw data manipulation with the 5 < ToT < 250 cut.
With this done, we can now finally produce the plot for the background rate for the poster. Continue in the parent section!
And now using the ToT cut:
ESCAPE_LATEX=true plotBackgroundRate lhood_crAll_tpx3_lnL80_tot_cut.h5 \ --centerChip 0 \ --combName "Tpx3" --combYear 2022 \ --showNumClusters --region crGold \ --showTotalTime --topMargin 1.5 \ --energyDset energyFromCharge \ --filterNoisyPixels \ --toaCutoff 20 --readToA \ --outfile tpx3_bckg_lnL80_tot_cut_noisy_filtered.pdf \ --outpath ~/org/Figs/statusAndProgress/Timepix3/Testing \ --useTeX --quiet
- Reconstruct all data with cuts
We will use these cuts:
tpx3ToACutoff = 100 # ToA cluster cutoff! # ToT related cuts. Removes any pixel below and above the given threshold # The range is *inclusive* # rmTotLow = 0 # this default excludes nothing # rmTotHigh = 11810 # for Tpx3 better: rmTotLow = 5 rmTotHigh = 250
in the ./../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/config.toml file.
Raw data parsing:
raw_data_manipulation -p tpx3_calibration.h5 -r calib -o raw_tpx3_calibration_tot_cut.h5 --tpx3 && raw_data_manipulation -p tpx3_background.h5 -r back -o raw_tpx3_background_tot_cut.h5 --tpx3
reconstruction -i raw_tpx3_calibration_tot_cut.h5 --out reco_tpx3_calibration_tot_cut.h5 && reconstruction -i raw_tpx3_background_tot_cut.h5 --out reco_tpx3_background_tot_cut.h5 && reconstruction -i reco_tpx3_calibration_tot_cut.h5 --only_charge && reconstruction -i reco_tpx3_calibration_tot_cut.h5 --only_fe_spec && reconstruction -i reco_tpx3_calibration_tot_cut.h5 --only_gas_gain && reconstruction -i reco_tpx3_calibration_tot_cut.h5 --only_energy_from_e && reconstruction -i reco_tpx3_background_tot_cut.h5 --only_charge && reconstruction -i reco_tpx3_background_tot_cut.h5 --only_gas_gain && reconstruction -i reco_tpx3_background_tot_cut.h5 --only_energy_from_e
Calculate likelihood values:
likelihood -f reco_tpx3_background_tot_cut.h5 \ --cdlYear 2018 --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --Fe55 reco_tpx3_calibration_tot_cut.h5 \ --computeLogL
likelihood -f reco_tpx3_background_tot_cut.h5 \ --cdlYear 2018 --cdlFile ~/CastData/data/CDL_2019/calibration-cdl-2018.h5 \ --lnL \ --signalEfficiency 0.8 \ --Fe55 reco_tpx3_calibration_tot_cut.h5 \ --region crAll \ --h5out lhood_crAll_tpx3_lnL80_tot_cut.h5
1.118.6. Generate plot of different windows + measurements
The data file for the measurements is ./resources/window_scan_transparencys.txt sent to me by Tobi via Discord.
import std / [sequtils, strutils, strformat] import ggplotnim proc suf(s: string): string = result = s.replace("_err", "") proc prepareWindowData(fname: string): DataFrame = let df = readCsv(fname, sep = ' ', quote = '"') let dfL = df.gather(df.getKeys().filterIt(it notin ["energy", "energy_err"]), "Type", "Transmission") .dropNan("Transmission") .mutate(f{string -> string: "Error" ~ (if "err" in `Type`: "Error" else: "Transmission")}) .mutate(f{string -> string: "Type" ~ suf(`Type`)}) var dfR = newDataFrame() for tup, subDf in groups(dfL.group_by("Error")): let sDf = subDf.arrange(["Type", "energy"]) dfR["energy"] = sDf["energy"] dfR["energy_err"] = sDf["energy_err"] dfR["Type"] = sDf["Type"] let c = tup[0][1].toStr dfR[c] = sDf["Transmission"] result = dfR.mutate(f{string -> string: "Material (data)" ~ ( if `Type` == "200nm": r"200 nm $\ce{Si_3 N_4}$" elif `Type` == "2um": "2 μm Mylar" elif `Type` == "300nm New": "Drop" elif `Type` == "300nm Old": r"300 nm $\ce{Si_3 N_4}$" else: "Drop")}, f{float: "energy" ~ `energy` / 1000.0}) .filter(f{idx("Material (data)") != "Drop"}) # 1. parse the window measurements from Tobi let dfR = prepareWindowData "~/org/resources/window_scan_transparencys.txt" # 2. compute the theoretical transmission values for SiN import xrayAttenuation, unchained # 2 thicknesses: 200 nm and 300 nm # 2 densities: 3.44 (default), 3.0 # 2 compounds: Si₃N₄ and SixNy with x/y = 1.0? let Si = Silicon.init() let N = Nitrogen.init() let Si₃N₄ = compound((Si, 3), (N, 4)) let SiN₁ = compound((Si, 1), (N, 1)) # instantiate Mylar let mylar = compound((C, 10), (H, 8), (O, 4)) proc genDf[L: Length](d: L, ρ: g•cm⁻³, c: AnyCompound): DataFrame = let Es = linspace(0.05, 15, 1000) let name = if c.name() == "C10H8O4": "Mylar" else: r"\ce{Si_3 N_4}" result = toDf({"Energy" : Es, "Material (theory)" : $d & " " & name }) .mutate(f{float: "Transmission" ~ transmission(c, ρ, d, `Energy`.keV).float}) var dfX = newDataFrame() for d in [200.nm, 300.nm]: dfX.add genDf(d, 3.44.g•cm⁻³, Si₃N₄) #dfX.add genDf(d, 3.g•cm⁻³, Si₃N₄) #dfX.add genDf(d, 3.44.g•cm⁻³, SiN₁) dfX.add genDf(2.μm, 1.4.g•cm⁻³, mylar) proc customTheme(): Theme = tomlTheme("~/org/resources/IAXO_poster/sin_window_theme.toml") #proc customTheme(): Theme = tomlTheme("~/org/resources/IAXO_poster/sin_window_theme_white_johanna.toml") proc genPlot(eMax: float, suffix: string, logPlot: bool) = var plt = ggplot(dfR.filter(f{float: idx("energy") <= eMax}), aes("energy", "Transmission", color = "Material (data)")) + geom_point(size = 1.5) + geom_line(data = dfX.filter(f{float: idx("Energy") <= eMax}), aes = aes("Energy", "Transmission", color = "Material (theory)")) + xlab("Energy [keV]") + geom_errorbar(aes = aes(yMin = f{`Transmission` - `Error`}, yMax = f{`Transmission` + `Error`}), size = 1.5) + themeLatex(fWidth = 0.251684477014, textWidth = 2383.92, width = 600, baseTheme = customTheme) if logPlot: plt = plt + scale_x_log10() plt + ggsave(&"~/org/Figs/IAXO_poster/window_{suffix}.pdf") #plt + ggsave(&"~/org/Figs/IAXO_poster/window_white_{suffix}.pdf") genPlot(4.0, "4keV", false) genPlot(3.0, "3keV", false) genPlot(15.0, "15keV_log10", true)
The initial code with all lines: ./Misc/sin_window_plot_IAXO_poster.nim
1.119.
[X]Johanna also wants a version of the window plot with white background. -> Done in the above as well now, with_whitesuffix[X]Johanna also wants this plot ./Code/CAST/axionPlusWindows/axionPlusWindow.nim in a version with larger fonts etc.
[X]Rewrite the above code usingxrayAttenuation[X]Make pretty for slides
-> See ./Code/CAST/axionPlusWindows/axionPlusWindow_xrayAttenuation.nim
1.120.
[X]create PRs for all TPA dependent libraries[ ]merge all PRs and tag new TPA version[X]Recover files for Tpx3 data on laptop[X]Reconstruct Tpx3 data[X]During parsing of raw tpx3 data, reading of data failed in one file -> FileData/DataTake_2022-05-19_17-36-56.h5-> That happened because due to a refactoring innimhdf5theHDF5BloscDecompressionErroris not raised as such anymore, but we get a regularerr < 0error fromH5Dreadinstead[X]Calibration data
[X]Recreate background rate plot[X]Create other Tpx3 plots for Johanna
1.120.1. Update the ToT calibration of the GridPix3 prototype detector
The file Markus sent us via Discord is ./../../../mnt/1TB/Uni/Tpx3Data/ToT_calibration_unit_fix/ToTCalib_2023-10-16_15-50-45.h5.
Ah, this is for the G6 W15 based GridPix detector. According to InGrid database that is the detector for the 'GridPix3RomePreliminary' data taking period (i.e. "Markus" detector).
Markus now also sent me the ToT calibration with updated parameters for the 'Tobi' chip
I created symlinks for these two files to ./../CastData/ExternCode/TimepixAnalysis/resources/ChipCalibrations/Tpx3_IAXO/ and file:///home/basti/CastData/ExternCode/TimepixAnalysis/resources/ChipCalibrations/Tpx3_Polarimetry/
and so:
./databaseTool --add ../resources/ChipCalibrations/Tpx3_IAXO ./databaseTool --add ../resources/ChipCalibrations/Tpx3_Polarimetry
updates the fit parameters and /interpreted/mean_curve -> ToTCalib.
Data is updated. I'm now waiting for confirmation from Markus, if the fit parameters are the only thing that meaningfully changed (or if the mean curve data should also be significantly different).
1.121.
[X]Rerun the raw data manipulation with the 5 < ToT < 250 cut. -> 1.118.5.2.1[X]Create background rate plot for poster[X]Create plots for Johanna
1.121.1. Create other Tpx3 plots for Johanna
Her current slides are .
We need to renew the following plots:
- occupancy map of Tpx3 background data (in counts) <-
plotData - histograms of all 55Fe runs (hits) in one plot <-
plotData - ToA length 55Fe vs background <-
plotData - background rate <-
plotBackgroundRate
When reconstructing the data a final time:
[X]extend the ToA cutoff from 50 pixels in theRawDatatpx3ToACutoff = 50field so that we can plot a longer range?[X]enable ToT cut? -> 1.118.5.2.1
We changed the ToA cutoff to 100 clock cycles and activated the ToT cut as mentioned (5<ToT<250)
- Generate plots with
plotData
- Occupancy plots via
--occupancy - 55Fe histogram plots via ?
- ToA length via
--ingrid
- Occupancy map
Important:
- We need to disable the masking regions we otherwise use for this
detector for the occupancy map!
That is, make sure the line
regionsfield looks like:regions = []in ./../CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml.
Now we just run:
T_MARGIN=1.75 USE_TEX=true WIDTH=600 HEIGHT=420 plotData \ --h5file reco_tpx3_background_tot_cut.h5 \ --runType rtBackground \ --occupancy \ --plotPath ~/org/Figs/Johanna_XDEP_Talk/
- We need to disable the masking regions we otherwise use for this
detector for the occupancy map!
That is, make sure the line
- 55Fe spectra in histogram form separated by run
First make sure to use the mask regions again:
regions = [1, 2] [MaskRegions.1] applyFile = ["reco_tpx3_background_tot_cut.h5", "reco_tpx3_calibration_tot_cut.h5", "lhood_tpx3_background_cast_cdl.h5"] applyDset = [] x = [150, 170] # mask x range from <-> to y = [130, 162] # mask y range from <-> to [MaskRegions.2] applyFile = ["reco_tpx3_background_tot_cut.h5", "reco_tpx3_calibration_tot_cut.h5", "lhood_tpx3_background_cast_cdl.h5"] applyDset = [] x = [125, 135] # mask x range from <-> to y = [110, 120] # mask y range from <-> to
And now run:
OVERLAP=6 CUSTOM_THEME=~/org/Figs/Johanna_XDEP_Talk/hits_theme.toml \ LINE_BREAK=true ESCAPE_LATEX=true DENSITY=true T_MARGIN_RIDGE=2.0 \ T_MARGIN=1.0 USE_TEX=true WIDTH=600 HEIGHT=400 \ plotData \ --h5file reco_tpx3_calibration_tot_cut.h5 \ --runType rtCalibration \ --cuts '("hits", 20.0, 300)' --applyAllCuts \ --ingrid --ingridDsets hits \ --separateRuns \ --plotPath ~/org/Figs/Johanna_XDEP_Talk/
- ToA length plot of 55Fe and background
And now run for 55Fe data:
CUSTOM_THEME=~/org/Figs/Johanna_XDEP_Talk/toa_length_theme.toml \ LINE_BREAK=true ESCAPE_LATEX=true T_MARGIN=1.0 \ USE_TEX=true WIDTH=600 HEIGHT=400 \ plotData \ --h5file reco_tpx3_calibration_tot_cut.h5 \ --runType rtCalibration \ --ingrid --ingridDsets toaLength \ --plotPath ~/org/Figs/Johanna_XDEP_Talk/
and for background data:
CUSTOM_THEME=~/org/Figs/Johanna_XDEP_Talk/toa_length_theme.toml \ LINE_BREAK=true ESCAPE_LATEX=true T_MARGIN=1.0 \ USE_TEX=true WIDTH=600 HEIGHT=400 \ plotData \ --h5file reco_tpx3_background_tot_cut.h5 \ --runType rtBackground \ --ingrid --ingridDsets toaLength \ --plotPath ~/org/Figs/Johanna_XDEP_Talk/
- Occupancy plots via
- Generate background rate plot with
plotBackgroundRate
See sec. 1.118.5.2 for the reconstruction of the data files.
CUSTOM_THEME=~/org/Figs/Johanna_XDEP_Talk/background_theme.toml plotBackgroundRate \ /mnt/1TB/Uni/Tpx3Data/lhood_crAll_tpx3_lnL80_tot_cut.h5 \ --centerChip 0 \ --combName "Tpx3" --combYear 2023 \ --title "GridPix3 IAXO prototype background rate" \ --showNumClusters \ --region crGold \ --showTotalTime \ --topMargin 1.5 \ --energyDset energyFromCharge \ --energyMin 0.0 \ --toaCutoff 20.0 --readToA \ --filterNoisyPixels \ --outfile background_rate_tpx3_cast_cdl.pdf \ --outpath ~/org/Figs/Johanna_XDEP_Talk/ \ --useTeX \ --textWidth 2383.92 \ --fWidth 0.251684477014 \ --quiet
1.122.
Over the last few days I wrote a few helper tools to modify HDF5 files:
./Misc/update_run_numbers.nim -> Updates all
runNumberattributes of the/runsor/reconstructiongroups (to update the outdatedrunNumberfield in Tpx3 data, for exampleupdate_run_numbers -f reco_tpx3_background_tot_cut.h5
)
- ./Misc/write_attribute.nim -> Allows to write a new attribute to a file
- ./Misc/delete_group.nim -> Deletes a single group
All these should be compiled via
nim c --out:bin foo.nim
because I added ./Misc/bin to my PATH.
1.122.1. Cross check total signal against g⁴aγ axion photon coupling
Igor told Cristina that in old notes he found that their total signal was expected to be about 1.4 counts at 6.6e-114.
Cristina gets very close to 3 counts in total signal expected at that value.
I added some code to mcmc_limit_calculation into the
--axionPhotonLimit sanity check to compare.
block: ## This is a cross check with Cristina's code. She gets about 3 counts of signal expected ## at ~6.6e-11^4 with 320h of data. Here we compute the same for our detector and setup, ## which yields a total signal of 1.47213 counts at the same coupling. The factor of 2 ## is pretty close to the 160h of tracking data we have. Our two detectors are pretty similar ## in general efficiency over the energy range of the axion-photon flux. let g_aγs = linspace(0.0, 5e-40, 1000) var ss = newSeq[float]() for g in g_aγs: ctx.coupling = g ss.add ctx.totalSignal() ggplot(toDf(g_aγs, ss), aes("g_aγs", "ss")) + geom_line() + xlab("g⁴_aγ [GeV⁻⁴]") + ylab("Total signal [counts]") + ggsave(SanityPath / "total_signal_vs_g4_aγ.pdf") ctx.coupling = 6.6e-11^4 log.infos("Total signal expected"): &"Total signal s_tot = {ctx.totalSignal()} at g_aγ = {ctx.coupling}"
The sanity log:
mcmc_limit_calculation sanity --limitKind lkMCMC --axionModel ~/phd/resources/readOpacityFile/solar_axion_flux_differential_g_ae_1e-13_g_ag_1e-12_g_aN_1e-15_fluxKind_fkAxionPhoton_0.989AU.csv --axionImage ~/phd/resources/axionImages/solar_axion_image_fkAxionPhoton_0.989AU_1492.93mm.csv --combinedEfficiencyFile ~/org/resources/combined_detector_efficiencies.csv --switchAxes --sanityPath /tmp/test_sanity/ --axionPhotonLimit
yields
- INFO:
=============Time=============- INFO: Total background time: 3158.01 h - INFO: Total tracking time: 159.899 h - INFO: Ratio of tracking to background time: 1 UnitLess - INFO:=============Total signal expected=============- INFO: Total signal stot = 1.47213 UnitLess at gaγ = 1.8974736e-41 - INFO: Saving plot: /tmp/testsanity/candidatessignaloverbackgroundaxionPhoton.pdf
1.123.
Tobi and me discussed again while I was at Uni after handing in my thesis about the time window the shutter remains open after the FADC trigger.
In my thesis I wrote that the window remains open 50 μs after the FADC trigger. That already seemed like a long time for me when writing it, but I forgot to check that number again before printing.
Tobi then reminded me yesterday :
tobi — Yesterday at 2:04 PM Hi, sag mal hattest du die Zeit die der Shutter nach dem FADC trigger offenbleibt mal nachgeschaut? Man musste das der Firmware als Parameter aus dem TOS mitgeben
Vindaar — Yesterday at 4:53 PM Ich schau morgen vormittag nochmal in Ruhe. Das man das mitgeben musste kann sein. Ist aber soweit ich weiß kein Parameter gewesen, den man typischerweise geändert hat?
tobi — Yesterday at 4:53 PM Ne den hatten wir einmal am anfang festgelegt und dann immer so benutzt, vll. ist der auch im Tos Fix In der Firmware wird der mit Funktion
20gesetztVindaar — Today at 11:03 AM Ist das ne Hex Zahl oder Dezimal? Ahh, jo, ne Hex Zahl glaube ich / default number of clock cycles the shutter remains open, after the FADC triggers / (200 clock cycles at 40 MHz == 5 mu s) #define DEFAULTFADCSHUTTERCOUNTON 200
Das ist die Konstante, die per Default an mode = 0x20 gesetzt wird Da das im Code glaube ich nie überschrieben wurde sind das wohl 5μs (statt den 50, die ich in meiner Arbeit geschrieben habe 🤭)
which includes me checking TOS this morning.
The relevant lines of code are:
./../CastData/ExternCode/TOS/include/fpga.hpp for the definition of the constant used to define the time in clock cycles,
// default number of clock cycles the shutter remains open, after the FADC triggers // (200 clock cycles at 40 MHz == 5 mu s) #define DEFAULT_FADC_SHUTTER_COUNT_ON 200
so 200 clock cycles @ 40 MHz == 5 μs.
./../CastData/ExternCode/TOS/src/fpga.cpp
_fadcShutterCountOn(DEFAULT_FADC_SHUTTER_COUNT_ON),
where the constant is set to the class attribute
[BROKEN LINK: No match for fuzzy expression: 647] where the attribute is written to the FPGA via i²c
if(FADCshutter){ // FADC trigger closes shutter on // Mode = 0x20 == 32 Mode = 0x20; tp->SetI2C(_fadcShutterCountOn); }
Relevant commit:
c3195bcdb0fd3b376201d473c1c4f0598a17b88d Author: Sebastian Schmidt <s.schmidt@physik.uni-bonn.de> AuthorDate: Fri Aug 12 09:11:25 2016 +0200 Commit: Sebastian Schmidt <s.schmidt@physik.uni-bonn.de> CommitDate: Fri Aug 12 09:11:25 2016 +0200
Parent: 07464cb included simple readout of FADC for a single frame Contained: master
working implementation of FADC. still missing proper filenames and multi chip support
[ ]Attempt to find out where the 50 μs came from
1.124. Meeting with Julia and Jaime about LLNL telescope
The meeting was very productive.
References that were brought up:
Our main talking points were about the difference between the expected focal length of 1500mm and my "measured" one of 1530mm, as well as the measurements that were done with the telescope at PANTER.
One interesting tidbit is that at PANTER the focal length was also not measured to be 1500mm, but slightly longer (about 1519 mm I think).
At PANTER the distance to the source is 128m according to Jaime. The references above should also contain some numbers! We don't know the spot size of the PANTER source though. For a realistic raytracing simulation of that, we need that. But Jaime mentioned that Mike (LLNL) used / always talked about using a point source. So to cross check that is what we can do. The fact that this assumes a point source should give us a result that is much closer to the raytracing result of the Sun in the focal spot that we had before (before the RNG sampling bug in the Sun was fixed), due to having rays from a very narrow region. I.e. using that we should reproduce something that is very similar to the plots mentioned under "best focus" on e.g. slide 13 in the second document linked above. 1.124.1
Another thing that was brought up and is also visible in the sets of slides is the energy dependence of the illumination of the mirror shells. We've seen this before, but we should check it again. I.e. can we reproduce the different illumination shown in the slides? 1.124.1
The next very important thing we learned about the plots shown in the thesis by Anders and JCAP paper is that the raytracing plots are always shown as log plots! I didn't notice that before! This explains why the "bow tie" shape is so much more prevalent in the results. 1.124.1
Jaime thinks reason for mismatch focal length mismatch is due to the
definition of xsep and the placement of the mirrors. In the end we
did agree (I think) that generally in theory xsep should be 4 mm in
the horizontal direction, but at what that ends up in the end depends
on how the layers were inserted. He mentioned that he thinks the
engineer placed the mirrors so that (at least for the second set of
mirrors, not sure about first) the mirrors are aligned on the front
(towards center of telescope / towards magnet). We currently align at
the center. We can check the impact of changing that. 1.124.1
Jaime will attempt to find out these things:
[ ]Find the Excel file that contains the numbers that were actually built by the engineer who assembled the telescope[ ]Find out the source size of the PANTER source.[ ]Find the rest of the PANTER data / send it to me
Next meeting:
1.124.1. Things we should do for the next meeting [/]
[ ]Implement an X-ray finger source like PANTER, i.e. 128 m and point size. -> check slides!! Then use this setup to simulate Al Kα and Ti Kα energies, make log plots of the images, compare with e.g. slide 13 in PDF, and calculate the HPD (half power diameter?). I.e. take the image, compute the sum of all rows / columns. Draw those as a line plot and width where the amplitude is half is the value for the HPD. Then use \[ \tan γ = \frac{\text{HPD}}{f} \] to calculate the apparent size as seen from the center of the optic. That gives us an angle. Compare those angles with the ones from table on slide 18 in the linked PDF
[ ]Check effect different energy on illumination of the shells. Check slides, take their energies and understand their setup and see if the illumination comes out to be the same. Maybe place anImageSensorat the end of the telescope.
[ ]Produce log plots of our raytracing images!
[ ]As Jaime believes the placement of the mirrors was aligned at the front and not center (as I currently do), it would be good to check the effect on the focal length if we move the mirrors such that alignment is at the front. However I'm pretty convinced this won't change anything (for that matter neither for a perfect telescope with xsep = 0). But it should be a simple thing to try. -> Move to align front mirrors, check images.
1.124.2. Other aspects
Jaime mentioned the focal length is always going to be different between visible light and X-rays, due to the fact that X-rays are not actually reflected at the surface, but rather inside the material at one of the interfaces.
However, the thicknesses of the layers are extremely small (O(<100 nm)), so any of these differences is much much smaller than the difference of just hitting a milli meter before on the mirror.
[ ]Question: What does the X-ray Fresnell equation math say about this? Does it actually make a difference? If so, how does one model this correctly?
1.124.3. Question for the next meeting
- Effective area - why so different?
[/]
Julia mentioned that they did a simulation, then measured the effective area at PANTER. From the difference and knowing that there would be hydrocarbons as the main source of contamination on the mirrors from the usage of epoxy. They added as much hydrocarbons to the mirror surface such that the raytracing result reproduced the real data as best as possible.
[ ]Understand what exactly is meant by hydrocarbons?[ ]Check if by adding hydrocarbons to the reflectivity calculations (on top of the multilayer!) the reflectivity actually gets worse, in particular at lower energies.
One thing we learned in this meeting is that I should use the JCAP effective area or what Jaime sent to Cristina. Namely, this file: resources/llnl_cast_nature_jaime_data/2016_DEC_Final_CAST_XRT/EffectiveArea.txt
[ ]Figure out what conditions this effective area file really corresponds to! Solar emission similar to the raytracing results of that same directory?
1.125. Raytracing calculations for next meeting
The notes about the raytracing calculations comparing to
are here: ./Doc/LLNL_TrAXer_comparison/llnl_traxer_raytracing_comparison.html.
1.126. Meeting with Jaime about LLNL telescope
The meeting today with Jaime was productive, but short.
Jaime shared the following presentation and data files with me by mail:
- ./Figs/statusAndProgress/LLNL_telescope/CAST_optic_concept.pptx
- file:///home/basti/org/resources/LLNL_telescope/cast20l4_f1500mm_asBuilt.txt
- file:///home/basti/org/resources/LLNL_telescope/cast20l4_f1500mm_asDesigned.txt
(more on these below)
We discussed

- ./Doc/LLNL_TrAXer_comparison/llnl_traxer_raytracing_comparison.html
- http://phd.vindaar.de/docs/LLNL_TrAXer_comparison/llnl_traxer_raytracing_comparison.html
The main conclusions are that everything looks pretty good now. It is nice to see that the focal spot moves about 20 mm back for the point source in finite distance. The shape of the focal spots is matching what it should match and looks almost identical to the LLNL raytracing and PANTER results.
Jaime assured me that the reason for the difference between the effective areas shown in the slides mentioned in my notes is because the yellow "best fit model" does not take into account that the telescope is actually larger than the magnet bore at CAST. And the green line mentioned in my notes on the "next" slide does take that into account. So it seems like:
- Start with a pretty high effective area for parallel light for the full telescope
- Lose some by taking into account non parallel light
- Lose some more by taking into account PMMA (hydrocarbon) contamination
- Lose even more by only considering the area of the magnet and not the whole telescope.
HOWEVER: I'm not yet convinced this is actually correct. But need to check this again if possible.
The final choice of the value to be used for the mirror imperfection is not done yet, it depends on getting the HPD values right.
This is the one thing a bit off in my results: the HPD values for the long axis are too large, about 300 arc seconds compared to about 200 arc seconds for the HPD (50%). The plots for this axis also seem to not quite match with the look of the plots for each image.
So maybe we have some bug in how we compute this? Check.
Also, in the .pptx file Jaime sent today Mike explicitly explains
what he means by HPD and it's not what I assumed it was, slide 10:
- Actually, more useful to consider the encircled energy function (EEF)
- Draw a circle around the PSF, integrate the flux, record the value
- Repeat process, building up cumulative distribution as a function of radius; normalize to unity
- It’s the integral of the PSF
- The half-power diameter (HPD) is just diameter where the EEF = 50%
and a bit more on slide 13:
ESTIMATES (just the optic) 50% encircled energy (the HPD) = 1 arcmin 80% encircled energy = 3 arcmin Extra contingency (e.g., in case the PSF is asymmetric or if there is misalignment) expand extraction region to circle with 4 arcmin diameter Now, RSS with size of the dominant axion producing region of the sun = 3 arcmin
Active diameter of Micromegas should be 5 arcmin 2.3 mm (compare with 3.5 mm used for ABRIXAS)
So this means we should also cross check our numbers if we use this approach!
The .pptx file also finally proves that the telescope length is
\(\SI{454}{mm}\) and each mirror (\(\SI{225}{mm}\) mirror length) has
\(\SI{2}{mm}\) separation to the center point of the optic, i.e. proving
\(x_{\text{sep}} = \SI{4}{mm}\), slide 4!
The two text files Jaime sent are super interesting to us: One of them is the telescope as it was designed and the other as it was actually built.
We can cross check those numbers with the numbers we use in our simulation and then see what changes if we use them!
As designed:
MirrorLength 225.00000 Gap 2.0000000 Thickness 0.210000 Spacing_perc 0.000000 FocalLength 1500.0000 10.3390 63.2412 60.9149 60.8322 53.8513 10.7690 65.8741 63.4511 63.3650 56.0936 11.2150 68.6075 66.0840 65.9942 58.4213 11.6780 71.4450 68.8173 68.7239 60.8379 12.1590 74.3908 71.6549 71.5576 63.3467 12.6580 77.4488 74.6006 74.4993 65.9511 13.1760 80.6233 77.6586 77.5532 68.6549 13.7130 83.9188 80.8331 80.7234 71.4617 14.2710 87.3398 84.1286 84.0144 74.3755 14.8500 90.8911 87.5496 87.4307 77.4003 15.4510 94.5775 91.1008 90.9771 80.5403 16.0740 98.4043 94.7872 94.6586 83.7999 16.7210 102.377 98.6139 98.4801 87.1836
As built:
MirrorLength 225.00000 Gap 2.0000000 Thickness 0.210000 Spacing_perc 0.000000 FocalLength 1500.0000 10.339 63.2384 60.9121 60.8632 53.8823 10.769 65.8700 63.4470 63.3197 56.0483 11.216 68.6059 66.0824 65.9637 58.3908 11.679 71.4175 68.7898 68.6794 60.7934 12.160 74.4006 71.6647 71.5582 63.3473 12.659 77.4496 74.6014 74.4997 65.9515 13.176 80.6099 77.6452 77.5496 68.6513 13.714 83.9198 80.8341 80.7305 71.4688 14.272 87.3402 84.1290 84.0137 74.3748 14.851 90.8910 87.5495 87.4316 77.4012 15.452 94.5780 91.1013 90.9865 80.5497 16.076 98.3908 94.7737 94.6549 83.7962 16.725 102.381 98.6187 98.4879 87.1914
Our numbers:
allR1: @[63.006, 65.606, 68.305, 71.105, 74.011, 77.027, 80.157,
83.405, 86.775, 90.272, 93.902, 97.668, 101.576, 105.632], #mapIt(it.mm),
allR5: @[53.821, 56.043, 58.348, 60.741, 63.223, 65.800, 68.474,
71.249, 74.129, 77.117, 80.218, 83.436, 86.776, 90.241], #.mapIt(it.mm),
allAngles: @[0.579, 0.603, 0.628, 0.654, 0.680, 0.708, 0.737, 0.767,
0.798, 0.830, 0.863, 0.898, 0.933, 0.970], #.mapIt(it.Degree),
The funny thing is, neither of these numbers matches our numbers, haha! We need to compute the angles from the radii R1 and R2 for example.
For example for layer 1:
import math let α = arcsin((63.2384 - 60.9121) / 225.0).radToDeg let α3 = arcsin((60.8632 - 53.8823) / 225.0).radToDeg echo α echo α3, " vs ", 3*α
This also means we need to have separate angles for each layer. But better we now use the radii as we have them instead of relying on the angles!
1.126.1. Calculate focal length based on Wolter equation for telescope data files
Let's use the code from ./Mails/llnlAxionImage/llnl_axion_image.html to compute the expected focal length in a similar manner, just using the radii as computed here.
import unchained, sequtils, math let R1s = @[63.006, 65.606, 68.305, 71.105, 74.011, 77.027, 80.157, 83.405, 86.775, 90.272, 93.902, 97.668, 101.576, 105.632].mapIt(it.mm) let αs = @[0.579, 0.603, 0.628, 0.654, 0.680, 0.708, 0.737, 0.767, 0.798, 0.830, 0.863, 0.898, 0.933, 0.970].mapIt(it.Degree) proc calcR3(r1, lMirror: mm, α: float): mm = let r2 = r1 - lMirror * sin(α) result = r2 - 0.5 * xSep * tan(α) import datamancer var df = newDataFrame() for i in 0 ..< R1s.len: let r1 = R1s[i] let α = αs[i].to(Radian).float let r3 = calcR3(r1, lMirror, α) let r1minus = r1 - sin(α) * lMirror/2 let fr3 = r3 / tan(4 * α) let fr1m = r1minus / tan(4 * α) #echo "Focal length at i ", i, " f = ", fr3, " using r1mid f_m = ", fr1m df.add (i: i, f_R3: fr3.float, f_R1m: fr1m.float) echo df.toOrgTable
import math, sequtils, datamancer const lMirror = 225.0 const xSep = 4.0 proc calcAngle(r1, r2: float): float = result = arcsin(abs(r1 - r2) / lMirror) proc calcR3(r1, lMirror: float, α: float): float = let r2 = r1 - lMirror * sin(α) result = r2 - 0.5 * xSep * tan(α) proc printTab(R1s, R2s, R4s, R5s: openArray[float]) = var df = newDataFrame() for i in 0 ..< R1s.len: let r1 = R1s[i] let r2 = R2s[i] let α = calcAngle(r1, r2) let r3 = calcR3(r1, lMirror, α) let r1minus = r1 - sin(α) * lMirror/2 let fr3 = r3 / tan(4 * α) let fr1m = r1minus / tan(4 * α) df.add (i: i, f_R3: fr3.float, f_R1m: fr1m.float) echo df.toOrgTable block AndersPhD: let R1s = @[63.006, 65.606, 68.305, 71.105, 74.011, 77.027, 80.157, 83.405, 86.775, 90.272, 93.902, 97.668, 101.576, 105.632] let αs = @[0.579, 0.603, 0.628, 0.654, 0.680, 0.708, 0.737, 0.767, 0.798, 0.830, 0.863, 0.898, 0.933, 0.970] proc calcR2(r1, α: float): float = r1 - sin(α.degToRad) * lMirror let R2s = toSeq(0 ..< R1s.len).mapIt(calcR2(R1s[it], αs[it])) echo "Using values from Anders PhD thesis" printTab(R1s, R2s, [], []) block AsDesigned: # `cast20l4_f1500mm_asDesigned.txt` let R1s = [ 63.2412, 65.8741, 68.6075, 71.4450, 74.3908, 77.4488, 80.6233, 83.9188, 87.3398, 90.8911, 94.5775, 98.4043, 102.377 ] let R2s = [ 60.9149, 63.4511, 66.0840, 68.8173, 71.6549, 74.6006, 77.6586, 80.8331, 84.1286, 87.5496, 91.1008, 94.7872, 98.6139 ] let R4s = [ 60.8322, 63.3650, 65.9942, 68.7239, 71.5576, 74.4993, 77.5532, 80.7234, 84.0144, 87.4307, 90.9771, 94.6586, 98.4801 ] let R5s = [ 53.8513, 56.0936, 58.4213, 60.8379, 63.3467, 65.9511, 68.6549, 71.4617, 74.3755, 77.4003, 80.5403, 83.7999, 87.1836 ] let diffs = [ 10.3390, 10.7690, 11.2150, 11.6780, 12.1590, 12.6580, 13.1760, 13.7130, 14.2710, 14.8500, 15.4510, 16.0740, 16.7210 ] echo "Using values from .txt file 'as designed'" printTab(R1s, R2s, R4s, R5s) block AsBuilt: # `cast20l4_f1500mm_asBuilt.txt` # These are the numbers from the "as built" text file let R1s = [ 63.2384, 65.8700, 68.6059, 71.4175, 74.4006, 77.4496, 80.6099, 83.9198, 87.3402, 90.8910, 94.5780, 98.3908, 102.381 ] let R2s = [ 60.9121, 63.4470, 66.0824, 68.7898, 71.6647, 74.6014, 77.6452, 80.8341, 84.1290, 87.5495, 91.1013, 94.7737, 98.6187 ] let R4s = [ 60.8632, 63.3197, 65.9637, 68.6794, 71.5582, 74.4997, 77.5496, 80.7305, 84.0137, 87.4316, 90.9865, 94.6549, 98.4879 ] let R5s = [ 53.8823, 56.0483, 58.3908, 60.7934, 63.3473, 65.9515, 68.6513, 71.4688, 74.3748, 77.4012, 80.5497, 83.7962, 87.1914 ] # this last one should be the difference between R5 and R1 let diffs = [ 10.339, 10.769, 11.216, 11.679, 12.160, 12.659, 13.176, 13.714, 14.272, 14.851, 15.452, 16.076, 16.725 ] echo "Using values from .txt file 'as built" printTab(R1s, R2s, R4s, R5s)
Using values from Anders PhD thesis
| i | fR3 | fR1m |
|---|---|---|
| 0 | 1501.1452 | 1529.7542 |
| 1 | 1500.7995 | 1529.4071 |
| 2 | 1500.2462 | 1528.8523 |
| 3 | 1499.554 | 1528.1586 |
| 4 | 1501.1365 | 1529.7394 |
| 5 | 1500.4047 | 1529.0057 |
| 6 | 1499.816 | 1528.4149 |
| 7 | 1499.4292 | 1528.026 |
| 8 | 1499.2931 | 1527.8876 |
| 9 | 1499.4644 | 1528.0564 |
| 10 | 1500.0058 | 1528.5952 |
| 11 | 1499.1816 | 1527.768 |
| 12 | 1500.579 | 1529.1623 |
| 13 | 1500.8171 | 1529.397 |
Using values from .txt file 'as designed'
| i | fR3 | fR1m |
|---|---|---|
| 0 | 1471.5582 | 1500.1664 |
| 1 | 1471.5794 | 1500.1861 |
| 2 | 1471.5244 | 1500.1297 |
| 3 | 1471.5363 | 1500.1398 |
| 4 | 1471.5242 | 1500.1259 |
| 5 | 1471.5125 | 1500.1123 |
| 6 | 1471.5293 | 1500.127 |
| 7 | 1471.5027 | 1500.0982 |
| 8 | 1471.5144 | 1500.1074 |
| 9 | 1471.501 | 1500.0913 |
| 10 | 1471.4966 | 1500.0841 |
| 11 | 1471.453 | 1500.0374 |
| 12 | 1471.2913 | 1499.8723 |
Using values from .txt file 'as built
| i | fR3 | fR1m |
|---|---|---|
| 0 | 1471.4905 | 1500.0987 |
| 1 | 1471.4842 | 1500.091 |
| 2 | 1471.4888 | 1500.094 |
| 3 | 1470.948 | 1499.5516 |
| 4 | 1471.7255 | 1500.3272 |
| 5 | 1471.5282 | 1500.128 |
| 6 | 1471.2753 | 1499.873 |
| 7 | 1471.5209 | 1500.1164 |
| 8 | 1471.5214 | 1500.1144 |
| 9 | 1471.4993 | 1500.0896 |
| 10 | 1471.5047 | 1500.0922 |
| 11 | 1471.2434 | 1499.8278 |
| 12 | 1471.6768 | 1500.2579 |
:shockedface: :explodinghead:
1.126.2. HPD calculation
Having now checked the calculation for the HPD, it really seems to be like that. I've now looked at the individual rows in different ways and the data really seems to be like that.
So either via groups of the DF by hand or by summing the Tensor
using t.sum(axis = ...) it gives the same result. Really seems to be
the data!
1.126.3. Update LLNL definition based on .txt files
Implemented the "as designed" .txt file into TrAXer now. As expected
the focal length moves 30 mm forward!
The shape of the signal does change slightly. It becomes a tiny bit larger when running with the same imperfect mirrors as before. With perfect mirrors, we'll check. Quick look seems to imply the spot has gotten smaller for perfect mirrors.
Anyway, this is clearly a good improvement, but our HPD (based on the sum) hasn't really changed a preliminary look seems to imply.
[ ]Insert plots for that tomorrow!
1.127. Meeting with Klaus
PhD:
[ ]Send mail to Igor asking if he wants to be 2nd corrector[ ]3rd: someone from theory[ ]4th:- http://matthias.hullin.net/ -> He does some raytracing stuff! Should be a very interesting match!
- https://pages.iai.uni-bonn.de/gall_juergen/ -> He does other computer vision stuff.
1.128. Meeting with Klaus
We discussed the unblinded data, candidates and the real data!
Our main takeaway is that it looks reasonable and we're happy with our results.
We discussed whether there even might be a chance of getting a competitive limit for the axion photon coupling.
The question is where we will want to publish a paper about the result.
Given that the paper will likely be out before (or at least not after) a detector paper, the limit paper needs a very short section about the detector.
[ ]Write a mail to Igor if he has any opinion / idea about where to publish[ ]Set up an Overleaf and give people access (e.g. Igor, Jaime, Julia and our group) to it. -> I think I'll pump out a quick draft of the paper before doing that!
For the paper having an idea where to publish makes writing it a bit easier because of the layout & size requirements.
Klaus mentioned that I won't have to write the entire paper myself!
The introduction should maybe mention the Xenon N ton result about axions as well as the tip of red giant branch paper!
Otherwise from my point I'll finish the thesis draft before properly writing the paper.
1.129. Meeting Klaus
- told him about the systematics regarding Sun ⇔ Earth
- about the numerical conversion point being ~0.3 cm behind window
- about the status of the talk
We agreed that this looks good.
[ ]Run the expected limits[ ]Update and finalize the slides of the talk, then send them to him[ ]Send email to Igor about talk about limit calculation. First propose the following dates:
If he's available on any of these dates, we'll do the talk on that date. Then I can invite people (starting with the list of people from the mail Wolfgang sent to us about the posters).
[ ]About the posters:- focus should be on axion (electron) instead of chameleon
- maybe ask Christoph about his original poster
[ ]write mail to Johanna, Markus, Tobi to discuss a possible poster, if they have ideas etc (maybe Christoph too)
1.130. Meeting Markus
I had a short meeting with Markus discussing the charge sharing effects in our detector.
Look into Markus MSc thesis.
He concluded that the charge sharing via the amplification avalanche affecting other pixels is only on the order of 1% or so. In a Mg photopeak it was ~0.62 pixels added to about 48.
However, the effect of UV photons is quite significant.
Their range scales with lower isobutane mixtures. At 5% it already has a significant tail to about 150 μm. In 2.3% that is only going to be more.
The interesting aspects are:
most likely energies of produced UV photons: dominant are a
10 and ~11 eV line and to lesser extent (up to 3 orders of magnitude less!) a few lines up to 15 eV. NIST has tables of the lines: https://www.physics.nist.gov/PhysRefData/Handbook/Tables/argontable2.htm and the more general page for the data: https://www.nist.gov/pml/handbook-basic-atomic-spectroscopic-data There are packages like ~PySCF: https://pyscf.org/ https://github.com/pyscf/pyscf/ to compute them in theory. BingChat provided this fun script that uses it:# I can try to generate a Python script to compute the UV emission energies for argon, # using the Hartree-Fock method and the PySCF library. However, this is not a trivial # task and I cannot guarantee the accuracy or completeness of the script. Please use # it with caution and verify the results with other sources. Here is the script: # Import PySCF library from pyscf import gto, scf # Define argon atom atom = gto.M(atom = 'Ar 0 0 0', basis = 'cc-pvdz') # Perform Hartree-Fock calculation mf = scf.RHF(atom) mf.kernel() # Get orbital energies and coefficients orb_energies = mf.mo_energy # in Hartree orb_coeffs = mf.mo_coeff # Define a function to convert Hartree to eV def hartree_to_ev(energy): return energy * 27.2114 # Define a function to compute UV emission energies for a given transition def uv_emission_energy(initial, final): # initial and final are tuples of (n, l) quantum numbers # n is the principal quantum number (1, 2, 3, ...) # l is the angular momentum quantum number (0 for s, 1 for p, 2 for d, ...) # Assume that the initial state is occupied and the final state is unoccupied # Find the orbital index that corresponds to the initial state initial_index = None for i in range(atom.nao): # Get the angular momentum of the orbital l = atom.ao_angular(i)[0] if l == initial[1]: # Get the occupation number of the orbital occ = mf.mo_occ[i] if occ > 0: # Get the principal quantum number of the orbital n = atom.ao_nuc_attr(i)[0] + l + 1 if n == initial[0]: # Found the initial state initial_index = i break if initial_index is None: print(f'Initial state {initial} not found') return None # Find the orbital index that corresponds to the final state final_index = None for i in range(atom.nao): # Get the angular momentum of the orbital l = atom.ao_angular(i)[0] if l == final[1]: # Get the occupation number of the orbital occ = mf.mo_occ[i] if occ == 0: # Get the principal quantum number of the orbital n = atom.ao_nuc_attr(i)[0] + l + 1 if n == final[0]: # Found the final state final_index = i break if final_index is None: print(f'Final state {final} not found') return None # Compute the UV emission energy as the difference between orbital energies uv_energy = orb_energies[final_index] - orb_energies[initial_index] # Convert to eV and return return hartree_to_ev(uv_energy) # Test the function for some transitions print(uv_emission_energy((3, 1), (4, 1))) # 3p -> 4p transition print(uv_emission_energy((3, 1), (5, 1))) # 3p -> 5p transition print(uv_emission_energy((3, 1), (6, 1))) # 3p -> 6p transition
- based on the gas composition the UV photons have different attenuation lengths. Ideally we could compute that, but our attenuation data only goes to 30 eV. How do we calculate the cross section at lower energies?
- Markus also saw a fraction that was compatible with what I see at CAST being added to the expected number of electrons.
Potentially look into Garfield++ for this as well.
It seems like if we modify our logic slightly to also activate pixels at larger distance than 1, we get closer to the real data. But our estimate is already quite good I'd say.
1.131. Meeting Klaus
[ ]fix up systematics used -> Easy[ ]mask spark regions in data -> Relatively easy (just cumbersome)[ ]rerun code then -> easy[ ]look at all relevant plots for the best 2-3 cases of the expected limits -> background rate, clusters, distributions of limits, some MCMC chains?[ ]Prepare slides for a talk about the data taking + limit calculation method now!
Other:
[ ]Can we understand why corners have so much more background with MLP?
1.132. Meeting Klaus
Good meeting. Discussed effective efficiency plots of networks trained only on generated data and applied to real. The one with only extra diffusion input neuron and the other one diffusion + gain.
Plots:
Note that these plots are very much work in progress. Our hacky gain
fix of gain * 0.85 is kind of in use, but not fully maybe… Was
used during training, but not during fake data gen I believe.
1.133. Meeting Klaus
Discussed
n
TODO:
[ ]Make same plot with only CDL runs that used FADC[ ]discuss with Markus on Thursday[ ]apply method in MLP data[ ]
Think about: Do we need to take out the background data we trained on? At least for background rate estimation?
[ ]?
Add linear fit. Run-3 results:
χ²/dof = 0.6180 p[0] = -0.0021 ± 0.0000 p[1] = 10.8441 ± 0.0574
Run-2 results:
χ²/dof = 0.3279 p[0] = -0.0014 ± 0.0000 p[1] = 9.0639 ± 0.0165
3 approaches:
- include all CDL runs in the fit
- use only the mean CDL gas gain and mean cut value of all CDL runs for the target
- ?
[X]makereadCalibDataaware oftfKind(try to find attribute) -> if attribute there and given a tfKind, only read data that passes this[ ]Implement computing the NN cut value for the (correct) CDL run numbers for 5.9 and 3 keV data to understand where the cut comes from
OHHHH I think the issue is just that the energy cut is crap in the CDL data, because of the bad mapping of charge there!
1.134. TODO Thesis TODOs for MLP / limits
[ ]Why does the MLP produce so much more background in the cluster center plot for Run-2 than for Run-3? What is it in the Run-2 data that makes it so much worse? Or rather is it really the bad calibration taking such a "toll"?[ ]Make Limit code more efficent in terms of number of candidates drawn! So that it's easier to compute limits for the cases without vetoes!
2. Uni:
2.1. Timepix3 extended
See the issues on the GitHub page for immediate TODOs: https://github.com/SiLab-Bonn/tpx3-daq/issues
2.1.1. TODO Quick TODO notes [/]
Move these elsewhere once time.
[ ]look at events that show up in the toaMin dataset in the plots of the likelihood resulting file that have values similar to that large peak that shows up due to noisy lines[ ]investigate why likelihood data in lhood plots for gold goes further to lower values than the full chip plot
2.1.2. STARTED background rate for Tpx3 data [2/3]
We need to implement some things in order to properly compute a background rate.
We need to:
- use different 55Fe runs to compute the energy calibration, same as in CAST
- gas gain of each run, then gas gain calib factor fit
From here there are 2 options:
- use only the two lines (escape + photo) to describe reference X-rays. Should be possible, but efficiency will vary wildly outside the given energies
- use the two lines to define reference at 2 energies. Then use CAST CDL data to extract qualitative shift in distributions away from those distributions. Rescale CDL to Tpx3 55Fe data and then apply that.
In order to achieve that we still have to do a few things:
[X]implement charge calibration (ToT calib) for Tpx3 chips[X]add Tpx3 chips to InGrid database[ ]refactorlikelihood.nimto allow replacing the current CDL logic and likelihood computation by something more modular. So that we can define some kind of "likelihood model" that gets input data & computes things… First step for that is ripping out theXrayReferenceFilelogic and computing things on the fly as required though. This also means the logic that currently is dependent on the specific CDL lines needs to be replaced. This will require thought.
2.1.3. Mapping 55Fe Tpx3 data to CDL data
Investigations and TODOs:
[X]look at events visible in 1.5 - 4.0 keV scan from:
basti at void in /t λ plotData --h5file /mnt/1TB/Uni/Tpx3Data/reco_tpx3_calibration.h5 --runType rtCalibration --chips 0 --chips 3 --config ~/CastData/ExternCode/TimepixAnalysis/Plotting/karaPlot/config.toml --applyAllCuts --cuts '("energyFromCharge", 1.5, 4.0)' --h5Compare /mnt/1TB/CAST/2017/CalibrationRuns2017_Reco.h5 --h5Compare /mnt/1TB/CAST/CDL_2019/calibration-cdl-2018.h5 --cdlDset 3.0 --ingrid --cuts '("eccentricity", 0.0, 2.0)'
which yields
where we can clearly see a big peak at almost 0 hits, which shouldn't happen given that the events are supposed to have > 1.5 keV. Means many events with very large charge values. Therefore: look at events of those!
- 1.5 keV < x < 4.0 keV
- hits < 20
-> events all look pretty normal
[X]implemented cuts on ToT low and high in raw data manipulation. Helped to clean up somewhat and improve the background.[ ]compute the comparison plots using the pre selection cuts!
For a correction of the CDL data to the Tpx3 data there are multiple options. First of all the distributions look more or less compatible. That is good news. There are minor differences though and there are many options to perform shifting / morphing of distributions.
[X]compute the mean of the eccentricity, fracRmsTrans, lengthDivRms distributions for CAST, Tpx3 & CDL. The mean can be used to compute an 'x' shift for each property at both energies. Possibly from there one can see if there is a linear relation between these shifts between 3 and 5.9 keV. If so one can use that relation to perform a linear shift at each energy. UPDATE: Done in ./../CastData/ExternCode/TimepixAnalysis/Tools/determineEffectiveEfficiency.nim[X]compute similarity scores between the different distributions (Kolmogorov Smirnov etc). Could be used to get quantitative statement about whether distributions seem 'compatible' UPDATE: Done in ./../CastData/ExternCode/TimepixAnalysis/Tools/determineEffectiveEfficiency.nim We use the KS test to determine when we find a good agreement between 55Fe Tpx3 and CAST CDL.[ ]theoretically: compute shift bin wise for each bin in the properties. Then see what that mapping from Tpx3 to CDL distribution looks like for each property and each of the 2 energies. Possibly generalize that similar to how current energy wise CDL morphing works.
- Effective software efficiency
UPDATE: Take the numbers shown here with a big grain of salt, among others because of ./Doc/StatusAndProgress.html section [BROKEN LINK: sec:effective_efficiency_tpx3]!
Computing the effective software efficiency of the Tpx3 55Fe data using:
./determineEffectiveEfficiency /mnt/1TB/Uni/Tpx3Data/reco_tpx3_calibration_tot_cut.h5 -r
(under
toolsin TPA)yields the following values for the effective efficiencies:
DataFrame with 3 columns and 18 rows: Idx Escapepeak Photopeak RunNumber dtype: float float int 0 0.9446 0.9362 0 1 0.9381 0.9328 2 2 0.9354 0.9355 3 3 0.9411 0.9296 4 4 0.9495 0.9288 5 5 0.9484 0.928 6 6 0.9427 0.9331 7 7 0.936 0.9282 8 8 0.9452 0.9343 9 9 0.9467 0.9271 10 10 0.9442 0.9282 11 11 0.9319 0.925 12 12 0.9397 0.9321 13 13 0.9404 0.9313 14 14 0.9361 0.9308 15 15 0.9408 0.9312 16 16 0.9464 0.9302 17 17 0.9512 0.9272 18this implies the efficiencies are too high, i.e. too few events are actually removed compared to what the likelihood reference dataset should suggest.
This is a motivation to indeed compute the mean of the distributions and perform a shift of the values.
UPDATE: After fixing up a couple of bugs in the code, the new numbers are now:
Idx Escapepeak Photopeak RunNumber dtype: float float int 0 0.9268 0.9327 0 1 0.9219 0.9288 2 2 0.9179 0.9335 3 3 0.928 0.9265 4 4 0.9316 0.9249 5 5 0.9364 0.9245 6 6 0.9298 0.9257 7 7 0.9152 0.9239 8 8 0.9336 0.9281 9 9 0.9359 0.9206 10 10 0.9268 0.926 11 11 0.9134 0.923 12 12 0.9259 0.9302 13 13 0.9228 0.9271 14 14 0.9226 0.9269 15 15 0.9213 0.9286 16 16 0.9261 0.927 17 17 0.9389 0.9236 18see plots:
and the shift between the means:
So: means are visible and the shift seems to be decreasing for larger energies.
UPDATE : See the section [BROKEN LINK: sec:effective_efficiency_tpx3] in
StatusAndProgress. See next section here.After having figured out what the reason is (more or less) for the too large software efficiencies, our approach of stretching / moving the variables seems sensible and produces efficiencies of the order of 80% (a bit lower).
The next steps are:
[ ]precisely find the required values for the transformations such that we reproduce 80% at 5.9 and 2.9 keV[ ]interpolate somehow between these energies[ ]implement that into the likelihood code & run it
Algorithm to determine correct read point for eccentricity:
- iteratively read data to point X for 55Fe data, rescale according to
formula, check percentile
~80%. Once close enough, use that read parameter. Might yield different means, but distributions not necessarily same.
- Solution to match 55Fe Tpx3 data to CAST CDL
After looking at the different distributions for the 3 properties of the 55Fe data and CDL data (see distribution plots in previous section indicating the means), we finally figured out an automatic way to perform a 'matching' of the distributions.
Essentially the distributions are equivalent, up to a stretching factor and different initial cuts on the eccentricity for each dataset.
The original cuts for the escape peak and photo peak are:
Escape:
let baseCut = Cuts(kind: ckXray, minPix: 3, cutTo: crSilver, maxLength: Inf, minRms: 0.1, maxRms: 1.1, maxEccentricity: Inf) let range0 = replace(baseCut): maxLength = 6.0 let range1 = replace(baseCut): maxEccentricity = 2.0 let range2 = replace(baseCut): maxEccentricity = 2.0 let range3 = replace(baseCut): maxEccentricity = 2.0 let range4 = replace(baseCut): maxEccentricity = 1.4 maxRms = 1.0 maxLength = 6.0 let range5 = replace(baseCut): maxEccentricity = 1.3 maxRms = 1.0 let range6 = replace(baseCut): maxEccentricity = 1.3 maxRms = 1.0 let range7 = replace(baseCut): maxEccentricity = 1.3 maxRms = 1.0 let ranges = [range0, range1, range2, range3, range4, range5, range6, range7] xray_ref = getXrayRefTable()
(Note:
replacesimply takes the argumentbaseCutand replaces the fields that are assigned) where the ranges correspond toresult = { 0: "C-EPIC-0.6kV", 1: "Cu-EPIC-0.9kV", 2: "Cu-EPIC-2kV", 3: "Al-Al-4kV", 4: "Ag-Ag-6kV", 5: "Ti-Ti-9kV", 6: "Mn-Cr-12kV", 7: "Cu-Ni-15kV" }.toOrderedTable()
with the corresponding (upper bound) energies:
result = @[0.4, 0.7, 1.2, 2.1, 3.2, 4.9, 6.9, Inf]
the escape peak (assuming 2.9 keV) thus corresponds to the Aluminum target / Aluminum filter at 4 kV, range 3, and the photo peak of course the Manganese target, range 6.
The data is read and these cuts are in principle applied, both for the 55Fe data as well as the CDL data, i.e. the eccentricity is filtered to 1.4 for the escape peak and 1.3 for the photo peak.
Comparing the distribution for the eccentricity, fig. 1 it seems the distributions are shifted to one another, but in such a way that the CDL data describes a squished distribution of the 55Fe data (or more data to larger eccentricities would be be required).
Figure 1: Comparison of the eccentricity for the 55Fe Tpx3 data with CAST CDL data shows a different mean. In particular it seems the distributions are shifted to one another, but in such a way that the CDL data describes a squished distribution of the 55Fe data (or more data to larger eccentricities would be required). The lines are KDEs with 2.5 times Silverman's rule as the bandwidth. The idea being if one cuts less of the 55Fe data, see fig. 2, the distributions would likely match ~exactly, barring having to stretch the CDL distribution.
Figure 2: Equivalent plot to the previous figure, but with a higher cut on the eccentricity. It is easy to imagine that stretching the CDL data to the range of the 55Fe data, the distributions would match. The idea out of the way, let's discuss the implementation of how to automate this.
We start by reading the 55Fe data with all cuts mentioned above applied, but the cut on the eccentricity. In an iterative process we then try different cuts on the eccentricity and stretch the CDL data until we get a good agreement via the Kolmogorov-Smirnov test.
Let \(X\) be the distribution of the target (55Fe data in our case; which varies per iteration by its upper bound) and \(Y\) the distribution we modify (CDL data). Then we perform the following modification to each element \(x\) in it:
\[ x' = \frac{x - \min{Y}}{\max{Y} - \min{Y}} \left( \max{X} - \min{X} \right) + \min{X} \]
where in actuality the
minandmaxare not taken to be the real minima or maxima of the distribution, but rather the 1 and 99 percentile of the data.After applying this operation to the CDL distribution, we use the Kolmogorov-Smirnov test to check for the similarity of the distributions. The Kolmogorov-Smirnov test checks the agreement between two 1D distributions, by taking the largest difference in the (empirical) cumulative distribution functions of the two distributions:
\[ \text{KS} = \sup_x \left| F_1(x) - F_2(x) \right| \]
where \(F_i\) is the (empirical) cumulative distribution function of each distribution and where we assume that they have the same binning and dropped the (typically denoted) subscript of the number of samples.
The (empirical) cumulative distribution function can simply be defined as
\[ F_i(x) = \frac{\text{number of elements in sample } \leq x}{n} \]
where \(n\) is the number of samples in total.
To find the best matching \(ε\), we then iteratively walk along the gradient of the best KS test match.
Let the start value for the eccentricity cut be \(ε_s\). Further let there be a step size \(Δε\), which is the initial modification we do to the cut. This step is dampened via an exponential decay based on the number of iterations \(n\):
\[ Δε' = Δε · \exp{- α n} \]
where α is a dampening factor.
We start by assuming a decrease in eccentricity:
\[ ε_1 = ε_s - Δε \]
and flip the sign of \(Δε\) each time the next KS test value is worse than the previous one
\[ \text{sgn}(Δε) = \begin{cases} \text{unchanged if} & \text{KS}_{n+1} < \text{KS}_n \\ \text{flipped if} & \text{KS}_{n+1} > \text{KS}_n \end{cases} \]
The iteration is stopped either if an absolute value \(\text{absKS}\) of the KS test is reached or if the change between the last and current iteration is smaller than some cutoff.
Implementation wise the code looks as follows:
const Δε = 0.1 const α = 0.1 const absKS = 0.005 const ΔKS = 1e-3 var n = 0 var sign = 1.0 while ks > absKS: # apply the cut let dfFe = dfFe.filter(f{`eccentricity` < ecc}) # filter CDL data to this peak var dfCdl = dataCdl.filter(f{`Peak` == peak}) # apply the stretching mutation dfCDL = fnTab.applyMutation(dfCdl, dfFe, peak, energy, genPlots = false) # compute KS ks = kolmogorovSmirnov(dfFe["eccentricity", float], dfCDL["eccentricity", float]) if ks < absKS or # stop early and not adjust `ecc` abs(ks - lastKs) < ΔKS: break if ks > lastKs: # flip sign if we're worse than before sign = -sign # compute new adjustment let adj = sign * Δε * exp(- n * α) ecc = ecc - adj lastKs = ks
Applying this method with the following parameters:
- \(ε_s = 1.9\)
- \(Δε = 0.1\)
- \(α = 0.1\)
- \(\text{absKS} = 0.005\)
- \(\text{ΔKS} = 0.001\)
yields the the paths through the KS / eccentricity phase space as shown in fig. 3 and fig. 4 for the photo and escape peak, respectively.
Figure 3: Path through the KS test / eccentricity space with the iteration number colored for the photo peak. The algorithm finds a minimum near \(\text{KS} = \num{0.011}\) at an eccentricity cutoff of \(ε = \num{1.3851}\). Figure 4: Path through the KS test / eccentricity space with the iteration number colored for the escape peak. The algorithm finds a minimum near \(\text{KS} = \num{0.0078}\) at an eccentricity cutoff of \(ε = \num{1.517}\). The final distributions then look as follows:
Eccentricity for photo and escape peak:
Length divided by transverse RMS
fraction of pixels in transverse RMS
Finally, the resulting effective efficiencies with this approach are then for the escape peak:
Escapepeak = 0.8056556605305023 Escapepeak based on data = 0.8005259569494497 Escapepeak based on unstretched data = 0.6564721924612837
and for the photo peak:
Photopeak = 0.811759739304908 Photopeak based on data = 0.8001147893236442 Photopeak based on unstretched data = 0.6664800944877408
where we can clearly see the method works as intended.
To apply this to all energies, a linear interpolation of the cutoffs will be applied in such a way as to cut at a certain amount higher than the regular cut would be and stretching the distributions accordingly.
The cut values are extrapolated based on a linear extrapolation of the relative extension at the 2.9 and 5.9 keV positions. In code this is implemented as:
proc lineParams(x: seq[float], ys: seq[(float, float)]): LineParams = let y1 = ys[1] let y0 = ys[0] let m = (y1[0] / y1[1] - y0[0] / y0[1]) / (x[1] - x[0]) let b = y0[0] / y0[1] - m * x[0] result = (m: m, b: b)
where the the y values are:
mins.add((pdf[dset, float].percentile(1), cdl[dset, float].percentile(1))) maxs.add((pdf[dset, float].percentile(99), cdl[dset, float].percentile(99)))
(one linear function is computed for the minima and maxima). The
pdfhere is the 55Fe data filtered to the correct eccentricity cutoff determined by the above algorithm andcdlis the CDL data for the corresponding peak. As such we compute a linear function for the relative increase of they(value of the property) compared to the CDL data. The idea being that the distribution of the properties themselves is very non-linear for each of the properties and thus a linear interpolation based on the actual values makes no sense. Instead we scale the real CDL min / max values to the extrapolated linear relative change found at the escape- / photopeak.The following is the data of the thus computed minima and maxima of each property:
cdlMin,cdlMax,dataMin,dataMax,dset,energy 1.04736456358612,3.151447443254324,1.052247479551602,3.475748896222421,eccentricity,0.277 3.390405227947017,10.54363594516984,3.263447568698237,10.10159300227821,lengthDivRmsTrans,0.277 0.0,0.5263157894736842,0.0,0.5248747593460514,fractionInTransverseRms,0.277 1.039201535181208,1.962778496181249,1.044028594069271,2.160871352727218,eccentricity,0.525 3.404328798783252,7.878455504206553,3.281593678299245,7.576559162515366,lengthDivRmsTrans,0.525 0.03736467236467239,0.5169855394883168,0.03012477048752635,0.5158325184709381,fractionInTransverseRms,0.525 1.033827048309434,1.928982990940981,1.038600224267751,2.117425084566234,eccentricity,0.93 3.666355039167723,7.98306629932565,3.542516592721439,7.724170534628716,lengthDivRmsTrans,0.93 0.07142857142857142,0.5,0.05834576972734867,0.4992993986437099,fractionInTransverseRms,0.93 1.022213770025473,1.716165977263773,1.026893790475074,1.876141813683324,eccentricity,1.49 4.004829218102024,7.908802061076143,3.882159754874499,7.716710401765532,lengthDivRmsTrans,1.49 0.16,0.4716981132075472,0.1330405103668262,0.471577911211998,fractionInTransverseRms,1.49 1.01782102383709,1.369673742659443,1.022376188181599,1.481050078529444,eccentricity,2.98 4.424848134617308,7.490022922475545,4.326359139876101,7.470368645164172,lengthDivRmsTrans,2.98 0.2424242424242424,0.4488188976377953,0.2110341050053968,0.4500735020758999,fractionInTransverseRms,2.98 1.013551171329737,1.285111101586735,1.017980120900679,1.373906411347694,eccentricity,4.51 4.629492795397957,7.570205235262263,4.566248440446261,7.718746344919934,lengthDivRmsTrans,4.51 0.2677595628415301,0.4375,0.2438152956077891,0.4400932399426343,fractionInTransverseRms,4.51 1.012890823149783,1.274739281515619,1.017220345206387,1.348767214623895,eccentricity,5.89 4.729015357614976,7.593209475103633,4.701080845591675,7.894558965980234,lengthDivRmsTrans,5.89 0.27,0.4322916666666667,0.2556111244019139,0.4360752561818784,fractionInTransverseRms,5.89 1.00972733247411,1.26392214645733,1.013893392614687,1.315617022796137,eccentricity,8.039999999999999 4.895430182532435,7.729834051210967,4.925652984896617,8.278244463372157,lengthDivRmsTrans,8.039999999999999 0.2823920265780731,0.4230769230769231,0.2832395029151455,0.4286419326599565,fractionInTransverseRms,8.039999999999999
and a piece of code to plot them:
import ggplotnim let df = readCsv("/t/data.csv") .gather(["cdlMin", "dataMin"], "typeMin", "mins") .gather(["cdlMax", "dataMax"], "typeMax", "maxs") ggplot(df, aes("energy", "mins", color = "typeMin")) + facet_wrap("dset", scales = "free") + facetMargin(0.5) + geom_point() + ggsave("/t/mins.pdf") ggplot(df, aes("energy", "maxs", color = "typeMax")) + facet_wrap("dset", scales = "free") + facetMargin(0.5) + geom_point() + ggsave("/t/maxs.pdf")
this results in the figure shown in fig. 6 and [BROKEN LINK: fig:effective_efficiency:extrapolated_max_values]
Figure 5: Minima for the extrapolated vs. the real 1 / 99 percentile values. Eccentricity is well behaved, the other two have an 'inversion'. In practice we take the larger / smaller of the extrapolation / data value. Figure 6: Maxima for the extrapolated vs. the real 1 / 99 percentile values. Eccentricity is well behaved, the other two have an 'inversion'. In practice we take the larger / smaller of the extrapolation / data value. We can see that only the eccentricity behaves "well" in the sense that the extrapolation is always larger for the data. For the other two properties the values at the photo and escape peak are getting smaller for lower energies, but at very low energies they become bigger again. Therefore, the extrapolation itself underestimates the required width. For that reason we take the
max/minof the real range values and the interpolation for each case to not restrict the distributions further than necessary.Computing the background rate with the method applied yields the background as shown in fig. 7 for the cases of no stretching + no morphing, stretching + no morphing and morphing / morphing case.
Figure 7: Comparison of background rate of the cases where no stretching and no morphing are used, only stretching and stretching + morphing. Effect is negligible.
2.1.4. Background rate below 2 keV
[X]of all events in the background rate left below 2 keV Tobi brought up a good idea: compute a histogram / bar graph of all pixels still visible in all the clusters. Are there some pixels that are over represented? How much ToT do they carry? -> This wasn't particularly useful. Not much out of the ordinary to see.
2.2. TimepixAnalysis / CAST data analysis extended
CK data: Run 1 2017/18: Run 2 2018/2: Run 3
2.2.1. Ray tracing
- look at https://github.com/McStasMcXtrace/McCode/blob/master/mcxtrace-comps/optics/Mirror_parabolic.comp and other files to see how to handle different geometries.
- trace of radians scene: https://github.com/mratsim/trace-of-radiance/blob/master/trace_of_radiance/scenes.nim This is the default ray tracing in one weekend book 1 scene.
- Physically Based Rendering: The book on ray tracing. https://www.pbr-book.org/3ed-2018/contents.html
My implementation of Ray Tracing in One Weekend:
Things to do next:
- implement Bounding boxes (RIOW book 2)
- implement rectangles (RIOW book 2) & cylinders (PBR)
- test a rudimentary CAST scene
- implement parabolic and hyperbolic shapes
- implement X-rays (i.e. different reflection / transmission behavior)
- implement light sources, model Axion emission from Sun as a light source
- done?
2.2.2. General
- Performance of gas gain slicing
Running the reconstruction to compute the gas gains using slices is extremely slow if there are already existing keys, which need to be removed.
In that case deleting all the attributes slows it down tremendeously.
Output of running over ./../../../mnt/1TB/CAST/2018_2/DataRuns2018_Reco.h5 with current code took
INFO Plotting polya of run: 265 and chip: 6 INFO Reconstruction of all runs in DataRuns2018_Reco.h5 with flags: {rfOnlyGasGain, rfReadAllRuns} took 27446.18325471878 seconds INFO Performed reconstruction of the following runs: INFO {240, 242, 244, 246, 248, 250, 254, 256, 258, 261, 263, 265, 267, 268, 270, 272, 274, 276, 278, 279, 281, 283, 285, 287, 289, 291, 293, 295, 297, 298, 299, 301, 303, 306} INFO while iterating over the following: INFO {240, 242, 244, 246, 248, 250, 254, 256, 258, 261, 263, 265, 267, 268, 270, 272, 274, 276, 278, 279, 281, 283, 285, 287, 289, 291, 293, 295, 297, 298, 299, 301, 303, 306}That's almost 8h! And the file for 2017 is still running for over a day now.
With the new changes using a compound datatype the runtime for the same operation:
INFO Reconstruction of all runs in DataRuns2018_Reco.h5 with flags: {rfOnlyGasGain, rfReadAllRuns} took 1514.196860313416 seconds INFO Performed reconstruction of the following runs: INFO {240, 242, 244, 246, 248, 250, 254, 256, 258, 261, 263, 265, 267, 268, 270, 272, 274, 276, 278, 279, 281, 283, 285, 287, 289, 291, 293, 295, 297, 298, 299, 301, 303, 306} INFO while iterating over the following: INFO {240, 242, 244, 246, 248, 250, 254, 256, 258, 261, 263, 265, 267, 268, 270, 272, 274, 276, 278, 279, 281, 283, 285, 287, 289, 291, 293, 295, 297, 298, 299, 301, 303, 306}which is less than half an hour! The speedup of ~20 is much smaller than the equivalent for the much bigger file for 2017/18, due to apparently non linear behavior wrt to number of attributes.
I'll stop the Run 2, 2017/18 data file after about 1 1/2 days of computation…
INFO Reconstruction of all runs in DataRuns2017_Reco.h5 with flags: {rfOnlyGasGain, rfReadAllRuns} took 6203.59544634819 seconds INFO Performed reconstruction of the following runs: INFO {76, 77, 78, 79, 80, 81, 82, 84, 85, 86, 87, 89, 90, 91, 92, 94, 95, 97, 98, 99, 100, 101, 103, 104, 105, 106, 107, 109, 111, 112, 113, 114, 115, 117, 119, 121, 123, 124, 125, 127, 146, 148, 150, 152, 154, 156, 158, 160, 162, 164, 166, 168, 170, 172, 174, 176, 178, 180, 182, 184, 186, 188} INFO while iterating over the following: INFO {76, 77, 78, 79, 80, 81, 82, 84, 85, 86, 87, 89, 90, 91, 92, 94, 95, 97, 98, 99, 100, 101, 103, 104, 105, 106, 107, 109, 111, 112, 113, 114, 115, 117, 119, 121, 123, 124, 125, 127, 146, 148, 150, 152, 154, 156, 158, 160, 162, 164, 166, 168, 170, 172, 174, 176, 178, 180, 182, 184, 186, 188}Compared to less than 2h.
2.2.3. STARTED extend IngridDatabase
For the current studies of the time dependency and different approaches to calculate the energy of clusters it would be extremely handy if the database was able to store data for BOTH run periods.
Extend it such that:
- chips are not in root group, but in a "run period" kind of group, which keeps track of from where to where it's valid (possibly with a list of run numbers it's valid for as well?)
- only have one file finally that can handle multiple versions of a chip!
- things done
- implemented adding of run groups, store runs and start / stop
- creates a group for each period at root level
- chips are now added to run period group
- things to do
- add chip name to list of chips found in run period group
- implement finding right run period for a given chip and run (or
timestamp)
- we have even the
dateTimefield in the attributes to use that to read - runs have the problem that there's ambiguity between run + chip, cause run numbers are not unique! timestamp + chip is though.
- we have even the
- don't need a group to store run period information. Can just iterate the root group for all run periods imo
2.2.4. STARTED binned vs time for gas gain
Mostly implementd now, but still needs:
- should all the gas gain information etc. really be stored in
attributes? they are ridiculously slow!
Could store all information in a single
gaindataset? First read and fit everything, then write it away? Then make polya fit a multidimensional database with one column per each interval used. - support for better gas gain vs calbration factor handling
- should at least use all individual gas gain values and take some kind of (possibly weighted mean) to plot for the fit
- possibly attempt to split also calbration spectra into sub intervals and fit each? again quite a bit of work, because we need to handle all the calibration factors etc then
- extend the plotting
plotTotalCharge...script to allow adding different datasets via a TOML file, so that one can plot different versions of the same thing in one file? Could also just export each dataframe as CSV and have a simple helper script to combine them.- flag to
--readTomlor something where additional files are stored with their corresponding keys. Then probably need alpha on datapoints finally though, so fix ggplotnim style for points!
- flag to
currently end 2018 energy calculation uses 2017 gasgain fit vs e calib factors from Ingrid Database, cause we store it there, but didn't change file!
See extension of database module for how to better handle this in future.
2.2.5. TODO Change energy calib to use closest two calibration runs
See status for more information on this task.
2.2.6. TODO finish flow chart of analysis
DEADLINE:
Currently we are building a flow chart representing the relevant parts of the analysis pipeline.
The file to generate the flow chart using GraphViz is found in: ./../CastData/ExternCode/TimepixAnalysis/resources/analysis_graph.nim
Things left to do:
- TODO general
- mention data layout? tree of nodes?
- run:
- general datasets
- chip groups
- chip nodes
- ?
- run:
- mention other kind of output that is generated? background rate etc?
- mention data layout? tree of nodes?
- TODO raw data manipulation
- raw data manipulation checks if run is good for 2014 data
- outputs HDF5 file, input for reco
- performs basic FADC conversion?
- creates occupancies of each run
- TODO reconstruction
- input is HDF5 from raw, outputs new HDF5
- FADC reco steps
- TODO log reader
- log reader needs to be added
- TODO CDL
- how CDL data processed
2.2.7. TODO likelihood
- likelihood requires log file information for tracking / non tracking
- cuts being applied and where
- logL calculation
- what property goes in how
- veto information used here:
- scints: < 300 or whatevs
- fadc: ?
- gridpix outer: ?
2.2.8. Roadmap to Axion Electron result
- How do I spend my time?
- Praktikum (currently ):
- 2 mornings a week, O(2 + 1 h)
- help students in between: ~2 h
- correct reports: 3 h
- IT Web:
- ~~ 5-10 h (partly in "free time")
- Tobi:
- lately few hours here and there looking at his code, providing input
- working with Johanna's code to give numbers for transparancy if filled with gas to different places in IAXO
- in the past: help Johanna, same as Tobi
- open source development
- mainly
ggplotnim - quite a few h per week, but hard to say, because only some of it during work
- mainly
- Praktikum (currently ):
- Problems
- covid work from home + fun with heart caused significant inefficiency in my work. less time properly concentrating. Working on fixing that, but hard.
- working less in general atm. Not 80 h of which possibly 30-40h are open source stuff
- What to do for Axion Electron limit
This assumes a "let's get to result" approach. Doesn't include possible "detours" due to other things to develop / fix.
- difference of Marlin vs. TPA:
- understand difference in efficiencies / ROC curves -> last point I personally would like to understand, because my code has some weird looking plots
- 1 - 2 weeks of semi efficient work (hard to put into numbers)
- Ray tracer:
- finish clean up / parallelization
- clean up: ~5 h
- parallelization done, but RNG not properly using multithread safe / well distributed numbers
- run for IAXO with gas
- make radius aware slices / image
- long term: invert ray tracer to start tracing from detector, use trace of radiance inspired code
- finish clean up / parallelization
- limit calculation:
- "just" need to decide on how to calc. Chi2? TLimit like? implement that. Doesn't seem too hard. Hard parts were TPA and ray tracer for expected signal
- look at Jaime's code for MM analysis -> using Chi2 method
- difference of Marlin vs. TPA:
2.2.9. TODO Ideas from Jochen based on StatusAndProgress.org
Talked to Jochen on round about.
He mentioned a few things that could be done.
- TODO understand left tail of fall time distribution of calibration data
In the fall time plot shown in ./Doc/StatusAndProgress.html we can see a long tail towards lower values. Jochen argued that this is likely not due to background, since the background should be significantly less than visible in numbers indicated by the tail.
Check that hypothethis.
- TODO create scatter plots of rise / fall time and energy
One should expect to see a more or less linear dependency of the rise and fall time on the energy. The more energy there is, the higher those times should be.
- TODO investigate a run from CDL with large variance
Jochen said he could think of one reason for the large variance. Basically a form of charge up. Potential sparks happening on the chip or near the chip (not necessarily even visible as events). Those would cause a charge up of the layer, resulting in a drop in voltage.
Lucian apparently created a scatter plot of the charge values per pixel once. Basically an occupancy of the (average?) charge values instead of the number of hits. A gradient was visible from top left to bottom right (although supposedly the top left contained sparks and the scale was the other way round in terms of where one would expect more charge depending on charge up).
This done for the
320 Cu Ni 15.0 323.23 21.81 run of the CDL data, because it has the largest variance and possibly do this for a binning by time too.
- TODO check ranges of CDL fits
On the fitted spectra of the CDL spectra it looked like the range for the fits was quite large. That could possibly lead to a shift towards one direction, if there are many bins with few entries.
This was supposed to be mitigated by using a maximum likelihood estimation fit instead of a normal chi square.
Check whether the range indicated by the drawn fit line is actually the one that was fitted in. If not maybe change the ranges.
- TODO filter out those SiPM events around 8 keV
And look at the count distribution of those events.
- TODO split veto of scintillators
Instead of always looking and applying the combined scintillators, split this and consider them individually. SiPM should only contribute near 8 keV?
- TODO check cuts for likelihood
I'm basically certain, but check again whether the cuts on the CDL data are also applied to the likelihood distributions when they are built.
If they are, Jochen says we might bias ourselves, because e.g. we already apply a cut on the eccentricity before we even create the distribution.
Yes, we do, but is it really a bias or simply well defined and desired?
Check the cuts for the likelihood then and also check what's cut aside from the likelihood for the actual background data.
NOTE: this is still correct as it is, because the original goal was to recover the Marlin analysis and not go ahead. If a change was made to this it'd be a change!
- TODO investigate broken cluster finder
First check the rate of broken events in Marlin.
This could be done by extracting all clusters as they are found by Marlin and then classify those. So in effect "use Marlin's cluster finder".
Then we would be able to see if we recover a background rate that's larger than it should be.
- TODO order of evaluation of FADC, veto cuts
Jochen said that the Zaragoza people always did their analys in a different order. Apparently they first threw out anything that triggered a veto and pretended those events never happened, i.e. they reduced the data taking time in that way.
Jochen said I should talk to the people next week and ask them if that's correct.
He was afraid that the calculation of the total time used to normalize the background rate would change if this was made, because less data was considered.
For myself: check in what way the total time used to normalize is actually calculated again. I know that I sum up the event duration of certain events. But of which events? Do I take all events? I don't think so, because obviously not all events are even part of the likelihood H5 files. Check that!
2.2.10. Tests [0/1]
Tests to write for TimepixAnalysis
2.2.11. CDL 2019 [0/1]
- TODO create FADC distributions for FADC data
Hendrik's talk only showed the rise and fall time distributions for normal background and Fe55 calibration data.
We should create the rise and fall time plots also for the different energy bins of the CDL calibration data. That should give us a better idea as to what the rise and fall time distributions look like for different energies (I think we did something like this once?). The main worry of the current cuts based on the different distributions is that the Fe55 data is more or less mono-energetic. This could result in a bias of the rise time to specific values.
In principle I would expect the rise time to only be dependent on the angle, size and density of the primary electron cluster that drifts towards the grid. A track with a larger angle to the grid should have a longer rise time, since the majority of the charge is angled away from the grid, whereas a track parallel to the grid should have a short rise time, comparable to an X-ray of similar density.
Can we find a way to determine whether FADC events within the "X-ray" like regime of the rise time are more often good tracks that don't actually look like X-rays at all? Maybe however, such tracks on average produce an even shorter rise time, because the whole length of the track results in an effectively higher density (since the problem is reduced to 1D; the distance of pixels from the grid at a specific time t).
2.2.12. TODO Understanding differences between CK and TimepixAnalysis
- DONE Check ToT calibration fit results
Compared the fit results in ./../../../mnt/Daten/CAST/2014_15_ChristophAnalysis/D03-W0063/calibdata/ToT-Calibration.eps to the results from the InGridDatabase and up they are the same up to 2 significant digits:
a = 0.3484 +- 7.8e-5 b = 58.56 +- 0.031 c = 1294.00 +- 3.08 t = -12.81 +- 0.058
NOTE: The fit parameters are the same as in this file: ./../../../mnt/Daten/CAST/2014_15/D03-W0063/calibrationData/calibdata/ToT-Calibration.eps
- Debug by comparing CDL data using Marlin vs. TimepixAnalysis
The CDL data stored in ./../../../mnt/Daten/CAST/CDL-reference/calibration-cdl.h5 can be used to debug the difference between Marlin and TP analysis output.
The first attempt was to extract the raw data from the above file and save it to a new H5 file. This is done in ./../CastData/ExternCode/TimepixAnalysis/Tools/DebugCkDiff/debugDiff.nim if the
--extractflag is being used on the calibration file.The resulting plots look like the following:
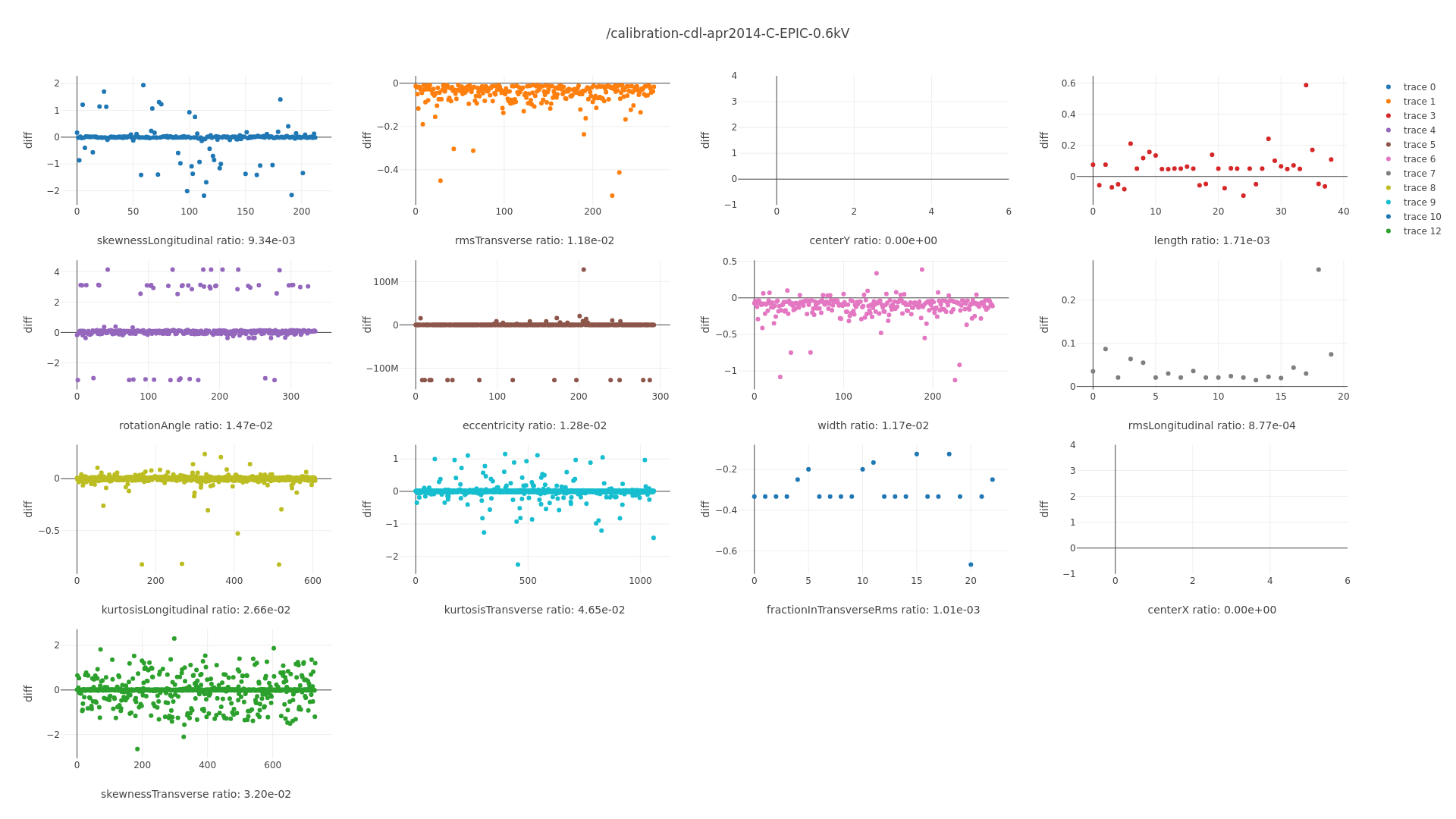
Figure 8: Difference between all geometric properties calculated by Marlin TPC and the TimepixAnalysis code To be noted is the following:
- from the first version of the
centerXdifference plot theapplyPitchConversiontemplate in ./../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/private/geometry.nim was fixed The plots do not show all data points, but only those that satisfy the following snippet:
cutVal = percentile(tpData, 95) * 0.01 let diff = zip(mFilter, tpFilter) --> map(it[1] - it[0]) --> filter(abs(it) > cutVal and abs(it) < 1e9)
so the difference needs to be larger than \(\SI{1}{\percent}\) of the variance.
Further analysis is problematic using the approach given by the
--extractoption however. That is because theChargeValuesVectorstored in the CDL H5 file is really the charge vector already and not the ToT values. These as a matter of fact are not even stored in the CDL file. Further complication is that each calibration group in the H5 file does not contain a single run. Instead each X-ray target is comprised of several runs, which are concatenated in the H5 file to one dataset.What can we do? We have all raw data runs in ./../../../mnt/Daten/CAST/2014_15/CalibrationRuns/. We first need to extract all runs corresponding to each X-ray target.
import nimhdf5, sequtils, sets, os, algorithm, shell, tables var h5f = H5file("/mnt/Daten/CAST/CDL-reference/calibration-cdl.h5", "r") var tab = initTable[string, HashSet[int]]() for grp in h5f: let runNumbers = h5f[grp.name / "RunNumber", float32] let runs = runNumbers.deduplicate.mapIt(it.round.int).toSet tab[grp.name] = runs discard h5f.close()
Now print the results for this buffer before we use it
import nimhdf5, sequtils, sets, os, algorithm, shell, tables var h5f = H5file("/mnt/Daten/CAST/CDL-reference/calibration-cdl.h5", "r") var tab = initTable[string, HashSet[int]]() for grp in h5f: let runNumbers = h5f[grp.name / "RunNumber", float32] let runs = runNumbers.deduplicate.mapIt(it.round.int).toSet tab[grp.name] = runs discard h5f.close() for key, val in tab: echo "X-ray source: ", key echo "Runs: ", val
Given these run numbers, we can now extract the correct folders.
import nimhdf5, sequtils, sets, os, algorithm, shell, tables var h5f = H5file("/mnt/Daten/CAST/CDL-reference/calibration-cdl.h5", "r") var tab = initTable[string, HashSet[int]]() for grp in h5f: let runNumbers = h5f[grp.name / "RunNumber", float32] let runs = runNumbers.deduplicate.mapIt(it.round.int).toSet tab[grp.name] = runs discard h5f.close() import shell, strutils const path = "/mnt/Daten/CAST/2014_15/" const pathCalib = "/mnt/Daten/CAST/2014_15/CalibrationRuns/" for key, val in tab: let dirname = "CDL_Runs" / key.strip(chars = {'/'}) shell: one: cd `$path` mkdir "-p" `$dirname` for r in val: let tocopy = $r & "-*" shell: one: cd `$pathCalib` cp -r `$tocopy` ".." / `$dirname`
Now perform raw data manipulation of all runs:
cd /mnt/Daten/CAST/2014_15/CDL_Runs/ for f in calib*; do echo "raw on " $f; /home/basti/CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/raw_data_manipulation $f \ --runType=calib \ --out=/mnt/1TB/CAST/CDL_Runs/$f.h5 \ --ignoreRunList; done
Continue with reconstruction
cd /home/basti/CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/ for f in /mnt/1TB/CAST/CDL_Runs/*.h5; do echo $f; ./reconstruction $f; ./reconstruction $f --only_charge; ./reconstruction $f --only_gas_gain; ./reconstruction $f --only_energy_from_e; done
This leaves us with the problem of having to create the plots for the difference for each property. We should approach the problem from the side of the Marlin data now. By iterating over all elements of the Marlin H5 file, getting the name of each group we can open the corresponding TimepixAnalysis H5 file. See the implementation for that in ./../CastData/ExternCode/TimepixAnalysis/Tools/DebugCkDiff/debugDiff.nim.
- from the first version of the
2.2.13. background rate
- background rate: binning like Christoph
for report:
- very preliminary
- w/o 2014 w/ Nim analysis
- background rate plot w/ 2018/2 data (same as above + in one?)
- bug in low energy?
2.2.14. FADC [0/2]
plots for riseTime and fallTime
- based on new data: 1 2018/2 calib + 1 2018/2 back
- nicely looking
- plus: plot only based on events passing logL cut
take
likelihoodoutfile and give to Hendrik to extracteventNumber-> for Run 2 and Run 3
- TODO calculate gas gain from FADC pulses
In theory it should be possible to extract knowledge about the gas gain from the FADC pulses.
Given that the pulse itself is actually produced by the fact that the primary electrons are amplified in the avalanche below the grid, the induced charge on the grid should be proportional to the gas gain!
Check this by creating the spectra of
- FADC min value individually for different Fe55 calibration runs
- each of the above creates a crude, inverted Fe55 spectrum. Caclculate the mean of the Fe55 K alpha peak and create a scatter plot between this and the gas gain calculated for the run. Is this correlated?
- TODO pluck FADC file to data calc from rawdatamanipulation
Need to pluck the calculation that was previously done in the raw data manipulation converting the FADC raw data to the
FadcDataobjects and put that intoreconstruction. For that refactor the calculations into a proc that goes intofadc_analysis.nim. - TODO change location minimum value of FADC pulses
Currently the minimum of the FADC pulses is taken to be the absolute minimum value of the given data tensor. That however does not make any sense, since we know that we have periodic noise on the data. This will just cause us to pick specific channels more likely.
Change the calculation to also use some percentile. 95% or something.
- TODO add test case for FADC data calculations
Test cases for both FADC conversion from raw to reconstructed as well as for the calculation of rise / fall times etc. still missing.
2.2.15. Scintillator [1/2]
- TODO create explanation for SiPM background histo distribution
[0/2]
The distribution of the SiPM trigger clock cycles for the background data is interesting.
See: ./../../schmidt/org/Figs/SPSC_Jan_2019/test/Run3/szinti1_run3.pdf
In principle, the data for the SiPM triggers should be dominated by muons, which traverse the detector orthogonally. We also expect the mean ionization of those muons to be ~2.67 keV/cm, resulting in ~8 keV deposition along the 3 cm drift distance in our detector.
The FADC has a trigger threshold of ~1.3 keV during the run 3 data taking campaign. This means of the 8 keV event 0.4875 cm of the track must be accumulated for the charge to sum up to an equivalent charge of 1.3 keV.
With a drift velocity of ~2 cm / µs, this is equivalent to s = v * t t = s / v t = 0.4875 cm / 2 cm / µs t = 0.24375 µs
At a clock frequency of 40 MHz, this results in n = t / T n = 0.24375 µs / 25 ns n = 9.75 clock cycles
The peak in the distribution however is rather at around 20 clock cycles. There may however be unknown delays in the whole readout change (I can't however think of a reason why there could be more systematic effects than a delay / offset; non linear behavior seems unlikely to me).
The question then is why the distribution is almost flat (assuming the 20 ck peak is the 8 keV peak). This means that we have almost as many other orthogonal events with much lower energy.
At around 60 clock cycles (= 1.5 µs) the whole track has drifted to the chip, assuming it is perfectly orthogonal. The size of the SiPM allows for shallow angles, which should explain the tail to ~ 70 clock cycles.
Thus, the edge at around 60 clock cycles must correspond to a deposited energy of around 1.3 keV (because the FADC triggered only after all the charge has drifted onto the grid).
- TODO Perform these calculations for the whole data …
and create a distribution of a thus calculated energy derived from the SiPM delay.
- TODO Extract the GridPix events, which correspond to …
the SiPM events in the distribution. Then
- take a look at those events, via properties and event display
- extract the energy calculated from the event and create a distribution of that energy. It should in theory be similar to the derived energy calculation of the above.
- TODO Understand grid discharge behavior
Considering the FADC's pulses, they have all rise times below 100 ns (1 GHz clock). This seems to contradict even allowing for orthogonal tracks accumulating charge quickly enough for the FADC to see a charge build up to trigger.
However, this may be an electronic effect, due to what happens on the way to / within the FADC? Maybe the rise time itself is not trustworthy. Plus, we have to consider the 100 ns integration time and 50 ns (?) differentiation time of the amplified input signal to the FADC.
An idea would be to simply consider the grid itself as a pure capacitor and check the typical time constants of that. Is it compatible with a charge build up of orthogonal events in the first place or not?
- TODO Perform these calculations for the whole data …
2.2.16. Outer chip histogram
both for Run 2 and Run 3
- normalize output of ./../CastData/ExternCode/TimepixAnalysis/Plotting/plotOuterChips/plotOuterChips.nim
- make subplots for background and calib in
plotOuterChips - filter by only using "blob" like events on
centerChip(ecc < 1.3, rmsTr < 1.2)
2.2.17. Define rudimentary FADC + scinti + outer chip cuts
apply them to result of likelihood
2.2.18. STARTED for SPSC meeting
- create histogram of # hits of outer chips for background and calibration in one plot
- if possible: same but only for event numbers that do pass the Likelihood cut!
2.2.19. TODO create following plots
Create the following plots:
- scatter of FADC: minvals / riseTime, fallTime
- create occupancy, properties plots with a cut on 4095 pixel events
- filter out events which are around the edge of chip (see occupancy, because there a A LOT of these), check where the centerX,Y of these is, and create plot of these cluster center locations
- rotation angle of calibration run: especially full range and escape peak, investigate the double feature of rotation angles. Peak around pi/2 and pi, which shouldn't be there
- plot FADC events and include argminvals, fallTime, riseTime. Use fallTime start and stop to plot these as ranges
2.2.20. TODO figure out memory leak of raw_data_manipulation
2.2.21. DONE until memory leak is found, replace…
calls to process single run by an exec process call to the raw data manipulation script for the individual run folder.
2.2.22. DONE write chip storage database file FEATURE
We need a database HDF5 file, which stores information about individual InGrids. The file should store the following:
- name of chip
- board its on
- number on that board
- FSR
- Threshold (+Means for completeness)
- ToT calibration
- SCurves
- potentially more misc info
for each individual chip. Mostly finished in ./../CastData/ExternCode/TimepixAnalysis/InGridDatabase/ CLOSED:
- DONE write module to handle that file
That Nim module needs the following:
- create base file
- add chip to file
- modify chip in file
The module will come with the database file and will provide procs to read the data conveniently. So that in other Nim programs one can import the installed `InGridDatabase` module and simply call, e.g.
getTot("<chipName>")
where we just get the chip name from our event files / data run HDF5 files.
- TODO implement copy into nimhdf5 and copy relevant chips…
to the data files
2.2.23. STARTED write generalized interface for the whole analysis as CLI program
Already started on my laptop as ./../CastData/ExternCode/TimepixeAnalysis/Analysis/ingrid/analysis.nim, but so far only barebones CLI (nothing implemented yet).
Update :
While no interactive frontend for the whole analysis was implemented, instead a helper tool to run the full analysis chain if desired. This is done in: ./../CastData/ExternCode/TimepixAnalysis/Analysis/ingrid/runAnalysisChain.nim
In principle this could be refactored into a whole that can be called as a libray. On that building an interactive frontend would be easy.
2.2.24. TODO continue actual analysis via new detector components ANALYSIS
2.2.25. TODO rewrite CNNs with Arraymancer
This should be relatively straight forward?
2.2.26. TODO remove FADC offset from scintillator triggers EXPERIMENTAL
When plotting the clock cycles, since the last SiPM veto trigger, the
result looks something like the following:
 There is a distribution around 10-50 clock cycles (25 ns each). That
means there is a delay of a max 60 * 25 = 1.5 µs between the last SiPM
trigger and the following FADC event, which closed the shutter.
There is a distribution around 10-50 clock cycles (25 ns each). That
means there is a delay of a max 60 * 25 = 1.5 µs between the last SiPM
trigger and the following FADC event, which closed the shutter.
We should compare this time with the time it took until the FADC closed its shutter. We can calculate that by considering the rotation of the cyclic FADC register. Maybe we can subtract that time from the clock cycles to get something more constant?
As a first try we can do the following: after temporal correction of the FADC data, take the minimum register of the FADC event. Calculate time of until end of window and subtract that from SiPM clock cycles:
func subtractFadcWindow(fMin, nClocks: int): int = # calc time until end of window # 2560 registers let tEnd = 2560 - fMin debugecho tEnd # FADC time should be in ns let fClockTime = 1 * 2560 # or rather tEnd is the time until the end of the window in ns # calc nClocks in ns let tClocks = nClocks * 25 # calc result from clock time result = int((tClocks - tEnd) / 25) # test it let nClocks = 42 let fMin = 2020#2560 - 880 echo "Time after correction ", subtractFadcWindow(fMin, nClocks)
Well, since I have no clue whether the example numbers even make any sense, it's hard to judge whether this might make some sense.
2.2.27. Fix CAST detector
- STARTED perform SCurves and TOT calibration
The SCurves for the 7 chips in principle work now. The data for them is in ./../CastData/data/SCurve/SeptemH/ and can be plotted using ./../CastData/Code/scripts/nim/plotSCurve/ Chip 0 and chip 3 look as expected. The other chips show a little bit of noise on the lines, especially chip 5 is very wiggly. To see whether this is a powering issue, I ran chip 5 individually, without the other chips. The data is in ./../CastData/data/SCurve/SeptemH/Chip5_single/ It can be seen that the issue still persists.
I'm wondering whether a wrong equalization of that chip might have this effect. Right now () I'm running another THSopt for chip 5 and will follow up with an equalization of it. THSopt resulted in:
THS = 65
Threshold equalization is now underway ().
- STARTED perform FADC calibration measurements
2.2.28. General [0/3]:
- Size of box at CAST under staircase
The size of the box is:
import strformat let depth = 0.59 height = 0.6 width = 0.8 vol = depth * height * width echo &"Measure: (h: {height}, w: {width}, d: {depth}" echo "Volume ", vol
- TODO give Jaime correct distances from detector to X-ray finger
- TODO chip names of Septem G and Septem H into wiki
- TODO expected axion electron limit w/ 30 times background suppression
Finish the calculation of an expected axion electron limit, with a potential 30 times reduction in background based on the new detector. Currently problematic since despite scaling background down, axion electron limit does not improve!
2.2.29. Detector:
- Hardware:
- STARTED finalize software
The software to readout and control the manipulator needs to be finished. The ./../CastData/ManipulatorController/PyS_manipController.py currently creates a server, which listens for connections from a client connecting to it. Commands are not final yet (use only "insert" and "remove" so far). Still need to:
- DONE separate server and client into two actually separate threads
- DONE try using nim client of chat app as the client. allows me to use nim, yay.
Note : took me the last two days to figure out, why the server application was buggy. See mails to Lucian and Fabian for an explanation titled 'Python asyncio'. Having a logger enabled, causes asyncio to redirect all error output from the asyncio code parts to land in the log file.
CLOSED: Python server is finished, allows multiple incoming connections at the same time, thanks to asyncio (what a PITA…). Final version is ./../CastData/ManipulatorController/PyS_manipController.py. Nim client works well as a client to control the server. See ./../CastData/ManipulatorController/nim/client.nim for the code currently in use.
- TODO implement some form of client into TOS.
- implement raw, simple client in C++, separate from TOS, simply compile it with it and call functions for this from TOS
- or simply check whether some client following some API is available on the system (put client into system PATH) and if so, use system calls to send commands via that client
Still need to implement client in C++ and finalize Python server, now that asyncio problem is identified.
- STARTED finalize software
- FADC:
- STARTED Ladungskalibration
[3/4]:
- WAITING TOS: FADC MODEREGISTER
Change the default FADC settings such that we use the 14 bit register in TOS, instead of the MODEREGISTER. Means changing sampling rate to bit 1 == 1, instead of 0. Include into ./../TOS/config/HFM_settings.ini and change default FADC settings. NOTE: currently implemented in ./../TOS/config/HFM_settings.ini as well as ReadHFMSettings and setFadcSettings. Not sure whether to actually use it so far.
- STARTED TOS: Change FADC trigger threshold
[2/3]
Change FADC Trigger Threshold of the registers from FADC ticks to mV. Should be done in ./../TOS/src/hvFadcManager.cpp and ./../TOS/src/console/userinterface.cpp. Should be easily done by: Check for FADC mode register (14 bit mode): fadcTriggerThresholdRegisterAll == 2048 -> in center of 12 bit register, 8192 in center of 14 bit register.
\(U = \frac{f_{\text{TTRA}}}{8192} - 1\)
or something like this. Step size of one tick for 12 bit:
step12 = 2 / 4096. step14 = 2 / 16384. return (step12, step14)
so roughly 0.5 mV and 0.12 mV. Using desired voltage calculate fTTRA value:
\(f_{\text{TTRA}} = (U + 1)8192\)
CLOSED:
- DONE Some tests of the functions defined above
We're defining some tests of the functions in ./../TOS/tests/HighLevelFunction_VME_tests/HighLevelFunction_VME_tests.cpp to test the new functions. To run these tests, in ./../TOS/TOS.pro change the following
CONFIG += debug CONFIG += tests
to
CONFIG += tests CONFIG += debug
since in a Qt project file, the last element is considered 'active'.
ticks to mV:
ticks = [2000, 8000, 1000] bit_mode14 = [False, True, False] def get_conversion_factor(bit_mode14): if bit_mode14 is True: return 8192. else: return 2048. pass Us = [] for tup in zip(ticks, bit_mode14): t, bm = tup cf = get_conversion_factor(bm) U = (t / cf - 1) * 1000 Us.append(int(U)) return Us
And mv to ticks:
mVs = [-50, 0, 400] def get_conversion_factor(bit_mode14): if bit_mode14 is True: return 8192. else: return 2048. pass ticks = [] for mV in mVs: cf = get_conversion_factor(True) cf2 = get_conversion_factor(False) tick = (mV / 1000. + 1) * cf tick2 = (mV / 1000. + 1) * cf2 ticks.append((int(tick), int(tick2))) return ticks
CLOSED:
- TODO change default FADC trigger in HFMsettings and write mV to output
Should change the default way the FADC trigger is set in the ./../TOS/config/HFM_settings.ini to use mV instead of FADC ticks (need to change ReadHFMSettings() for that as well). Then, enable possibility to write not only FADC ticks into output files, but also mV values for each pulse.
- DONE Some tests of the functions defined above
- STARTED Zeitkalibration
[0/2]
- First correct the temporal correction in ./../TOS/scripts/PyS_eventDisplay/septemModule/septemClasses.py, as to not roll the WHOLE data array read from the FADC, but rather each channel individually!
- Finally convert FADC channels to time intervals. How to do: see CAEN manual p. 15 (cf. with ./../TOS/src/High-Level-functions_VME.cc ?)
- TODO Verstärkung des Hauptverstärkers -> FADC ticks
Do a proper calibration of the dependency of main amplifier settings to the depth of pulses seen on the FADC ticks, such that a functional dependency between Uout = Aampl * Tfadc w/ Aampl ^= total amplification factor of main amplifier Tfadc ^= FADC ticks on output can be created. NOTE: Expected measurement time: ~10 h
- TODO clean up waitcondidtions.cpp
Clean up the call to GetAllData() refered to above. Such that either the channel variable is removed (and replaced by a properly named variable, which is needed for the function call), or simply write an overloaded GetAllData(void), which simply calls GetAllData(int) with 4 as the argument. Otherwise find a use for the channel variable and keep it.
- STARTED Investigate FADC noise in our lab
We connected the CAST detector with all components, except the HV was turned off. That way we could take data with the FADC.
Then we connected the LEMO connector for the grid to another intermediate board, which was empty. By itself, this intermediate board did pick up noise from the lights.
We went ahead and built a Faraday cage around it from a box, wrapped in aluminum foil. This got rid of the noise.
Then we connected the HV cable for the center chip. In a ramped and non-ramped setting, the noise was not reproducible, with the board wrapped in the box.
We also connected the VME crate with one of Jochen's power filters. It did not seem to help to get rid of the noise (we did see some noise, at some point with box in place, but then we probably left some hole in the box while closing it).
We still saw general noise, if the aluminum box was touching the normal lab power supply and one then "massaged" the aluminum.
- STARTED Ladungskalibration
2.2.30. Software:
- TOS:
- STARTED HV control via TOS
[1/2]
finalize the HV control to be done via TOS again. Need to incorporate the SiPM especially and change the channel numbers.
- TODO implement good default values for FSR
- TODO Run to HDF5 data
Implement run data automatically to be stored in HDF5 binary files. No need to write tons of small .txt files.
- TODO w/ HDF5
[0/3]:
- calc. rough energy via active pixels
- prepare CNNs for energy ranges (convert CUDA to normal Theano weights, runnable on CPU)
- feed events during run into correct CNN and create background rate plot on the fly. Done via separate Python script. Maybe write background rate plot into same HDF5 file on the fly? If HDF5 supports multiple openings at same time, maybe somewhat sketchy.
- TODO w/ HDF5
- TODO write tests for TOS
- write test, which hands certain temperature to temperature watcher thread to see, whether it would actually call the shutdown function of the HFM
- write test for calculation of CheckOffsetFullMatrix (especially getting the minimum value of the map defined in the function) See ./../ProgrammingTesting/C/test_minimum_of_map/min_in_map.cpp as an example that the code works.
- TODO new TOS functions
- Threshold fn, uma + load thresholds, SetMatrix and read out
- lf all (automatically load all default FSR filenames)
- auto calibration
- start a default run, which is defined by some file, e.g. CASTrun.txt defines run needed at CAST
- TODO define data format used to store chip settings
Define some data format to store all settings related to a single chip in one file:
- FSR values
- Threshold matrix (incl. uniform matrix)
- whatever calibrations?
Possibilities coming to mind:
- JSON
- XML (srsly?)
- HDF5 (overkill?)
- … gotta be something better out there
- STARTED HV control via TOS
- PySeventDisplay:
- Additional scripts
- Extract scintillator triggers
./../CastData/Code/scripts/nim/extract_scinti_triggers/ is now a pretty handy tool to read the scintillator triggers from a Run folder or a H5 file and then plot the histogram of the triggers.
- plot SCurves
./../CastData/Code/scripts/nim/plotSCurve/ This folder contains a simple tool to plot the data for an SCurve.
- Extract scintillator triggers
2.2.31. TODO IAXO gas phase
DEADLINE:
Talk to Tobi again about IAXO gas phase calculations.
2.2.32. TODO Investigate charge calibration offset in charge over time
TODO: investigate the offset seen in the calbration charge vs the background charge. Why consistently higher?
3. CAST data taking extended
This section covers all the important tasks related to the current data taking period at CAST.
3.1. STARTED Analyze FADC noise [1/2]
3.1.1. DONE script to plot FADC noise against time
Script to check for FADC noise is finished in first version. ./../CastData/Code/FADC_noise_analysis/fadc_noise_analysis.nim The findPeaks function still needs to be modified and it must be made easier to actually look at the data afterwards?
Still need to make sure that we actually get the correct noise (currently many normal events captured as well. So how many real noise events are missed?)
The script creates output files in ./../CastData/Code/FADC_noise_analysis/out which contain the name of the noisy FADC file and the date of that event.
Use ./../CastData/Code/FADC_noise_analysis/PyS_noise_histogram.py to plot a time evolution of these output files.
3.1.2. STARTED calculate dead time caused by FADC noise
We need to calculate the dead time caused by FADC noise. For that we have to walk over the data and calculate the effective data taking time over e.g. 5 minute intervals.
Howver, it is important to be able to focus on specific times. Might think about the following. Noise analysis script which can be given:
- a date (e.g. as "12/12/17") as a command line argument
- a date interval ( "12/12/17-06:00", "12/12/17-09:00" ) as two command line arguments
- a flag to only focus on a run (requries a date)
which then creates the data needed for the plot of noise / effective dead time vs. time.
3.2. TODO correct findPeaks function
Remove uneccessary comparisons (many done twice) and add feature to also check for peaks.